Abstract
Myotonic dystrophy type 1 (DM1) is a genetic disorder linked to a (CTG)n repeat expansion in the 3′ untranslated region of the DMPK gene. Upon transcription in the nucleus, the CUG repeats form a stable RNA stem-loop that sequesters the RNA-binding protein MBNL1 from its normal function in the cell. MBNL1 regulates the alternative splicing of many pre-mRNAs, and upon MBNL1's sequestration, the alternative splicing of many genes is mis-regulated, leading to disease symptoms. MBNL1 is known to bind directly to at least 3 of the pre-mRNAs that it regulates, but how MBNL1 binding mechanistically regulates alternative splicing is unclear. Here, we demonstrate that MBNL1 controls the splicing of exon 5 in the cardiac troponin T (cTNT) pre-mRNA by competing directly with the essential splicing factor U2AF65 for binding at the 3′ end of intron 4. When U2AF65 is prevented from binding to the pre-mRNA, the U2 snRNP can no longer be recruited and the following exon is skipped. Furthermore, MBNL1 and U2AF65 appear to compete by binding to mutually exclusive RNA structures. When bound by splicing factors, the 3′ end of intron 4 can form either a stem-loop or a single-stranded structure. MBNL1 binds a portion of the intron as a stem-loop, whereas U2AF65 binds the same region in a single-strand structure. Mutations that strengthen the stem-loop decrease U2AF65 binding affinity and also repress exon 5 inclusion, independently of MBNL1. Thus, U2AF65 binding can be blocked either by MBNL1 binding or by the stabilization of RNA secondary structure.
Keywords: muscleblind-like 1, myotonic dystrophy, alternative splicing
Alternative splicing is a fundamentally important process that many organisms use to increase proteomic diversity. Many diseases are known to arise from the mis-regulation of alternative splicing (1). Myotonic dystrophy (DM) is an example of a disease where the alternative splicing of many pre-mRNAs is mis-regulated. The mis-splicing seen in DM is thought to be caused, at least in part, by the mis-localization of the splicing factor MBNL1 (muscleblind-like 1) (2–4). MBNL1 is known to interact directly with 3 pre-mRNAs that are mis-regulated in DM, the cardiac troponin T (cTNT) (5, 6), fast troponin T (7), and SERCA1 (8). It is unclear how many more of the mis-regulated pre-mRNAs observed in DM may also be directly bound and regulated by MBNL1.
We have recently shown that MBNL1 binds a stem-loop within intron 4 of the cardiac troponin T (cTNT) pre-mRNA. This stem-loop is located directly upstream of exon 5, which is mis-regulated in this disease (5, 6). The mechanism used by MBNL1 to repress exon 5 remains undetermined. One mechanism might entail a direct binding competition between MBNL1 and other splicing factors. One of the best articulated models of the regulation of alternative splicing is sex determination in Drosophila melanogaster, where the protein Sex Lethal (Sxl) competes with the splicing factor U2AF65 at the 3′ end of certain introns [reviewed in ref. 9]. U2AF65 is thought to be 1 of the first splicing factors to bind an intron and thus, helps define the 3′ end of the intron. In concert with other proteins that bind near the 3′ end, U2AF65 is responsible for helping to recruit the U2 snRNP to the branch-point sequence (10–12). When Sxl inhibits U2AF65 binding and the subsequent recruitment of the U2 snRNP, the spliceosome selects another 3′ splice site, or the intron is retained if another 3′ splice site cannot be defined (13, 14).
We hypothesized that MBNL1 may act through a similar mechanism to compete with U2AF65. U2AF65 is a potential competitive target of MBNL1 because a putative U2AF65 binding site appears to be in the loop portion of the stem-loop which MBNL1 binds (5). We found that MBNL1 does compete with U2AF65 for binding of a region within intron 4. This competition with U2AF65 is functionally important, because recruitment of the U2 snRNP is reduced by MBNL1. Furthermore, we found that MBNL1 and U2AF65 compete by binding mutually exclusive RNA structures.
Results
U2AF65 Recognizes a Canonical Sequence Within Intron 4 of the cTNT pre-mRNA.
We have shown that MBNL1 binds a stem-loop located at the 3′ end of intron 4 (Fig. 1 A and B) (5). A putative polypyrimidine tract (py-tract) for intron 4, the likely U2AF65 binding site, exists primarily within the loop portion of this stem-loop (Fig. 1 A and B). This loop contains an uninterrupted sequence of 5 uracil and 7 cytosine residues and is 25 residues upstream of the 3′ splice site and 9 residues downstream of a consensus branch-point sequence.
Fig. 1.
U2AF65 binds the putative polypyrimidine tract in the cTNT intron 4. (A) Schematic of the 3′ end of intron 4 of the cTNT pre-mRNA. The cTNT 50mer used for binding studies is indicated, and the bases in the stem-loop structure. The 3′ splice site, putative polypyrimidine tract and putative branch-point are also indicated. (B) Structures of tested RNAs. (C) Binding curves of tested RNAs.
The affinities of U2AF65 for regions of this intron were determined by using an electrophoretic mobility shift (gel shift) assay. U2AF65 bound to a 50-nucleotide sequence (cTNT 50mer) containing the putative py-tract with an affinity (Kd) of 310 ± 30 nM (Fig. 1C, see supporting information (SI) Fig. S1 for representative gel shifts). Truncated versions of this RNA that retained the putative py-tract, but removed another run of pyrimidines nearer to the 3′ splice site, showed an unchanged binding affinity for U2AF65 (cTNT 32mer, Fig. 1 B and C). Mutations that replaced the putative py-tract nearly abolished U2AF65 binding (UUCG Loop, Fig. 1 B and C). Based on its location, the sequence of the region, and the binding data and the truncations, it is likely that this region is indeed the py-tract and comprises the binding site of U2AF65 within this intron.
MBNL1 and U2AF65 Bind Competitively to the 3′ End of Intron 4.
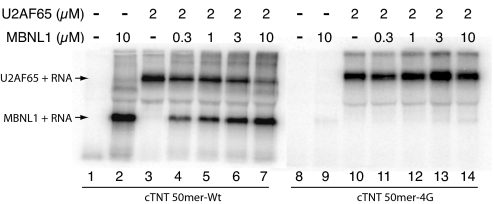
To determine whether MBNL1 competes with U2AF65 for binding at this intronic site, a UV cross-linking assay was used. While holding U2AF65 concentration constant, increasing the concentration of MBNL1 strongly reduced U2AF65 cross-linking (Fig. 2). If the 2 proteins could bind to the RNA at the same time, it was expected that a ternary complex of higher molecular weight would be observed. However, because U2AF65 cross-linking was significantly decreased and little or no apparent ternary complex was observed, it appears that MBNL1 and U2AF65 bind competitively to the cTNT 50mer-Wt sequence.
Fig. 2.
MBNL1 and U2AF65 competitively bind intron 4 of the cTNT pre-mRNA. MBNL1 and U2AF65 were UV cross-linked to the cTNT 50mer-Wt RNA and run on a denaturing gel.
A mutated RNA (cTNT 50mer-4G, Fig. 2) was used to determine whether nonspecific binding of MBNL1 affected U2AF65 binding. In this RNA, 4 guanosine residues in the stem were mutated that are required for MBNL1 binding (5) and cross-linking (6), whereas U2AF65 binding and cross-linking remain intact. As expected, cross-linking of this RNA to U2AF65 was not affected by MBNL1, showing that MBNL1 specifically competes with U2AF65 only for the wild-type RNA sequence.
MBNL1 Inhibits “A Complex” Formation on Intron 4.
The role of U2AF65 is to define the 3′ end of an intron and help recruit the U2 snRNP to the branch-point (10–12), forming “A complex” (15, 16). If U2AF65 does not bind, recruitment of the U2 snRNP to the branch-point is usually compromised (11, 12). We therefore predicted that the competition of MBNL1 with U2AF65 would inhibit the formation of the A complex on this intron.
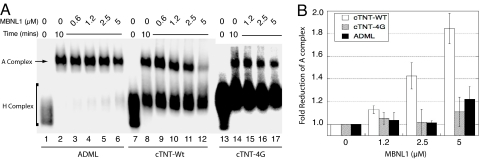
An RNA construct was made that contained human cTNT exon 5, together with 100 nts of the 3′ end of the RNA upstream of intron 4. Sequences from a constitutively spliced intron from chicken troponin I (TNI) were placed upstream of that sequence (see Fig. S2A for diagram). Radiolabeled RNA was incubated with HeLa nuclear extract, allowing splicing complexes to form on the RNA substrate. A time course shows that this cTNT pre-mRNA forms A complex well (Fig. S2B). The ADML pre-mRNA substrate served as a positive control because it forms A complex robustly (Fig. S2B). In the absence of ATP, no complex band is formed on the cTNT-Wt pre-mRNA, indicating that this is A complex (Fig. S2C). At later time points, larger complexes (B and C) are seen on the ADML pre-mRNA, but the cTNT pre-mRNAs are not robust enough to form these complexes in vitro before RNA degradation.
The 10-min time point was chosen to assay the effect of MBNL1 on A complex formation because RNA degradation was minimal and complex formation was optimal on the cTNT pre-mRNA (Fig. S2B). Complex formation on the cTNT-Wt pre-mRNA was reduced by nearly 2-fold with increasing concentrations of MBNL1, whereas little effect was seen on the ADML pre-mRNA or on the mutant cTNT-4G pre-mRNA (Fig. 3 A and B). This indicates that MBNL1 reduces the formation of A complex only on its specific pre-mRNA target, and does not perturb A complex formation in general.
Fig. 3.
MBNL1 reduces “A complex” formation specifically on intron 4 of the cTNT pre-mRNA. (A) A complex formation on the cTNT-Wt pre-mRNA, ADML and cTNT-4G pre-mRNAs. (B) Quantitation showing the effect of MBNL1 on A complex formation on cTNT-Wt, cTNT-4G and ADML pre-mRNAs.
Stabilization of the Stem-Loop Reduces U2AF65 Binding Affinity and Represses Exon 5 Inclusion in Vivo.
We previously observed that mutations that increased the number of base-pairs in the stem-loop repressed exon 5 inclusion, independently of MBNL1 (5). This led us to hypothesize that these mutations might repress the inclusion of exon 5 by reducing the affinity of U2AF65 for the py-tract, in a manner similar to the repression mechanism used by MBNL1. To investigate this possibility, we tested the affinity of U2AF65 for the cTNT intron 4 py-tract using stem-loops with increased stability.
A correlation was seen between the stability of the stem-loop and the binding affinity of U2AF65, with stronger stems nearly abolishing U2AF65 binding (Fig. 4 A and B). The Stem Strengthen (SS) mutation stabilized the stem by adding one base pair to each end of the stem, increasing the Tm of the stem-loop by 5 °C, and reduced U2AF65 binding nearly 10-fold (Fig. 4 A and B). The No Mismatch (NM) mutation increased the stability of the stem by 10 °C and reduced U2AF65 binding to the point where a Kd value could not be determined (Fig. 4 A and B). It is interesting that the mutation that more strongly stabilized the stem structure had a larger effect on binding. This result suggests that U2AF65 binds the py-tract in a single-stranded region of this RNA. As a negative control, a mutation (UUCG loop, UL) was made that removed the py-tract from the loop to fully abolish U2AF65 binding (Fig. 4 A and B).
Fig. 4.
Mutations that stabilize the stem-loop structure reduce U2AF65 binding affinity and repress exon 5 inclusion in vivo. (A) Schematic of different mutations, the thermal stability of each mutation and the binding affinity of U2AF65 for each RNA. (B) Binding curve of tested mutations. (C) Representative RT-PCR data from in vivo splicing of cTNT mutant series. (D) Quantitation of RT-PCR data, and data from CUG960 cotransfection experiments.
Furthermore, we found a strong correlation between U2AF65 binding affinity and exon 5 inclusion in vivo (Fig. 4 C and D). The NM, SS, UL mutations all showed minimal exon 5 inclusion using an in vivo splicing assay. This repression was independent of MBNL1, because coexpression of expanded CUG repeats (which would sequester MBNL1) had no affect on exon 5 inclusion for these mutations (Fig. 4D). These results indicate that inclusion of exon 5 can be regulated either by competition of MBNL1 with U2AF65 at the 3′ end of the intron, or by the formation of RNA secondary structures that reduce the affinity of U2AF65 for its binding site.
U2AF65 and MBNL1 Bind Distinct RNA Structures.
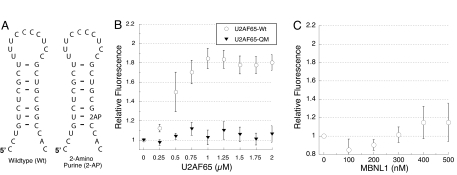
To determine the effect of MBNL1 or U2AF65 binding on the stem-loop structure, a fluorescence-based assay was used. The fluorescent adenosine analogue 2-amino purine (2-AP) was used to replace a guanosine residue that forms a “wobble base-pair” with a uracil residue near the base of the stem (Fig. 5A). 2-AP was chosen as a spectroscopic probe for this purpose because its fluorescence is strongly quenched when it is in a base-paired structure (here with uracil), whereas its fluorescence strongly increases when it is in a single-stranded environment. The guanosine residue near the base of the stem was selected for replacement as no adenosine residues were available in the stem and replacement of this position had no effect on MBNL1 or U2AF65 binding or the stability of the stem.
Fig. 5.
U2AF65 destabilizes the stem-loop upon binding, whereas MBNL1 binds the stem-loop structure. (A) Schematic of fluorescent RNA used to probe cTNT structure. (B) Effects of wild-type or Quad Mutant (QM) U2AFA65 on cTNT fluorescent probe. (C) Effect of MBNL1 on the fluorescent cTNT probe.
Titration of U2AF65 to the RNA increased the fluorescence of the probe nearly 2-fold (Fig. 5B). In general, the fluorescence of a 2-AP probe has been shown to increase 2- to 3-fold when 2-AP switches from a base-paired to a single-stranded environment (17). Therefore, this result suggests that U2AF65 binds this RNA in a single-stranded structure. U2AF65 was titrated to a point where the RNA should be approximately 85% bound, based on the Kd determined from the gel shift assay. A U2AF65 protein that contains 4 mutations which inhibit RNA binding (U2AF65–Quad Mutant or QM (18)) had no effect on the fluorescence of the probe (Fig. 5B).
In contrast to U2AF65, titration of MBNL1 (to a concentration at which 85% of the RNA should be bound) had no significant effect on the fluorescence of the 2-AP probe (Fig. 5C). This result indicates that the stem structure remains intact upon MBNL1 binding. However, a recent crystal structure of 2 zinc fingers from MBNL1 with a six nucleotide RNA suggests that at least 2 of MBNL1's zinc fingers bind RNA sequences in a single-stranded structure (19). This crystal structure was only obtained with a minimal RNA sequence that was not long enough to form a double-stranded structure, so it is unclear whether a longer RNA sequence would be in a different structure. It might be possible that MBNL1 does bind the cTNT stem as single-stranded RNA, but the protein quenches the 2-AP probe so that the fluorescence does not increase. However, this is unlikely, because 2-AP probes have not been seen to be strongly quenched by direct protein binding when compared with base stacking and base-pairing in a stem (17, 20).
The recent crystal structure argues that it is probable that MBNL1 opens at least 1 or 2 bases in the stem. However, this fluorescence data suggests that other portions of the stem remain structured. We therefore conclude that at least a portion of the stem remains intact upon MBNL1 binding, whereas U2AF65 binds to a completely single-stranded structure.
Discussion
MBNL1 Regulates the cTNT Exon 5 Through Competition with U2AF65.
The competition of MBNL1 with U2AF65 represents an example of an alternative splicing factor that regulates splicing primarily through modulation of an RNA structural element and not through direct occlusion of another splicing factor's binding site. The competition of MBNL1 with U2AF65 does not constitute a novel mechanism, because other alternative splicing factors have been shown to compete directly with U2AF65. The alternative splicing factor Sex Lethal (Sxl) has been shown to compete with U2AF65 in the tra and msl-2 pre-mRNAs (13, 14), whereas the Polypyrimidine Tract Binding Protein (PTB) has been observed to compete with U2AF65 for sequences within the α- or β-tropomyosin pre-mRNAs (21, 22).
Sxl and PTB both have binding specificities similar to that of U2AF65, because all 3 proteins primarily prefer long runs of single-stranded uracil residues (21). When competitively binding with U2AF65, both Sxl and PTB are considered to occlude the U2AF65 binding site, and sterically inhibit U2AF65 from accessing its binding site. However, MBNL1 has a different binding specificity from U2AF65, and binds adjacent sequences primarily outside of the py-tract. We cannot rule out steric contributions to the competition between MBNL1 and U2AF65, but the fluorescence assay strongly suggests that these proteins bind this RNA in different conformations (Fig. 5). Both the differential binding sites of MBNL1 and U2AF65, and the role that RNA secondary structure can play in modulating U2AF65 binding, suggest that these splicing factors compete for recognition of the 3′ end of intron 4 largely through binding mutually exclusive RNA structures (see Fig. 6 for model).
Fig. 6.
Model of regulation of cTNT exon 5 by MBNL1 and RNA structure. (A) Initial recognition of intron 4 by splicing factors. U2AF65 binds the intron in a single-stranded structure, but U2AF65 binding is inhibited if it cannot destabilize the stem because of MBNL1 binding or mutations that stabilize the stem-loop. (B) U2 snRNP recruitment to all 3′ splice sites where U2AF65 is present. (C) Spliceosomal recruitment and splicing. (D) mRNA splice products.
The Role of MBNL1 in Regulating Alternative Splicing by Modulating RNA Secondary Structure.
It is well documented that RNA structural elements alone can regulate alternative splicing. For instance, stable structures have been shown to inhibit U1 snRNP binding to the 5′-splice site following exon 7 of both the SMN1 and SMN2 pre-mRNAs (23). Other structural elements that encompass exon 6B in chicken β-tropomysin have been shown to also inhibit binding of all of the U snRNPs, promoting the skipping of that exon (24). At present, it is unclear whether these examples also involve alternative splicing factors that regulate these RNA structures, as MBNL1 appears to regulate secondary structures within the cTNT pre-mRNA.
Other alternative splicing factors have been postulated to regulate RNA structural elements, but there is no clear evidence to show that these factors modulate RNA structure in a way that directly regulates alternative splicing. For example, the splicing factor hnRNP A1 has been shown to inhibit binding of splicing factors ASF/SF2 and SC35 when they regulate splicing of the tat pre-mRNA from HIV-1 (25). Binding of hnRNP A1 is thought to alter the secondary structure of the pre-mRNA in a way that affects splicing (26), but this has not been demonstrated. Similarly, ribosomal protein L32 from S. cerevisiae binds a structured RNA element near the 5′-splice site of an intron within its own pre-mRNA, leading to intron retention (27). However, this structured binding site still allows for recruitment of the U1 snRNP, showing that RNA structures do not always inhibit binding of the U snRNPs (28). It is still unclear how L32 causes intron retention if it does not inhibit binding of U1.
In regards to the competition between MBNL1 and U2AF65, it has been demonstrated with a crystal structure that U2AF65 binds the py-tract in a single-stranded structure (29). In this structure, 7 uridine residues bind along 2 RNA recognition motifs of U2AF65, leaving the 3′ and 5′ ends spatially far from each other. This crystal structure sheds light on why U2AF65 must bind the cTNT intron in a single-stranded form, because it is unlikely that the stem could form with the loop in such an extended structure. At present, MBNL1 is an alternative splicing factor that has been shown to regulate alternative splicing on the level of RNA structure. It is likely that other examples will follow as it is becoming increasingly apparent that RNA structure plays important roles in controlling pre-mRNA splicing.
Materials and Methods
Cloning and Protein Purification.
MBNL1 and U2AF65 were purified as described (5, 18), and are described in SI Text.
RNA Synthesis, Labeling, and Purification.
The RNA substrates used for the Complex formation assay were transcribed with T7 polymerase, off linearized Puc19 plasmid. During transcription, RNAs were radiolabeled using [α-32P]CTP. All other RNA substrates were ordered from IDT DNA, and 5′ end-labeled using [γ-32P]ATP.
Cross-Linking Assay.
RNA and proteins were incubated at room temperature for 10 min, then on ice for 10 min, under the same conditions as used in the gel shift assay. Samples were placed on a pre-chilled block and exposed to UV for 1 min 20 seconds (1 Joule/cm2), 3.5 cm from the light source using a FB-UVXL-1000 lamp (Fisher Scientific). Samples were then incubated in protein denaturing buffer for 2 min at 95°C and were resolved on a 10% denaturing SDS/PAGE gel. Gels were then dried and autoradiographed.
Gel Shift Assay.
Final conditions for the RNA-U2AF65 binding experiments were 175 mM NaCl, 5 mM MgCl2, 20 mM Tris (pH 7.5), 1.25 mM BME, 12.5% glycerol, 2 mg/ml BSA, and 0.1 mg/ml Heparin. Prior to incubation, RNA substrates were snap annealed in 66 mM NaCl, 6.7 mM MgCl2, and 27 mM Tris (pH 7.5). Protein was then added to the RNA, in a 10 μL volume and incubated for 10 min at room temp before 3–5μL were loaded on a prechilled 5% acrylamide (37.5:1), 0.5x TB gel, and run for 1 hr at 175 volts, at 4 °C. Gels were dried and autoradiographed.
For binding curves, gels were quantified using ImageQuant (Molecular Dynamics). The percentage of RNA bound was determined by taking the ratio of RNA:Protein complex to total RNA, per lane. Binding curves were graphed and apparent Kd values were determined with KaleidaGraph (Synergy) software using the following equation: y = ((m2 + m1 + m0) − ((−m2 − m1 − m0)2 − (4 × m1 × m0))0.5/(2 × m1), where y = % RNA bound, m2 = Kd, m1 = total RNA concentration and m0 = protein concentration. This equation assumes a 1:1 interaction between the RNA and protein, which allows only an apparent Kd to be determined for RNAs containing more than one binding site. To determine the standard error of the apparent Kds, 3–5 binding titrations were performed with each substrate and the apparent Kd values determined for each titration separately, prior to averaging. The error bars on the binding curve were obtained by averaging the individual titration points and calculating the standard deviation.
Complex Formation Assay.
HeLa nuclear extract was prepared (30) and complex formation was performed (31) as previously described. In short, HeLa extract was added to a solution with RNA and buffer and incubated at 30 °C for the appropriate time point. The sample volume was 12 μL, and contained 25% HeLa extract, 55 mM KOAc, 30 mM KCl, 14.5 mM Hepes (pH 8.0), 10% glycerol, 0.12 mM EDTA (pH 8.0), 0.5 mM DTT, 3.2 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate, 0.7 units/μL of RNase Inhibitor (Ambion). After the appropriate time point, samples were incubated with 4 μL of heparin (4 mg/mL), and incubated at room temp for 2 minutes. ADML and cTNT-wt samples were then loaded onto a prechilled 1.8% agarose gel (1x TG), and run at 75 volts for 1.5 hours, at 4 °C, in a 1x TG solution. To better resolve A complex, cTNT-4G samples had 80 mM KOAc, and were run on a 1.2% agarose gel (1x TG), for 4.5 hours at 40 volts, 4 °C. The gels were then fixed in 10% acetic acid, 10% methanol solution for 10 min. Gels were dried, autoradiographed, and quantified using ImageQuant (Molecular Dynamics). To determine the fold reduction, the percentage of complex formed in the absence of MBNL1 was divided by the percentage complex formed in the presence of MBNL1.
2-Amino Purine Fluorescence.
RNA (Dharmacon RNA Technologies) was diluted to 1 μM concentration in the binding buffer (175 mM NaCl, 20 mM NaCl, 5 mM MgCl2) and snap annealed. All reactions were performed at room temperature, in the binding buffer, using an l-formate Jobin-Yvon Horiba Fluoromax fluorimeter. A 3 mm wide Spectrosil microcell cuvette (Starna Cells, Inc) was used. The 2-amino purine was excited at 312 nm, and spectra were collected from 310 to 420 nm. The fluorimeter slits were 3 nm, with an integration time of 0.2 seconds. Spectra collected with buffer and protein was used to subtract out background. Three spectra were collected and averaged for every titration point, with 3–4 titrations performed to determine average values and standard deviations. To calculate relative fluorescence, the value at 375 nm was used.
In Vivo Splicing.
HeLa cell transfections, and RT-PCR was done as described in ref. 5, and are described in the SI Text.
Supplementary Material
Acknowledgments.
We thank members of the Berglund, von Hippel, and Barkan labs (especially Kausiki Datta and Alice Barkan) for experimental advice and help with equipment. This work was supported by National Institutes of Health Grants AR053903 (to J.A.B.) and GM-15792 (to P.H.v.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900342106/DCSupplemental.
References
- 1.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nature Biotech. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 2.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 3.Cho DH, Tapscott SJ. Myotonic dystrophy: Emerging mechanisms for DM1 and DM2. Biochim Biophys Acta. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Nykamp KR, Swanson MS. Toxic RNA in the nucleus: Unstable microsatellite expression in neuromuscular disease. Prog Mol Subcell Biol. 2004;35:57–77. doi: 10.1007/978-3-540-74266-1_3. [DOI] [PubMed] [Google Scholar]
- 5.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho TH, et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Y, et al. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hino S, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum Mol Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 9.Penalva LO, Sanchez L. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev. 2003;67:343–359. doi: 10.1128/MMBR.67.3.343-359.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaur RK, Valcarcel J, Green MR. Sequential recognition of the pre-mRNA branch point by U2AF65 and a novel spliceosome-associated 28-kDa protein. RNA. 1995;1:407–417. [PMC free article] [PubMed] [Google Scholar]
- 11.Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 12.Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 13.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 14.Valcarcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 15.Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci USA. 1990;87:8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed R, Griffith J, Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 17.Datta K, Johnson NP, von Hippel PH. Mapping the conformation of the nucleic acid framework of the T7 RNA polymerase elongation complex in solution using low-energy CD and fluorescence spectroscopy. J Mol Biol. 2006;360:800–813. doi: 10.1016/j.jmb.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 18.Henscheid KL, Voelker RB, Berglund JA. Alternative modes of binding by U2AF65 at the polypyrimidine tract. Biochemistry. 2008;47:449–459. doi: 10.1021/bi701240t. [DOI] [PubMed] [Google Scholar]
- 19.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi-Brunozzi HY, Stephens OM, Beal PA. Conformational changes that occur during an RNA-editing adenosine deamination reaction. J Biol Chem. 2001;276:37827–37833. doi: 10.1074/jbc.M106299200. [DOI] [PubMed] [Google Scholar]
- 21.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 22.Lin CH, Patton JG. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 23.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirand-Pugnet P, Durosay P, Clouet d'Orval BC, Brody E, Marie J. beta-Tropomyosin pre-mRNA folding around a muscle-specific exon interferes with several steps of spliceosome assembly. J Mol Biol. 1995;251:591–602. doi: 10.1006/jmbi.1995.0458. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 26.Damgaard CK, Tange TO, Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA. 2002;8:1401–1415. doi: 10.1017/s1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eng FJ, Warner JR. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 28.Vilardell J, Warner JR. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 29.Sickmier EA, et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 31.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.