Abstract
Increased encephalization, or larger brain volume relative to body mass, is a repeated theme in vertebrate evolution. Here we present an extensive sampling of relative brain sizes in fossil and extant taxa in the mammalian order Carnivora (cats, dogs, bears, weasels, and their relatives). By using Akaike Information Criterion model selection and endocranial volume and body mass data for 289 species (including 125 fossil taxa), we document clade-specific evolutionary transformations in encephalization allometries. These evolutionary transformations include multiple independent encephalization increases and decreases in addition to a remarkably static basal Carnivora allometry that characterizes much of the suborder Feliformia and some taxa in the suborder Caniformia across much of their evolutionary history, emphasizing that complex processes shaped the modern distribution of encephalization across Carnivora. This analysis also permits critical evaluation of the social brain hypothesis (SBH), which predicts a close association between sociality and increased encephalization. Previous analyses based on living species alone appeared to support the SBH with respect to Carnivora, but those results are entirely dependent on data from modern Canidae (dogs). Incorporation of fossil data further reveals that no association exists between sociality and encephalization across Carnivora and that support for sociality as a causal agent of encephalization increase disappears for this clade.
Keywords: Akaike Information Criterion, allometry, encephalization, Mammalia, phylogeny
Grounded in the concept that increased encephalization represents more neurons per unit body mass, which in turn implies higher potential cognitive function, there often is an assumption that greater encephalization equates in some way to greater intelligence (1, 2). Indeed, increased encephalization has been linked to greater behavioral flexibility and adaptability to novel environments (3–5), and has been observed in the evolution of multiple amniote clades, including certain carnivoran subgroups (6, 7), primates (8–10), cetaceans (11), and birds (12). Yet the brain is energetically expensive to maintain, requiring by mass nearly an order of magnitude more energy than other somatic tissues (13, 14), and encephalization has been shown to decrease in response to reduced predation pressure (e.g., see ref. 15) and domestication (e.g., see ref. 16), suggesting selective benefits to eliminating excess brain volume when cognitive demands are reduced. A variety of hypotheses have sought to explain potential benefits that might offset the cost of increased encephalization, including trade-offs relative to other metabolically expensive tissues (14, 17), constraints imposed by basal metabolic rate (18), or key innovations (11) and interspecific (19) or intraspecific (20) interactions. However, due to difficulties in measuring brain volumes in extinct taxa (21), these hypotheses are often based on little or no data from the fossil record.
The mammalian order Carnivora presents a model system for studying encephalization, as it possesses a well resolved phylogeny (22–24), an extensively sampled fossil record (25, 26), endocranial volumes for most extant taxa (27–29), a morphometric model for estimating endocranial volumes in fossil taxa (21), and body mass estimates for fossil taxa (26, 30). Herein, we develop a comprehensive view of the evolutionary history of encephalization across 289 terrestrial species (including 125 extinct species) of Carnivora, providing an extensive sampling of fossil and living taxa for both major subclades: Caniformia and Feliformia.
Results
Akaike Information Criterion (AIC) model selection recovered 4 optimal models (OM) within 2 log-likelihood units of the highest score (Table 1). There is broad agreement among the OM with differences primarily in estimates of allometric slopes. The most conspicuous feature is a basal Carnivora allometry grouping of nonfelid feliforms and stem canids in 3 of the 4 OM (Fig. 1). The best-supported model combines slopes for crown Canidae (wolves, foxes, jackals, etc.) and the basal Carnivora allometry, reconstructing encephalization increases between parallel allometries. The second model further combines the slope of Ursidae (bears) with the basal Carnivora and stem Canidae allometries. The third does not combine slopes, reconstructing distinct but similar slopes for the basal allometry, crown Canidae, and Ursidae. The fourth model does not reconstruct a basal regression, but instead combines slopes for nonfelid feliforms and stem and crown Canidae, although the stem canid and nonfelid feliform intercepts are similar.
Table 1.
Encephalization parameter values for the 4 models within 2 log-likelihood units of the best model
| Model | Name | Slope | Intercept | n | K | ESS | AICc | LnL |
|---|---|---|---|---|---|---|---|---|
| 1 | Felidae (includes Prionodon) | 0.5870 | 2.5795 | 39 | 19 | 11.98 | −879.90 | 0.00 |
| Basal regression—Stem Canidae and nonfelid Feliformia | 0.6375† | 2.3508 | 105 | |||||
| Crown Canidae | 0.6375† | 2.6696 | 35 | |||||
| Amphicyonidae | 0.5564 | 2.3879 | 13 | |||||
| Ursidae | 0.6551 | 2.6014 | 16 | |||||
| Mephitidae with archaic musteloids | 0.7090* | 2.0859 | 27 | |||||
| Musteloids (without skunks)—modern group (<10 Ma) | 0.7090* | 2.5279 | 54 | |||||
| 2 | Felidae (includes Prionodon) | 0.5870 | 2.5795 | 39 | 18 | 12.06 | −879.46 | −0.22 |
| Basal regression—Stem Canidae and nonfelid Feliformia | 0.6375† | 2.3508 | 105 | |||||
| Crown Canidae | 0.6375† | 2.6696 | 35 | |||||
| Amphicyonidae | 0.5564 | 2.3879 | 13 | |||||
| Ursidae | 0.6375† | 2.6799 | 16 | |||||
| Mephitidae with archaic musteloids | 0.7090* | 2.0859 | 27 | |||||
| Musteloids (without skunks)—modern group (<10 Ma) | 0.7090* | 2.5279 | 54 | |||||
| 3 | Felidae (includes Prionodon) | 0.5870 | 2.5795 | 39 | 20 | 11.95 | −877.63 | −1.14 |
| Basal regression—Stem Canidae and nonfelid Feliformia | 0.6387 | 2.3485 | 105 | |||||
| Crown Canidae | 0.6302 | 2.6848 | 35 | |||||
| Amphicyonidae | 0.5564 | 2.3879 | 13 | |||||
| Ursidae | 0.6551 | 2.6014 | 16 | |||||
| Mephitidae with archaic musteloids | 0.7090* | 2.0859 | 27 | |||||
| Musteloids (without skunks)—modern group (<10 Ma) | 0.7090* | 2.5279 | 54 | |||||
| 4 | Felidae (includes Prionodon) | 0.5870 | 2.5795 | 39 | 21 | 11.91 | −876.15 | −1.88 |
| Nonfelid Feliformia | 0.6340† | 2.3404 | 67 | |||||
| Stem Canidae | 0.6340† | 2.3875 | 38 | |||||
| Crown Canidae | 0.6340† | 2.6769 | 35 | |||||
| Amphicyonidae | 0.5564 | 2.3879 | 13 | |||||
| Ursidae | 0.6551 | 2.6014 | 16 | |||||
| Mephitidae with archaic musteloids | 0.7090* | 2.0859 | 27 | |||||
| Musteloids (without skunks)—modern group (<10 Ma) | 0.7090* | 2.5279 | 54 |
Slope, intercept, and additional model statistics are given for each group.
* and † indicate slopes that have been set as parallel. n, group sample size; K, number of estimated parameters for the entire model; ESS; total summed error residuals; AICc, model AICc score; LnL, model log-likelihood rescaled such that the best model has LnL = 0.
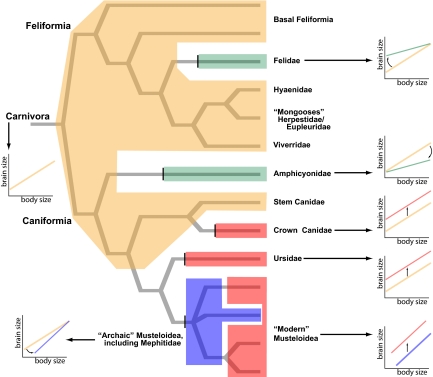
Fig. 1.
The evolution of carnivoran encephalization mapped onto the branching pattern of the phylogeny presented in Fig. S3. Schematic representations of the regression lines are included for each transformation in encephalization allometries. A tan polygon encloses the basal Carnivora allometry. Other polygons enclose derived allometries coded by color: Green indicates a lowering of the allometric slope, blue indicates an increase in slope (although intercepts vary), and red indicates an increase in intercept with constant slope. Arrows indicate the direction of change (increase/decrease) in relative brain size associated with (i) change in allometric slope in Amphicyonidae, Felidae, and “archaic” musteloids, and (ii) shifts in intercept among parallel allometries in Canidae, Uridsae, and “modern” musteloids. See Results for discussion.
Within Caniformia, the OM reconstruct distinct allometries for Ursidae and the extinct Amphicyonidae (“bear-dogs”) (Fig. 1), separate crown canids from their stem lineage, and demonstrate independent encephalization increases for ursids, living canids, and Musteloidea (skunks, red panda, weasels, raccoons, and allied taxa) (6, 7). Irrespective of whether Ursidae and crown Canidae possess identical or merely similar slopes, the estimated intercepts are similar, and because both diverge independently from the basal allometry, these increases must have been independently derived in each clade. Parallel allometries are also reconstructed for “archaic” and “modern” musteloids, documenting a similar increase between these groups (Fig. 1). Increased encephalization in musteloids probably represents multiple transformations among subclades, and it would be preferable to identify parallel changes in monophyletic lineages, but small sample size for fossil musteloids currently precludes finer partitioning within the group. Nevertheless, for 7 model pairs differing only in parsing musteloids by families [Mephitidae (skunks), Mustelidae (weasels, otters), and Procyonidae (raccoons)] versus archaic/modern partitions (see SI Text), models with distinct musteloid families averaged 8.68 log-likelihood units worse than the optimal models, strongly supporting an archaic/modern dichotomy. Notably, fossil and extant Mephitidae are grouped with archaic musteloids; 7 models separating a distinct mephitid allometry averaged 2.95 log-likelihood units worse than corresponding models including skunks with archaic musteloids. Therefore, it is important to note that lower encephalization for skunks relative to other living musteloids (6) is a retention of, rather than a reversal to, the ancestral musteloid condition.
In contrast to the dynamic caniform pattern, all Feliformia [civets, cats, hyaenas, mongooses, and Madagascar's carnivorans (Eupleridae)], except Felidae (cats), conform to a single allometry (Fig. 1). Four models partitioning feliform subclades averaged 4.91 log-likelihood units worse than corresponding models separating only Felidae. Additionally, no support exists for further division within Felidae; 9 models separating stem and crown felids averaged 3.10 log-likelihood units worse than corresponding single-allometry models for Felidae. The data thus indicate remarkably conservative brain–body size scaling for the Feliformia.
The OM reconstruct independent and significant encephalization increases for Canidae, Ursidae, and Musteloidea (38%, 39%, and 55%, respectively; log-likelihoods >3.0), in addition to 3 changes in slope from the basal Carnivora allometry: decreases for Amphicyonidae and Felidae and an increase for archaic musteloids. Although amphicyonids are not significantly smaller-brained than taxa in the basal regression (binomial P = 0.38, log-likelihood = 0.35), the intersection for these regressions (≈1.6 kg) falls well below the minimum amphicyonid body mass. A Mann–Whitney test of body masses, partitioned by smaller- or larger-than-expected brain volume, is significant (1-tail P = 0.034). Felidae are significantly larger-brained than the basal Carnivora allometry (binomial P = 0.67, log-likelihood = 2.21), and the intersection falls at the large end of observed felid body masses (≈93 kg). Therefore, although similar slopes are reconstructed for both clades (Table 1), Amphicyonidae deflect the slope such that larger species exhibit lower encephalization, whereas Felidae increase encephalization among smaller taxa (Fig. 1). Archaic musteloids are significantly less encephalized than the basal Carnivora allometry (binomial P = 0.22, log-likelihood = 4.41). Their intersection falls above the archaic musteloid body mass range (≈41 kg), and therefore the increased slope represents decreased encephalization for smaller archaic musteloid taxa.
Discussion
These well resolved and well supported optimal models of carnivoran encephalization permit evaluation of predictions made by the social brain hypothesis (SBH) (20). The SBH proposes that group living (“sociality”) increases cognitive demand on individuals, imparting selective pressure for increased encephalization. Recently, Perez-Barberia, et al. (31) found statistically significant associations between larger-than-expected brain volume and sociality for living taxa in 3 mammalian orders: Primates, Carnivora, and Artiodactyla. This presents a potential explanatory mechanism for the patterns observed in this study; for example, living canids are characterized by relatively large brains and highly social behaviors (7, 31), therefore the observed encephalization increase between stem and crown taxa in the Canidae might signal the origin of complex social behaviors in this clade.
However, if sociality is viewed as the causal agent for increased encephalization in mammals, then sociality also should be widespread among musteloids and bears, which exhibit encephalization increases similar to Canidae. Instead, musteloids are predominantly nonsocial, and all bears are solitary. Similarly, in contrast to the predictions of the SBH, increased encephalization is observed for small cats, yet those taxa are almost exclusively solitary. Furthermore, among taxa comprising the basal Carnivora allometry, the Hyaenidae and Herpestidae contain both social and nonsocial species. A Mann–Whitney test comparing larger- and smaller-than-expected brain volume, relative to the basal allometry, against sociality for hyaenas and mongooses is not significant (1-tail P = 0.154; Figs. S1 and S2). Therefore, the SBH does not adequately explain the evolutionary history of encephalization in Carnivora. Invoking the SBH for modern Carnivora would require a number of unparsimonious explanations for why some social taxa are not more encephalized than closely related solitary taxa, and why some groups in which wholesale shifts to higher encephalization are observed do not display concomitant increases in the incidence of sociality.
Closer inspection of the carnivoran data presented by Perez-Barberia, et al. (31) reveals that Canidae alone are responsible for the statistical significance; reanalysis of the same data, but excluding canids, removes the encephalization–sociality significance for the Carnivora (Fisher's Exact Test, P = 0.167). Brain architecture, and therefore expansion of certain structures within the brain, surely must influence higher cognitive function and social behavior. For example, the prorean gyrus has been implicated in mediating social behaviors among canids (32), and this region undergoes an expansion that roughly corresponds to the timing of the shift to increased encephalization in crown Canidae (7, 33). However, the data are ambiguous as to whether brain-size increase played a direct causative role in canid social evolution (reversing the vector of the SBH) or whether crown canids simply coopted the evolution of larger brains to enhance their social behaviors. In either case, the association of increased encephalization and highly social behaviors appears to be restricted to the Canidae among modern Carnivora and cannot be generalized to the entire order. The idea that sociality played a causative role in the expansion of relative brain size is not valid for this clade as a whole, and social structures for fossil carnivorans cannot be inferred simply from relative brain-size arguments.
The substantially increased sampling of body mass and endocranial-volume data presented here, particularly for fossil taxa and Feliformia, documents a complex set of evolutionary changes in encephalization allometries for the Carnivora. Because well supported phylogenies are now available, these transformations in brain–body size scaling can be localized to specific branches of the carnivoran evolutionary tree, documenting independent increases and decreases in encephalization across the order. Moreover, this increased sampling for living and fossil Carnivora now provides sufficient data to permit robust tests of hypotheses of potential mechanisms underlying brain-size evolution, documenting that explanations other than sociality must be sought for the multiple brain-size increases and decreases observed during the evolutionary history of this clade.
Methods
Data.
We compiled data on endocranial volume and body mass estimates for 289 species spanning the entire order Carnivora. In total, we surveyed 164 extant and 125 fossil carnivoran taxa (183 caniforms and 106 feliforms). All data for fossil Feliformia were new to this study, and we substantially augmented data for fossil Caniformia with additional data for 5 previously reported species (6, 7) and by adding 23 species not included in previous analyses.
Endocranial volume closely approximates actual brain volume in most extant mammals including Carnivora (1, 27, 34), permitting accurate representation of brain volumes among extant taxa as well as direct comparison to fossil taxa. Endocranial volume data for extant taxa were taken primarily from the literature (27–29), augmented with estimates for 13 extant species by using a morphometric model that estimates endocranial volume from 3 external measurements of the neurocranium (21). Endocranial volume estimates for fossil taxa were derived primarily from the application of this model to fossil cranial specimens. These data were augmented with volume estimates derived from direct volumetric measurement of fossil endocasts (1, 35–37) and from one virtual endocast calculated from computed tomographic scans for the fossil bear, Ursus deningeri (38). Body mass estimates for extant taxa were obtained primarily from a worldwide compendium of mammalian adult body masses (39). Body mass data for fossil taxa were obtained from the Neogene of the Old World (NOW) database (40) or were calculated from measurements of the lower first molar (30) or basal skull length (41). Endocranial volume and body mass data are reported in Table S1.
Model Selection and Hypothesis Testing.
To test hypotheses of the evolution of encephalization allometries, we constructed a composite cladogram of the Carnivora, synthesized from numerous phylogenetic analyses performed on this clade (e.g., see refs. 22 and 24 and Fig. S3). We explicitly tested models of change in encephalization allometries with respect to this phylogeny. Each evolutionary model then represents a unique configuration of allometries across the phylogeny.
When considering evolution of these scaling relationships across a phylogeny, the most parsimonious model for any set of taxa is always a single allometry (Fig. 2B). We subsequently compared models with increasingly complex structures, treating allometric regressions as characters evolving across the branching pattern of the phylogeny (e.g., see ref. 7 and Fig. 2C). In evaluating more complex models, we then combined slopes (i.e., defined parallel slopes) for allometries of taxa adjacent to one another on the cladogram, treating the intercept as the evolving character (Fig. 2D). Multiple-allometry models can describe encephalization more accurately than a single allometry for all Carnivora, and increasing the number of regressions tended to decrease overall residual variance because groups of related taxa are described by more precisely tuned scaling relationships. However, this reduction was achieved at the cost of increasing overall model complexity (42–44). When parallel regressions were hypothesized, a suboptimal slope was fit to each individual regression, increasing overall residual variance but simultaneously reducing the number of estimated parameters.
Fig. 2.
Schematic representation of the search strategy for carnivoran encephalization allometries. (A) Hypothetical body size and brain volume data for a set of taxa are given. There is a clear, if noisy, allometric relationship between the variables. (B) A single allometry is fit through the variables. This represents the simplest possible hypothesis relating the scaling of brain volume to body mass for any set of taxa. (C) A more complicated hypothesis is proposed, with 3 allometries proposed for distinct groups. Here it is assumed that the sets of taxa are proposed relative to a phylogenetic hypothesis. Note that the estimated slopes for 2 of the allometries (thin and dashed lines) are very similar to one another. (D) A more parsimonious hypothesis is proposed where the slopes for 2 allometries are defined as equal to one another, the 2 allometries differing only in a phase shift of their intercepts.
We estimated slope and intercept values for carnivoran encephalization allometries by using major-axis (Type II) regression (45), following methods detailed in previous analyses (46, 47). We calculated model likelihoods by using the small-sample corrected Akaike Information Criterion (AICc), following refs. 7, 43, 44, and 48. AICc optimizes model goodness-of-fit to the data, while simultaneously incorporating penalties for increased model complexity. From normal distribution theory, log-likelihood differences >2 can be interpreted as falling outside 2 standard deviation (SD) confidence limits (43, 49, 50), and we therefore adopt this point as the cutoff for significant differences in support between models. For each model structure we calculated the parameter values that maximized the likelihood (50, 51), starting with the most parsimonious single All-Carnivora model (incorporating all carnivoran taxa throughout the history of this clade). Multiple allometry models, and models combining slopes to form parallel regressions, then were tested iteratively across the range of observed slopes, and the slope value that minimized total residual variance (sum of residual variance for all of the combined-slope taxa) was proposed as the slope for the set of combined allometries. In total, we tested 137 distinct models of evolutionary change in encephalization allometries. Model descriptions, slope and intercept parameter values for each reconstructed allometry, AICc scores, and rescaled model log-likelihoods are reported in Table S2.
Comparison of Degree of Encephalization Across Allometries.
Comparing the degree of encephalization across allometries is straightforward when slopes are equal; one computes the difference in intercepts (e.g., 1), which can readily be transformed into a percentage change. Among the OM, there were 3 such shifts in intercepts between parallel encephalization allometries, all of them increases: Ursidae and Canidae relative to the basal Carnivora allometry, and modern Musteloidea relative to archaic musteloids. However, differencing of intercepts is not valid across allometries of different slopes; a decrease in slope for one regression relative to another lowers encephalization for large-bodied taxa and/or raises it for small-bodied taxa, depending on the position of the intersection of the allometries. The converse holds true for a slope increase. In 3 instances the OM reconstruct changes in slopes from the basal carnivoran allometry: Amphicyonidae, Felidae, and archaic Musteloidea.
We calculated expected brain volumes for taxa in each group that displayed a transformation relative to a baseline allometry for that clade. For the 3 shifts in intercepts with parallel slopes, we compared members of the derived group relative to the plesiomorphic allometry (e.g., modern musteloids relative to archaic musteloids). The 3 changes in slope each involved transformations from the basal Carnivora allometry, and that allometry was used as the baseline. We tallied larger-than- and smaller-than-expected observed brain volumes relative to the baseline allometry, coding larger-than expected as 1 and smaller-than-expected as 0, and we calculated the binomial proportion of larger brains. A proportion less than 0.5 indicates an excess of smaller than expected brain volumes or a general decrease in encephalization, and the opposite is true for proportions greater than 0.5. We calculated the log-likelihood difference between (i) the observed proportion and (ii) a proportion of 0.5 (48, 50), testing the hypothesis that the members of derived groups fall significantly above or below the baseline allometry, and therefore whether there was a significant increase or decrease in the degree of encephalization associated with the transformation in encephalization allometry.
Supplementary Material
Acknowledgments.
We thank W. Simpson, W. Stanley, J. Meng, C. Norris, J. Galkin, R. Purdy, W. Joyce, D. Brinkman, M. Benoit, P. Tassy, C. Sagne, A. Currant, X. Wang, S. McLeod, and G. Takeuchi for access to collections, and M. Spaulding, G. Bever, and D. Azzolini for assisting in data collection. P. Wagner, A. Wyss, M. Norell, and 2 anonymous reviewers improved the manuscript by providing helpful comments. This project was supported by National Science Foundation, American Museum of Natural History Collections Study Grants (DEB-0608208 to J.A.F. and DEB-0614098 to J.J.F.), the Brown Family Foundation Graduate Fellowship, and the University of Michigan Society of Fellows.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901780106/DCSupplemental.
References
- 1.Jerison H. Evolution of the Brain and Intelligence. New York: Academic; 1973. [Google Scholar]
- 2.Herculano-Houzel S. Encephalization, neuronal excess, and neuronal index in rodents. Anat Rec. 2007;290:1280–1287. doi: 10.1002/ar.20598. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre L, Reader SM, Sol D. Brains, innovations and evolution in birds and primates. Brain Behav Evol. 2004;63:233–246. doi: 10.1159/000076784. [DOI] [PubMed] [Google Scholar]
- 4.Sol D, Duncan RP, Blackburn TM, Casey P, Lefebvre L. Big brains, enhanced cognition, and response of birds to novel environments. Proc Natl Acad Sci USA. 2005;102:5460–5465. doi: 10.1073/pnas.0408145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratcliffe JM, Brock-Fenton M, Shettleworth SJ. Behavioral flexibility positively correlated with relative brain volume in predatory bats. Brain Behav Evol. 2006;67:165–176. doi: 10.1159/000090980. [DOI] [PubMed] [Google Scholar]
- 6.Finarelli JA, Flynn JJ. The evolution of encephalization in caniform carnivorans. Evolution. 2007;61:1758–1772. doi: 10.1111/j.1558-5646.2007.00131.x. [DOI] [PubMed] [Google Scholar]
- 7.Finarelli JA. Testing hypotheses of the evolution of brain-body size scaling in the Canidae (Carnivora, Mammalia) Paleobiology. 2008;34:35–45. [Google Scholar]
- 8.Sears KE, Finarelli JA, Flynn JJ, Wyss AR. Estimating body mass in New World “monkeys” (Platyrrhini, Primates) from craniodental measurements, with a consideration of the Miocene platyrrhine, Chilecebus carrascoensis. Am Mus Nov. 2008;3617:1–29. [Google Scholar]
- 9.Kay RF, Ross C, Williams BA. Anthropoid Origins. Science. 1997;275:797–804. doi: 10.1126/science.275.5301.797. [DOI] [PubMed] [Google Scholar]
- 10.Martin RD. Primate Origins and Evolution: A Phylogenetic Reconstruction. London: Chapman and Hall; 1990. [Google Scholar]
- 11.Marino L, McShea DW, Uhen MD. Origin and evolution of large brains in toothed whales. Anat Rec A. 2004;281:1247–1255. doi: 10.1002/ar.a.20128. [DOI] [PubMed] [Google Scholar]
- 12.Nealen PM, Ricklefs RE. Early diversification of the avian brain: Body relationship. J Zool. 2001;253:391–404. [Google Scholar]
- 13.Mink JW, Blumenschine RJ, Adams DB. Ratio of central nervous system to body metabolism in vertebrates—its constancy and functional basis. Am J Physiol. 1981;241:R203–R212. doi: 10.1152/ajpregu.1981.241.3.R203. [DOI] [PubMed] [Google Scholar]
- 14.Aiello LC, Wheeler P. The expensive-tissue hypothesis—the brain and the digestive-system in human and primate evolution. Curr Anthrop. 1995;36:199–221. [Google Scholar]
- 15.Palombo MR, Kohler M, Moyà-Solà S, Giovinazzo C. Brain versus body mass in endemic ruminant artiodactyls: A case studied of Myotragus balearicus and smallest Candiacervus species from Mediterranean Islands. Quat Int. 2007;182:160–183. [Google Scholar]
- 16.Kruska DCT. In: The Evolutionary Biology of Intelligence, Nato ASI series in Ecology G17. Jerison H, Jerison I, editors. Berlin: Springer; 1988. pp. 211–250. [Google Scholar]
- 17.Isler K, van Schaik CP. Costs of encephalization: The energy trade-off hypothesis tested on birds. J Hum Evol. 2006;51:28–243. doi: 10.1016/j.jhevol.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Isler K, van Schaik CP. Metabolic costs of brain size evolution. Biol Lett. 2006;2:557–560. doi: 10.1098/rsbl.2006.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerison H. Brain evolution: New light on old principles. Science. 1970;170:1224–1225. doi: 10.1126/science.170.3963.1224. [DOI] [PubMed] [Google Scholar]
- 20.Dunbar RIM. The social brain hypothesis. Evol Anthropol. 1998;6:178–190. [Google Scholar]
- 21.Finarelli JA. Estimation of endocranial volume through the use of external skull measures in the Carnivora (Mammalia) J Mammal. 2006;87:1027–1036. [Google Scholar]
- 22.Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA. Molecular phylogeny of the Carnivora (Mammalia): Assessing the impact of increased sampling on resolving enigmatic relationships. Syst Biol. 2005;54:317–337. doi: 10.1080/10635150590923326. [DOI] [PubMed] [Google Scholar]
- 23.Yu L, Zhang YP. Phylogeny of the caniform Carnivora: Evidence from multiple genes. Genetica. 2006;127:65–79. doi: 10.1007/s10709-005-2482-4. [DOI] [PubMed] [Google Scholar]
- 24.Wesley-Hunt GD, Flynn JJ. Phylogeny of the Carnivora: Basal relationships among the carnivoramorphans, and assessment of the position of “Miacoidea” relative to crown-clade Carnivora. J Syst Palaeo. 2005;3:1–28. [Google Scholar]
- 25.Wesley-Hunt GD. The morphological diversification of carnivores in North America. Paleobiology. 2005;31:35–55. [Google Scholar]
- 26.Finarelli JA. Hierarchy and the reconstruction of evolutionary trends: Evidence for constraints on the evolution of body size in terrestrial caniform carnivorans (Mammalia) Paleobiology. 2008;34:553–562. [Google Scholar]
- 27.Gittleman JL. Carnivore brain size, behavioral ecology, and phylogeny. J Mammal. 1986;67:23–36. [Google Scholar]
- 28.Gittleman JL. Carnivore life history patterns: Allometric, phylogenetic, and ecological associations. Am Nat. 1986;127:744–771. [Google Scholar]
- 29.Gittleman JL. Carnivore olfactory bulb size: Allometry, phylogeny and ecology. J Zool. 1991;225:253–272. [Google Scholar]
- 30.Legendre S, Roth C. Correlation of carnassial tooth size and body weight in recent carnivores (Mammalia) Hist Biol. 1988;1:85–98. [Google Scholar]
- 31.Perez-Barberia FJ, Shultz S, Dunbar RIM. Evidence for coevolution of sociality and relative brain size in three orders of mammals. Evolution. 2007;61:2811–2821. doi: 10.1111/j.1558-5646.2007.00229.x. [DOI] [PubMed] [Google Scholar]
- 32.Radinsky L. Outlines of canid and felid brain evolution. Ann NY Acad Sci. 1969;167:277–288. [Google Scholar]
- 33.Lyras GA, Van der Geer AAE. External brain anatomy in relation to the phylogeny of Caninae (Carnivora: Canidae) Zool J Linn Soc. 2003;138:505–522. [Google Scholar]
- 34.Radinsky L. Relative brain size: A new measure. Science. 1967;155:836–838. doi: 10.1126/science.155.3764.836. [DOI] [PubMed] [Google Scholar]
- 35.Radinsky L. Brains of early carnivores. Paleobiology. 1977;3:333–349. [Google Scholar]
- 36.Radinsky L. Evolution of brain size in carnivores and ungulates. Am Nat. 1978;112:815–831. [Google Scholar]
- 37.Radinsky L. Endocasts of amphicyonid carnivorans. Am Mus Nov. 1980;2694:1–11. [Google Scholar]
- 38.Garcia N, Santos E, Arsuaga JL, Carretero JM. Endocranial morphology of Ursus deningeri Von Reichenau, 1904, from the Sima de Los Huesos (Sierra de Atapuerca) Middle Pleistocene site. J Vert Paleo. 2007;27:1007–1017. [Google Scholar]
- 39.Smith FA, et al. Body mass of late Quaternary mammals. Ecology. 2003;84:3403. [Google Scholar]
- 40.Fortelius M. University of Helsinki; 2003. [Accessed July, 2003]. Neogene of the Old World Database of Fossil Mammals (NOW) Version 4.0. Available at www.helsinki.fi/science/now/ [Google Scholar]
- 41.Van Valkenburgh B. In: Body Size in Mammalian Paleobiology: Estimation and Biological Implications. Damuth J, MacFadden BJ, editors. New York: Cambridge Univ Press; 1990. pp. 181–206. [Google Scholar]
- 42.Anderson DR, Burnham KP, Thompson WL. Null hypothesis testing: Problems, prevalence, and an alternative. J Wildl Manage. 2000;64:912–923. [Google Scholar]
- 43.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. New York: Springer; 2002. [Google Scholar]
- 44.Burnham KP, Anderson DR. Multimodel inference: Understanding AIC and BIC in model selection. Soc Meth Res. 2004;33:261–304. [Google Scholar]
- 45.Sokal RR, Rohlf FJ. Biometry. New York: W. H. Freeman; 1995. [Google Scholar]
- 46.Martin RD. Relative brain size and basal metabolic-rate in terrestrial vertebrates. Nature. 1981;293:57–60. doi: 10.1038/293057a0. [DOI] [PubMed] [Google Scholar]
- 47.Pagel MD, Harvey PH. How mammals produce large-brained offspring. Evolution. 1988;42:948–957. doi: 10.1111/j.1558-5646.1988.tb02513.x. [DOI] [PubMed] [Google Scholar]
- 48.Finarelli JA. Mechanisms behind active trends in body size evolution in the Canidae (Carnivora: Mammalia) Am Nat. 2007;170:876–885. doi: 10.1086/522846. [DOI] [PubMed] [Google Scholar]
- 49.Royall RM. Statistical Evidence: A Likelihood Paradigm. New York: Chapman and Hall; 1997. [Google Scholar]
- 50.Edwards AWF. Likelihood: Expanded Edition. Baltimore: The Johns Hopkins Univ Press; 1992. [Google Scholar]
- 51.Wagner PJ. In: Deep time—Paleobiology's perspective. Erwin DH, Wing SL, editors. Lawrence, KS: Paleontological Society; 2000. pp. 341–371. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.