Abstract
Tre-2, BUB2, CDC16, 1 domain family member 4 (TBC1D4) (AS160) is a Rab-GTPase activating protein implicated in insulin-stimulated glucose transporter 4 (GLUT4) translocation in adipocytes and myotubes. To determine whether loss-of-function mutations in TBC1D4 might impair GLUT4 translocation and cause insulin resistance in humans, we screened the coding regions of this gene in 156 severely insulin-resistant patients. A female presenting at age 11 years with acanthosis nigricans and extreme postprandial hyperinsulinemia was heterozygous for a premature stop mutation (R363X) in TBC1D4. After demonstrating reduced expression of wild-type TBC1D4 protein and expression of the truncated protein in lymphocytes from the proband, we further characterized the biological effects of the truncated protein in 3T3L1 adipocytes. Prematurely truncated TBC1D4 protein tended to increase basal cell membrane GLUT4 levels (P = 0.053) and significantly reduced insulin-stimulated GLUT4 cell membrane translocation (P < 0.05). When coexpressed with wild-type TBC1D4, the truncated protein dimerized with full-length TBC1D4, suggesting that the heterozygous truncated variant might interfere with its wild-type counterpart in a dominant negative fashion. Two overweight family members with the mutation also manifested normal fasting glucose and insulin levels but disproportionately elevated insulin levels following an oral glucose challenge. This family provides unique genetic evidence of TBC1D4 involvement in human insulin action.
Keywords: glucose transport, insulin resistance, AS160
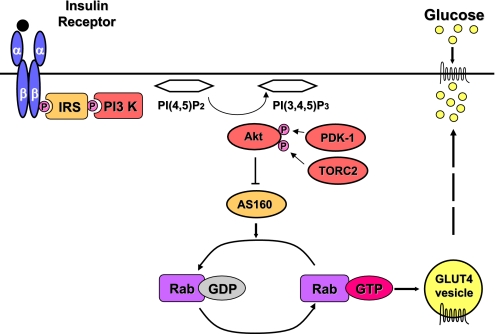
Blood glucose levels are very precisely regulated in healthy people despite enormous variation in the timing and magnitude of carbohydrate ingestion. Postingestive insulin-stimulated glucose uptake into skeletal muscle and adipose tissue is an essential element of this tight metabolic regulation. Glucose transporter 4 (GLUT4) selectively mediates insulin-stimulated glucose transport into myotubes and adipocytes. The importance of this metabolic step in vivo was demonstrated by generating muscle and adipose tissue-specific GLUT4 knockout mice, both of which are insulin resistant (1). Human physiological studies suggest that defects in insulin-stimulated glucose transport in muscle constitute the earliest abnormality in insulin-resistant people (2). GLUT4 is the main insulin-responsive hexose transporter. Insulin rapidly increases the amount of GLUT4 in the plasma membrane by causing specialized GLUT4-containing vesicles to move from an intracellular location to the plasma membrane where the vesicles dock and then fuse with the plasma membrane (3). This process is referred to as GLUT4 translocation. Whilst impaired GLUT4 translocation appears to be common to many different insulin resistance-inducing metabolic insults (4), precisely how insulin triggers GLUT4 vesicle movement and subsequent membrane fusion remains incompletely understood. AKT2 is clearly activated in response to insulin and triggers GLUT4 membrane translocation (5), but until recently the link between proximal insulin signaling steps [insulin receptor substrate (IRS)–phosphatidylinositol 3 kinase (PI3K)–protein kinase B (AKT)] and GLUT4 translocation was unknown. Tre-2, BUB2, CDC16, 1 domain family member 4 (TBC1D4) is a 160-kDa AKT substrate with a GTPase-activating protein (GAP) domain and several AKT phosphorylation sites (6, 7). Insulin-induced activation of AKT2 leads to phosphorylation of these sites. Considerable evidence indicates that phosphorylation of TBC1D4 suppresses its GAP activity, thereby promoting elevation of the GTP-bound form of selected Rabs and GLUT4 translocation (7–9) (Fig. 1). In the basal state, TBC1D4 appears to maintain these Rabs in their GDP form and so prevents GLUT4 translocation. TBC1D4 knockdown in 3T3L1 adipocytes increases basal cell membrane GLUT4 levels, indicating that TBC1D4 Rab-GTPase activity is required for full intracellular retention of GLUT4 storage vesicles (8, 9). Knockdown of TBC1D4 also inhibits insulin-stimulated GLUT4 translocation by about 25% by a currently unknown mechanism (8).
Fig. 1.
Schematic illustration of insulin-stimulated GLUT4 translocation. Binding of insulin (black circle) to its tetrameric receptor (2 α- and 2 β-subunits) leads to tyrosine phosphorylation (P) of insulin receptor substrate (IRS) proteins and the recruitment of phosphatidylinositol 3 kinase (PI 3-kinase), which catalyzes the conversion of PI(4,5)P2 to PI(3,4,5)P3. PI(3,4,5)P3 generation results in the phosphorylation of AKT2 (protein kinase B, beta) by 3-phosphoinositide-dependent protein kinase 1 (PDK1) and transducer of regulated cAMP response element-binding protein 2 (TORC2), causing its activation. AKT2 phosphorylates TBC1D4 (AS160) suppressing its GAP activity and resulting in active GTP-bound Rab proteins that promote glucose transporter 4 (GLUT4) vesicle translocation. PI(4,5)P2, phosphotidylinositol 4,5 bisphosphate; PI(3,4,5)P3, phosphotidylinositol 3,4,5 trisphosphate; GEF, guanine exchange factor; GTP, guanosine triphosphate; GDP, guanosine diphosphate; TBC1D4, Tre-2, BUB2, CDC16, 1 domain family member 4 (also known as AS160, AKT substrate of 160 kDa.
Although rare, monogenic syndromes of severe insulin resistance/type 2 diabetes can provide unique insight into the function of the affected gene/protein in humans as well as offering clues to understanding more common polygenic disease (10). To determine whether missense/nonsense mutations within TBC1D4 might alter glucose transport and predispose carriers to insulin resistance, we screened all of the coding regions of TBC1D4 in a cohort of 156 severely insulin-resistant patients. Patients were selected for the study if they had acanthosis nigricans, a cutaneous manifestation of extreme hyperinsulinaemia, and/or a fasting insulin level of greater than 150 pmol/L, or if already diabetic at the time of referral, if they required more than 200 units of insulin daily.
Results
Identification of a Premature Stop Mutation in TBC1D4.
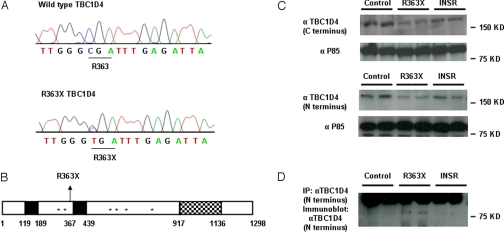
We identified a heterozygous substitution of thymine for cytosine at nucleotide position 1,087 in exon 3, resulting in the substitution of a premature stop codon (TGA) for arginine (CGA) at codon 363 (Fig. 2A) in a female patient with acanthosis nigricans. The mutation is predicted to truncate the protein at amino acid 363 (R363X), resulting in loss of the proposed GAP domain and several AKT phosphorylation sites (Fig. 2B) (6, 7). It was absent in 200 ethnically matched control alleles. The proband was also screened for mutations in other genes previously implicated in GLUT4 translocation. No mutations were identified in SLC2A4 (solute carrier family 2 member 4/GLUT4), TBC1D1, VAMP2 (vesicle-associated membrane protein 2), STX4 (synatxin4), SNAP23 (synaptosome-associated 23-kDa protein), RAB10, and LNPEP [leucyl/cystinyl aminopeptidase/insulin-regulated aminopeptidase (IRAP)].
Fig. 2.
Identification of the R363X TBC1D4 mutation. (A) Wild-type TBC1D4 and R363X TBC1D4 mutant sequence traces. The heterozygous substitution of thymine for cytosine results in the substitution of a stop codon for arginine (R) at amino acid 363. (B) Schematic representation of TBC1D4 showing the position of the R363X premature stop mutation, phosphotyrosine binding domains (PTB, solid black regions), AKT phosphorylation sites (*), and RAB GTPase activating protein (GAP) domain of TBC1D4 (checked region). (C) TBC1D4 expression levels (160 kDa) as determined by immunoblot analysis with both a C-terminal and N-terminal TBC1D4 antibody in EBVL lysates from the proband (R363X) compared with protein expression in cells from one healthy control (control) and one severely insulin-resistant subject with a loss-of-function mutation in the INSR gene (INSR). Τhe lysates were reprobed with an antibody raised against the P85 subunit of PI3K to confirm equal loading. (D) Expression of the truncated TBC1D4 protein in EBVL lysates from the proband confirmed by immunoprecipitation (IP) and subsequent immunoblotting with an N-terminal TBC1D4 antibody. The unexpectedly large size of the truncated protein (about 80 kDa) is the result of the slower mobility of the nonreduced form (see Fig. S1).
As TBC1D4 is detectable in lymphocytes and the proband was unwilling to have a muscle or fat biopsy, we documented reduced expression of full-length TBC1D4 protein using both a C-terminal and an N-terminal TBC1D4 antibody in lysates from her Epstein Barr virus transformed lymphocytes (EBVLs) (Fig. 2C). We then confirmed that the truncated TBC1D4 protein was expressed in the EBVLs by immunoprecipitating and then immunoblotting with an antibody raised against the N terminus of TBC1D4 (Fig. 2D).
Cellular Characterization of the R363X TBC1D4 Truncation Mutant.
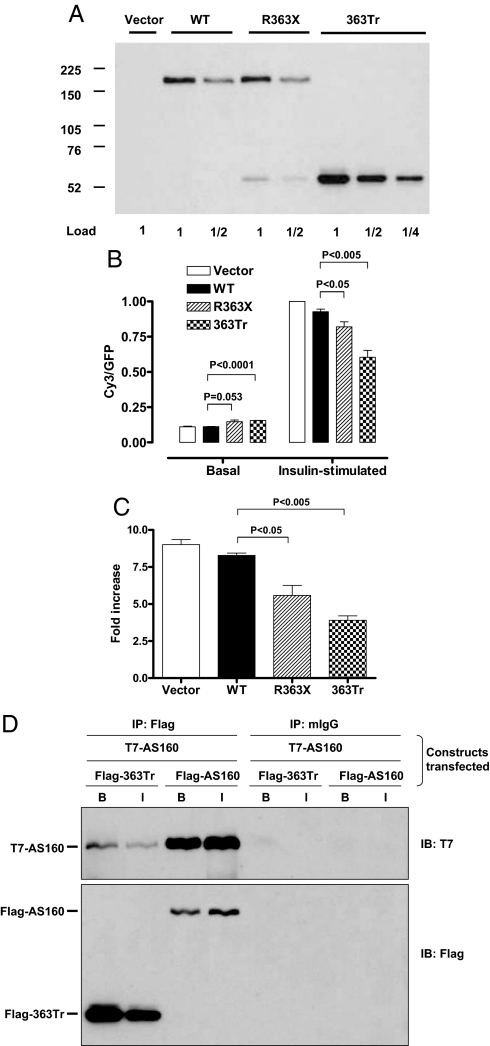
When overexpressed in 3T3L1 adipocytes by ≈10-fold over endogenous TBC1D4 (Fig. 3A), mutant TBC1D4 (R363X) increased GLUT4 at the cell surface in the basal state by 31% and inhibited the amount of GLUT4 at the plasma membrane in the insulin-stimulated state by 12%, relative to the wild-type TBC1D4 (Fig. 3B). The relatively modest effect of this premature stop mutant is probably accounted for by substantial readthrough of the premature stop mutation; the signal from the Flag-tagged readthrough product was about 4 times as strong as that of the Flag-tagged truncated version (Fig. 3A). To more effectively assay the impact of the truncated protein, we also generated a vector that terminates the protein at position R363 (363Tr). Overexpression of this truncated protein (363Tr) by ≈30-fold over the endogenous TBC1D4 increased cell surface GLUT4 in the basal state by 38% and reduced insulin-stimulated cell surface GLUT4 by 36%, relative to the wild-type TBC1D4 (Fig. 3B). Presentation of the data in Fig. 3B as the ratio of GLUT4 at the cell surface in the insulin-stimulated state to that in the basal state shows that both the mutant and 363Tr TBC1D4 proteins significantly reduce the fold increase in cell surface GLUT4 (Fig. 3C). When coexpressed with wild-type TBC1D4, the truncated protein binds to full-length TBC1D4, providing a plausible mechanism whereby the heterozygous truncated variant could potentially interfere with its wild-type counterpart in a dominant negative fashion (Fig. 3D).
Fig. 3.
Cellular analysis of GLUT4 translocation. (A) In vitro expression levels of TBC1D4 protein in 3T3L1 adipocytes (full-length TBC1D4 at 160 kDa and truncated protein at 52 kDa) transfected with empty P3XFLAG CMV10 vector (vector), wild-type TBC1D4 vector (WT), R363X vector (R363X), or the artificially generated 363 truncated protein (363Tr) vector as determined by immunoblotting with anti-Flag. We have also immunoblotted these samples with an antibody against a conserved peptide in mouse and human TBC1D4. Since the ectopic human TBC1D4 has a slightly lower mobility than the endogenous mouse TBC1D4, it was possible to estimate the relative amounts of each; the ectopic wild-type and R363X full length TBC1D4 were ≈10-fold more abundant than the endogenous TBC1D4 (data not shown). Also then by comparison of the anti-Flag signals of full-length R363X and 363Tr, we estimate that the 363Tr was overexpressed by about 30-fold in the transfected cells, relative to endogenous TBC1D4. (B) Cell surface GLUT4 levels, normalized to a value of 1.0 for the vector control in the insulin-stimulated state. Values are expressed as means ± SE for 4–6 independent measurements of the Cy3/GFP ratio in cells with and without 30-min treatment with 160 nM of insulin. (C) Data from Fig. 2B expressed as the ratio of cell surface GLUT4 in the insulin-stimulatd state to that in the basal state. (D) Immunoblots (IB) showing coimmunoprecipitation (IP) of T7-tagged TBC1D4 from basal (B) and insulin-stimulated (I) 293 cells with Flag-tagged 363Tr or full-length TBC1D4. To control for nonspecific immunoprecipitation, lysates were also immunoblotted after immunoprecipitation with irrelevant mouse Ig and protein G-agarose (mIgG).
Clinical Phenotyping and Family Cosegregation Studies.
The proband presented at the age of 11 with nuchal, groin, and axillary acanthosis nigricans. At birth she weighed 2.4 kg [-0.8 BMI SDS (body mass index standard deviation score)]. She remained slim during early childhood but from the age of 6 years, her weight increased and at the time of presentation her BMI SDS was +2.7. Physical examination was otherwise normal. Her fasting glucose and insulin levels fell within age-, gender-, and BMI-matched ranges, but following an oral glucose challenge, she displayed dramatic hyperinsulinemia (Table 1 and Fig. 4A). After puberty and a period of weight loss (age 23 years; BMI SDS +1.4), her acanthosis nigricans resolved. Although her glucose tolerance had now normalized, she was still overweight and again displayed an elevated peak-to-fasting insulin ratio of 14 (supporting information (SI) Table S1).
Table 1.
Clinical and biochemical characteristics of the proband and her mother
| Proband | Reference ranges, mean (95% CI)* | Mother | Reference ranges, mean (95% CI)* | |
|---|---|---|---|---|
| Age (years) | 11 | 41 | ||
| Sex | F | F | ||
| BMI (kg/m2) | 27 | 37 | ||
| BMI SDS | 2.7 | 3.1 | ||
| Basal glucose (mg/dL) | 97.3 | 84.1 (83.2; 85.0) | 81.1 | 90.8 (86.7; 95.0) |
| 120 min glucose (mg/dL) | 158.6 | 102.1 (100.1; 104.2) | 137.0 | 103.6 (95.0; 112.3) |
| Basal insulin (pM) | 101 | 94 (85; 103) | 115 | 80 (70; 92) |
| 120 min insulin (pM) | 6237 | 460 (413; 508) | 1720 | 367 (283; 452) |
| 120 min/basal insulin ratio | 62 | 4.8 (4.7; 4.8)† | 15 | 4.7 (3.9; 5.4)† |
| HbA1C | 5.3% | |||
| Triglyceride (mg/dL) | 88.5 | 115 | <177 | |
| Cholesterol (mg/dL) | 146.7 | 169.9 | <193 | |
| HDL Cholesterol (mg/dL) | 61.8 | 46.3 | >38.6 |
To convert the values for glucose into millimoles per liter multiply by 0.0555. To convert the values for insulin to microinternational units per milliliter divide by 6.945. To convert the values for triglycerides to millimoles per liter multiply by 0.0113. To convert the values for cholesterol and HDL to millimoles per liter multiply by 0.0259.
*Age-, gender-, and BMI-matched control data for the proband were derived from oral glucose tolerance tests performed in 215 nondiabetic Caucasian girls (23) [mean and 95% CI for: age 11.46 years (11.13; 11.79); BMI, 29.1 kg/m2 (28.3; 29.9)]. Gender- and BMI-matched data for the mother were derived from 31 nondiabetic Caucasian adult women participating in the Isle of Ely study (24) (BMI range 35–40 kg/m2).
†In control subjects, peak insulin values usually occur at 30 or 60 min. Definitive peak insulin data were not available as plasma insulin was only measured at 30 and 120 min after the oral glucose load in the adolescent controls and at 30, 60, and 120 min in the adult controls; however, in these control samples, the mean ratios for 30 min and 120 min to basal insulin concentrations were 9 and 5, respectively, in the gender- and BMI-matched adolescents; and the mean ratios for 60 and 120 min to basal insulin concentrations were 7 and 5, respectively, in gender- and BMI-matched adults.
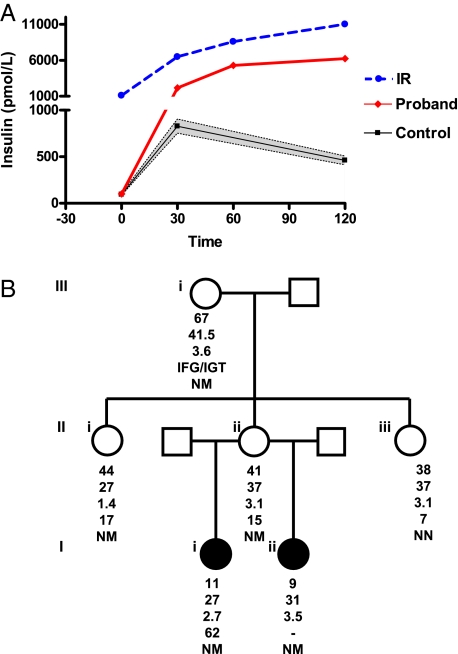
Fig. 4.
Phenotypic characterization of the R363X TBC1D4 kindred. (A) Plasma insulin levels (pmol/L) before (0 min) and after (30, 60, and 120 min) a 75-g oral glucose challenge. Black squares and error bars represent mean and 95% CI for plasma insulin levels during oral glucose tolerance tests performed in 215 nondiabetic Caucasian girls (23) (control; mean and 95% CI for: age, 11.46 years (11.13; 11.79); BMI, 29.1 kg/m2 (28.3; 29.9). Red diamonds denote data derived from the proband (proband) at age 11 years and blue circles, data from a 13-year-old lean female (IR) with type A insulin resistance because of a dominant negative mutation in the insulin receptor (INSR A1135E), which causes severe hepatic and peripheral insulin resistance. (B) Proband's (Ii) family pedigree. The squares represent male family members and the circles female family members; solid symbols denote family members with acanthosis nigricans. Below each symbol, age (years) is given, followed by the BMI (kg/m2), the BMI standard deviation score (SDS), the peak-to-basal insulin ratio, and the genotype, with N denoting the normal (wild-type) allele and M the mutant allele. IFG, impaired fasting glucose; IGT, impaired glucose tolerance.
The heterozygous R363X mutation was present in the proband's half sister (Iii), mother (IIii), grandmother (IIIi), and a maternal aunt (IIi) (Fig. 4B). Like the proband, her half sister presented with weight gain (BMI SDS +3.5) and acanthosis nigricans at the age of 9. She elected not to have blood sampling but did provide a buccal swab for genotyping. Her mother, a 41-year-old woman, was obese (BMI SDS +3.1) but had no acanthosis nigricans. Following an oral glucose load she too manifested a disproportionate rise in insulin, reflected by an elevated peak-to-fasting insulin ratio of 15 (Table 1). On repeat testing, a year later, an elevated peak-to-fasting insulin ratio was again found (Table S1). When first tested, the proband's affected aunt (Fig. 4B, IIi), an overweight 44-year-old woman (BMI 27 kg/m2), displayed a similarly elevated peak-to-fasting insulin ratio of 17. Following diet-induced weight loss (4.5 kg), her BMI fell to 24 kg/m2, and her peak-to-fasting insulin ratio normalized to 7 (Table S1). The only unaffected member tested (IIiii), an obese 38-year-old woman (BMI 37 kg/m2), manifested normal glucose tolerance and a normal peak-to-fasting insulin ratio of 7 (Fig. 4B and Table S1). The proband's grandmother (Fig. 4B, IIIi) also carried the mutation. She had a peak-to-fasting insulin ratio of 4, but at the time of testing she had an elevated fasting glucose and impaired glucose intolerance, indicative of significant β-cell dysfunction (11). Since the peak-to-fasting insulin ratio is dependent on the secretory capacity of the β-cells, her peak-to-fasting insulin ratios are difficult to interpret.
Discussion
Both insulin resistance and β-cell failure are believed to result from complex biochemical and gene–environment interactions. Whereas considerable progress has already been made in understanding monogenic β-cell disorders and, more recently, genomewide association studies have identified several common variants thought to be primarily associated with β-cell dysfunction, progress in understanding the genetic basis of insulin resistance remains slow (12, 13). This probably reflects the complexity of the insulin signaling cascade, the fact that insulin sensitivity is determined by insulin action in several different tissues and the number of factors, such as body weight, diet, physical activity, puberty, and old age, which modify insulin sensitivity. All previously described monogenic disorders in the insulin signaling cascade in humans result from proximal defects in the pathway, the vast majority being the result of mutations in the insulin receptor (14). We have also reported 1 kindred with autosomal dominant fasting and postprandial hyperinsulinemia because of a heterozygous AKT2 mutation (15). Although we and others have screened several genes (including GLUT4) implicated in GLUT4 trafficking, the premature stop TBC1D4 variant (R363X) is the first mutation that significantly impairs GLUT4 translocation.
The prematurely truncated TBC1D4 protein, which lacks the Rab-GTPase domain and AKT phosphorylation sites, slightly increases basal GLUT4 cell membrane levels. Expression of the truncation mutant may displace some wild-type TBC1D4 from the GLUT4 vesicles. Since the truncated protein lacks a GAP domain, one might expect an increase in GTP-bound Rab substrate proteins and consequently increased basal GLUT4 translocation. The inhibition of GLUT4 at the cell surface in the insulin-stimulated state caused by overexpression of the R363X and 383Tr is qualitatively consistent with the concept that the R363X mutation inhibits GLUT4 translocation in humans. However, it should be noted that quantitatively the degree of inhibition would be expected to be small when the amount of the truncated protein is expressed at about the same level as the wild-type protein, as would be the case in the human heterozygote, since ≈30-fold overexpression in 3T3L1 adipocytes decreased the amount of GLUT4 at the cell surface by 34%. Thus, it remains possible that some unknown role of TBC1D4 in other tissues contributes to the human phenotype. TBC1D4 mRNA is widely expressed in human tissues with highest expression in skeletal muscle and white adipose tissue (16, 17). Exactly how the truncated mutant inhibits insulin-stimulated GLUT4 translocation remains unclear. One possibility is that association of the truncated mutant with wild-type TBC1D4 reduces the suppression of its GAP activity that normally occurs when TBC1D4 is phosphorylated in response to insulin. As a result, insulin stimulation leads to a smaller increase in GTP-bound Rabs and thus some inhibition of GLUT4 translocation. Alternatively, it is possible that phosphorylated full-length TBC1D4 also has a positive role in GLUT4 translocation. This notion is consistent with observations made in TBC1D4 knockdown experiments; knocking down TBC1D4 in 3T3L1 adipocytes caused a 3-fold increase in GLUT4 at the cell surface in the basal state and a 25% reduction in cell surface GLUT4 levels in the insulin-treated state (8).
We have found that TBC1D4 dimerises with itself (Fig. 3D). The fact that the truncated protein can associate with full-length TBC1D4 offers a potential mechanism whereby relatively low-level expression of the variant might impair GLUT4 translocation. This is important for 2 reasons: first, the variant is heterozygous and second, at least in the 3T3L1 adipocytes, we observed considerable readthrough of the premature stop codon. Previous studies suggest that TGA stop codons, which is what the patients in this kindred have, appear to be the leakiest of all stop codons (18, 19), but whether or not this is occurring in vivo and to what extent it might be occurring is difficult to quantify accurately. Nevertheless, immunoblots of lysates from the proband's transformed lymphocytes suggest that total TBC1D4 expression levels are reduced and that the truncated TBC1D4 protein is expressed. Alternatively, given the increased basal GLUT4 levels with reduced insulin-induced GLUT4 translocation seen in TBC1D4 knockdown 3T3L1 adipocytes (8), it is also plausible that the phenotype may simply be explained by reduced TBC1D4 expression in vivo.
Two aspects of the human R363X phenotype were particularly striking: first, the proband presented with a very modestly elevated fasting insulin level of 101 pmol/L, but a massively elevated postglucose insulin peak of 6,237 pmol/L. We hypothesize that this selective preservation of fasting glucose and insulin levels with marked postprandial hyperinsulinaemia might reflect (i) preserved insulin action in the liver being primarily responsible for maintaining fasting glucose levels, (ii) increased basal (fasting) membrane GLUT4 levels, and (iii) impaired insulin-stimulated GLUT4 translocation. In vivo, the tendency to increase basal GLUT4 membrane expression does not appear to be sufficient to cause hypoglycemia as none of the carriers manifest this problem, but supraphysiological doses of insulin are required to compensate for the postprandial defect in insulin-stimulated GLUT4 translocation. Within a cohort of over 200 probands with severe insulin resistance, we have only identified 3 other probands with similarly disproportionate postglucose hyperinsulinemia. We have yet to identify causative genetic variants in these patients.
Second, the phenotype observed in this kindred appears to be very subtle in lean carriers (in keeping with the apparently subtle in vitro effects of the truncated mutant) but particularly sensitive to secondary modifiers such as puberty and obesity. Puberty is a well-recognized but ill-understood cause of insulin resistance. Intriguingly, it also appears to cause predominantly peripheral as opposed to hepatic insulin resistance (20). Although the kindred described herein is small, the proband's phenotype was most apparent during puberty and improved significantly following the completion of puberty and weight loss. Following puberty and significant weight loss the proband's acanthosis nigricans resolved and her insulin levels fell considerably although she was still overweight and had a high peak-to-basal insulin ratio. Her aunt normalized her fasting and postglucose insulin levels after reducing her BMI to 24 kg/m2, whereas her mother, who remained obese, maintained elevated peak insulin levels. The fact that GLUT4 levels are known to be specifically reduced in adipose tissue in obese states might explain the predisposition of this phenotype to be exacerbated by obesity (21).
A limitation of this study is the absence of direct in vivo measurements of GLUT4 expression and of glucose transport in adipose tissue and skeletal muscle. Family members declined our invitation to undergo hyperinsulinemic euglycemic clamps which, whilst still not providing direct measurements of in vivo glucose transport, might at least have indicated impaired peripheral glucose disposal.
In summary, we have described a family in which the proband, who presented with acanthosis nigricans and extreme postingestive hyperinsulinemia, was found to have inherited a heterozygous premature stop mutation in TBC1D4. In vitro, the mutant protein binds wild-type TBC1D4 and inhibits GLUT4 translocation. Thus, it is plausible to suggest that this family may represent the first inherited disorder of GLUT4 translocation in humans. Of clinical importance is the fact that reliance on biochemical assessments of insulin resistance that incorporate only fasting insulin would not have suggested a diagnosis of a state of severe insulin resistance in the proband, when she presented with acanthosis nigricans, and could have led to subsequent misdirected investigation. This family demonstrates that clinical syndromes of severe isolated postprandial hyperinsulinemia, sufficient to produce the classical dermatological marker of severe insulin resistance, exist in the human population and that their recognition requires measurement of circulating insulin levels in the postprandial and fasting states.
Methods
Genetic Studies.
Each participant, or a parent in the case of children under 16 years, provided written informed consent; minors provided oral consent. All clinical studies were conducted in accordance with the principles of the Declaration of Helsinki and were approved by the National Health Service Research Ethics Committee.
Genomic DNA was isolated from leukocytes derived from whole blood from 156 probands with severe insulin resistance and the coding regions and splice junctions of TBC1D4 (AS160) were amplified by PCR and sequenced. EBV-transformed lymphocytes were lysed in 50 mmol/L Hepes (pH 7.4), 150 mmol/L NaCl, 10 mmol/L ethylenediamine tetraacetate, 0.5 mmol/L phenylmethylsulfonylfluoride, 2.5 mmol/L benzamidine, 1 μg/mL leupeptin, antipain, pepstatin, and 1% Triton X-100. Protein concentration was determined using the Bradford dye-binding procedure (Bio-Rad Laboratories). Cell lysates were subjected to 8% reducing SDS/PAGE, proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore), and membranes were probed with a rabbit polyclonal antibody raised against the C terminus of TBC1D4 and a horseradish peroxidase-conjugated secondary antibody (Abcam). Antibody binding was detected by enhanced chemiluminescence (Amersham ECL Western blotting system, GE Healthcare).
To demonstrate expression of the truncated protein, we obtained a custom-made affinity-purified rabbit antibody raised against the N terminus of TBC1D4 (21st Century Biochemicals). EBVL lysates were immunoprecipitated with this N-terminal TBC1D4 antibody using the Exactacruz F kit from Santa Cruz Biotechnology (catalog no. sc-45043) as per manufacturer's instructions. Protein A agarose beads (Sigma, catalog no. P3476–5ML) were used to bind the N-terminal anti-TBC1D4 antibody. Bead-antibody-protein complexes were washed 4 times in lysis buffer and then finally in PBS. The immunoprecipitated protein was eluted in 50 μL of 2× nonreducing electrophoresis buffer by gentle pipetting at room temperature. Nonreducing buffer was used because the mobility of the reduced truncated protein is approximately the same as that of the antibody heavy chain. The supernatant was run on an 8% SDS gel and immunoblotted with the N-terminal TBC1D4 antibody as described above except that a HRP-conjugated secondary antibody from the Exactacruz kit was used for detection. The truncated R363X protein is detected at just above the 50-kDa marker when dissolved in reduced electrophoresis buffer. To determine its molecular weight in a nonreducing buffer, protein lysates from 3T3L1 adipocytes overexpressing a myc-tagged 363Tr were dissolved in 2× nonreducing buffer and immunoblotted with an antibody raised against the myc epitope. This confirmed that the truncated protein was detectable just above the 75-kD marker (Fig. S1).
Cellular Studies of Mutant TBC1D4 Function.
Human wild-type TBC1D4 (AS160) in the P3XFLAG CMV10 vector (6) was used to generate a Flag-tagged mutant construct (R363X) using the QuikChangeTM site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocols. To evaluate the biological effects of the truncated mutant, a second construct was created in this vector with no coding DNA beyond the stop codon identified in the proband (designated 363Tr). All constructs were fully sequenced.
The relative amount of GLUT4 at the cell surface in 3T3L1 adipocytes was assayed by transfecting cells through electroporation with a plasmid for expression of HA-GLUT4-GFP and measuring the HA-GLUT4-GFP at the cell surface by quantitative single-cell immunofluorescence, as previously described (7). In this method, HA-GLUT4-GFP at the cell surface is labeled with anti-HA and Cy3-conjugated secondary antibody; the fluorescence intensities for Cy3 and GFP in individual cells are quantitated, and the relative amount of HA-GLUT4-GFP at the cell surface is expressed as the ratio of Cy3 to GFP, to correct for different levels of expression of the HA-GLUT4-GFP protein.
The association of TBC1D4 with itself and with the 363Tr form was examined by cotransfecting human embryonic kidney 293E cells with plasmids for expression of N-terminal T7-tagged human TBC1D4 and either N-terminal Flag-tagged TBC1D4 or the 363Tr with Lipofectamine 2000. The cells were treated with insulin (1 μM for 10 min) or left unstimulated, then lysed in 40 mM Hepes, 150 mM NaCl, pH 7.4 containing 1.5% nonionic detergent octaethyleneglycol dodecyl ether and protease and phosphatase inhibitors. The lysate was cleared by centrifugation at 12,000 × g for 15 min and the portions of the supernatant were immunoprecipitated with anti-Flag agarose or irrelevant mouse Ig and protein G-agarose. SDS samples of the immunoprecipitates were immunoblotted for the T7 and Flag epitopes.
Glucose Tolerance Tests and Biochemical Assays.
Whole body insulin sensitivity was assessed with a 2-hour, 75-g oral glucose tolerance test. After insertion of an antecubital i.v. line, blood samples were collected at −30, 0, 10, 30, 60, 90, and 120 min for determination of plasma glucose, insulin, c-peptide, and free fatty acids. Insulin, leptin, and adiponectin were measured using customized autoDELFIA immunoassays as previously described (22).
Statistical Analysis.
GLUT4 translocation data are expressed as means ± SE. Differences between vectors were compared with use of the unpaired Student's t test. All reported P values are from 2-sided tests, and P values of less than 0.05 were considered to indicate statistical significance.
Supplementary Material
Acknowledgments.
We thank the proband and her family for their participation. The plasmid for expression of T7-tagged TBC1D4 was kindly provided by Dr. Mitsunori Fukuda of Tohoku University. This work was supported by grants from the Wellcome Trust (to R.K.S., S.O., and D.B.S.), Medical Research Council (to S.D., C.L., and N.J.W.), British Heart Foundation (to J.J.R.), GlaxoSmithKline (to D.B.S.), the National Institute for Health Research Cambridge Biomedical Research Centre, Raymond & Beverly Sackler scholarship (to S.D.), and Grant DK25336 from the National Institutes of Health (to G.E.L.). No potential conflict of interest relevant to this article was reported.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900909106/DCSupplemental.
References
- 1.Minokoshi Y, Kahn CR, Kahn BB. Tissue-specific ablation of the GLUT4 glucose transporter or the insulin receptor challenges assumptions about insulin action and glucose homeostasis. J Biol Chem. 2003;278:33609–33612. doi: 10.1074/jbc.R300019200. [DOI] [PubMed] [Google Scholar]
- 2.Shulman GI. Unraveling the cellular mechanism of insulin resistance in humans: New insights from magnetic resonance spectroscopy. Physiology (Bethesda) 2004;19:183–190. doi: 10.1152/physiol.00007.2004. [DOI] [PubMed] [Google Scholar]
- 3.Huang S, Czech MP. The Glut4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Hoehn KL, et al. IRS-1 independent defects define major nodes of insulin resistance. Cell Metab. 2008;7:421–433. doi: 10.1016/j.cmet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng Y, Ramm G, Lopez JA, James DE. Rapid activation of Akt2 is sufficient to stimulate Glut4 translocation in 3T3L1 adipocytes. Cell Metab. 2008;7:348–356. doi: 10.1016/j.cmet.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Kane S, et al. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase activating protein (GAP) domain. J Biol Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 7.Sano H, et al. Insulin stimulated phosphorylation of a Rab GTPase activating protein regulates Glut4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 8.Eguez L, et al. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab. 2005;2:263–272. doi: 10.1016/j.cmet.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Sano H, et al. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 10.O'Rahilly S, Barroso I, Wareham NJ. Genetic factors in type 2 diabetes: The end of the beginning? Science. 2005;307:370–373. doi: 10.1126/science.1104346. [DOI] [PubMed] [Google Scholar]
- 11.Abdul-Ghani MA, Tripathy D, Defronzo RA. Contributions of beta-cell dysfunction and insulin resistance to the pathogenesis of impaired glucose tolerance and impaired fasting glucose. Diabetes Care. 2006;29(5):1130–1139. doi: 10.2337/diacare.2951130. [DOI] [PubMed] [Google Scholar]
- 12.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry JR, Frayling TM. New gene variants alter type 2 diabetes risk predominantly through reduced beta-cell function. Curr Opin Clin Nutr Metab Care. 2008;11(4):371–377. doi: 10.1097/MCO.0b013e32830349a1. [DOI] [PubMed] [Google Scholar]
- 14.Krook A, O'Rahilly S. Mutant insulin receptors in syndromes of insulin resistance. Baillieres Clin Endocrinol Metab. 1996;10:97–122. doi: 10.1016/s0950-351x(96)80330-2. [DOI] [PubMed] [Google Scholar]
- 15.George S, et al. A family with severe insulin resistance and diabetes due to a mutation in Akt2. Science. 2004;304:1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagase T, et al. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5(1):31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 17.Treebak JT, et al. Potential role of TBC1D4 in enhanced post-exercise insulin action in human skeletal muscle. Diabetologia. 2009;52(5):891–900. doi: 10.1007/s00125-009-1294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci USA. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bidou L, et al. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 2004;11:619–627. doi: 10.1038/sj.gt.3302211. [DOI] [PubMed] [Google Scholar]
- 20.Amiel SA, et al. Insulin resistance of puberty: A defect restricted to peripheral glucose metabolism. J Clin Endocrinol Metab. 1991;72:277–282. doi: 10.1210/jcem-72-2-277. [DOI] [PubMed] [Google Scholar]
- 21.Shepherd PR, Kahn BB. Glucose transporters and insulin action-implications for insulin resistance and diabetes mellitus. N Engl J Med. 1999;341:248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 22.Semple RK, et al. Elevated plasma adiponectin in humans with genetically defective insulin receptors. J Clin Endocrinol Metab. 2006;91:3219–3223. doi: 10.1210/jc.2006-0166. [DOI] [PubMed] [Google Scholar]
- 23.Druet C, et al. Insulin resistance and the metabolic syndrome in obese French children. Clin Endocrinol (Oxf) 2006;64:672–678. doi: 10.1111/j.1365-2265.2006.02526.x. [DOI] [PubMed] [Google Scholar]
- 24.Loos RJ, et al. TCF7L2 polymorphisms modulate proinsulin levels and beta-cell function in a British Europid population. Diabetes. 2007;56:1943–1947. doi: 10.2337/db07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.