Abstract
Marek’s disease virus type 1 (MDV-1) shows a strict dependency on the direct cell-to-cell spread for its propagation in cell culture. As MDV-1 shows an impaired nuclear egress in cell culture, we wished to address the characterization of capsid/tegument genes which may intervene in the maturation of intranuclear capsids. Orthologs of UL17 are present in all herpesviruses and, in all reported case, were shown to be essential for viral growth, playing a role in capsid maturation and DNA packaging. As only HSV-1 and PrV UL17 proteins have been characterized so far, we wished to examine the role of MDV-1 pUL17 in virus replication. To analyze MDV-1 UL17 gene function, we created deletion mutants or point mutated the open reading frame (ORF) to interrupt its coding phase. We established that a functional ORF UL17 is indispensable for MDV-1 growth. We chose to characterize the virally encoded protein by tagging the 729 amino-acid long protein with a repeat of the HA peptide that was fused to its C-terminus. Protein pUL17 was identified in infected cell extracts as an 82 kDa protein which localized to the nucleus, colocalizing with VP5, the major capsid protein, and VP13/14, a major tegument protein. By using green fluorescent protein fusion and HA tagged proteins expressed under the cytomegalovirus IE gene enhancer/promoter (PCMV IE), we showed that MDV-1 pUL17 nuclear distribution in infected cells is not an intrinsic property. Although our results strongly suggest that another viral protein retains (or relocate) pUL17 to the nucleus, we report that none of the tegument protein tested so far were able to mediate pUL17 relocation to the nucleus.
Keywords: Marek’s disease virus, UL17, nuclear localization
1. Introduction
Marek’s disease (MD) is a T cell lymphoproliferative disease of chickens caused by an oncogenic herpesvirus, Marek’s disease virus (MDV) [4–6]. Marek’s disease virus belongs to the Alphaherpesvirinae subfamily and is the type species of the genus Mardivirus [2, 9, 21, 38]. The horizontal spread of the virus from infected to naïve birds relies on the production of so-called cell-free virus in the feather follicle epithelium [3], whereas the spread of MDV in cell culture is restricted to a direct transmission of the virus from an infected cell to its uninfected neighbours, referred to as direct cell-to-cell spread [10]. Tegument or glyco-proteins which are implicated in the cell-to-cell spread are essential for MDV [14, 28, 33] or varicella zoster virus (VZV) [36], and dispensable for most other herpesviruses whereas MDV [14] or VZV [7] do not need a functional VP16, which plays a pivotal role in Herpes Simplex virus 1 (HSV-1) egress [22, 42]. To improve our knowledge on the roles of MDV tegument proteins in virus replication, we chose to focus our studies on the proteins which contribute to the inner layer of the tegument and interact with the capsid. In this group of proteins, pUL17 protein orthologs are found in alpha-, beta- and gamma-herpesviruses [17, 26, 40]. In HSV-1 the UL17 gene encodes a 78 kDa protein which is essential for virus replication and required for cleavage and packaging of the viral DNA [15, 26]. HSV-1 pUL17 has been assigned a role in targeting the capsids to intranuclear replication compartments where the viral DNA is packaged and cleaved [30] and has been shown to recruit pUL25 [32]. In Pseudo-rabies Virus (PrV) the cleavage/packaging activity is conserved but, unlike that of HSV-1 [31], pUL17 was not detected as a tegument associated protein [18].
To characterize MDV pUL17 we constructed deletion and HA-epitope- or monomeric-Red-Fluorescent-Protein (mRFP)-tagged mutants by using Bacterial Artificial Chromosome (BAC) clones both of the highly pathogenic RB-1B MDV-1 strain [24] and of the cell-adapted Bac20 MDV-1 virus [27]. We could show that MDV-1 pUL17 is essential for viral replication, and that this 82 kDa protein localizes to the nucleus of infected cells, colocalizing partially with the major capsid protein (VP5) and a tegument protein, VP13/14. The localisation of pUL17 as fluorescent dots seen in virus infected cells is not an intrinsic property of the protein nor a result of its interaction with tegument proteins.
2. Materials and methods
2.1. Cells and viruses
Chicken embryo skin cells (CESC) culture and propagation of viruses on CESC have been described [13, 14]. DF-1 cells [16] were cultivated in the same conditions as CESC. COS-7 cells were maintained in William’s E medium (Invitrogen, Cergy Pontoise, France) with 5% FCS (Invitrogen).
2.2. Bacteria, plasmids and BAC
Top 10 Escherichia coli (Invitrogen) were used for plasmids pEGFP-C1 (Clonetech, St. Germain-en-Laye, France), pFuse (InvivoGen, Toulouse, France), pEPKan-S and pBAD-I-SceI [35], or their derivatives and the derivatives of pGEM-T plasmids (Promega, Charbonnières-les-Bains, France). The π dependent plasmid pSU311 and its derivative pSU311-2X-HA-KanR were maintained in E. coli strain MA6931 and MA 6717 pyr+, respectively [39]. BAC amplification and Red recombination were performed in E. coli EL250 [20] containing either pRB-1B (MDV-1 strain RB-1B BAC) [24] or Bac20 (MDV-1 strain 584Ap80c BAC) [27]. Antibiotics were used at the following concentrations: chloramphenicol, 34 μg/mL; kanamycine or ampicillin, 50 μg/mL and zeocin 25 μg/mL.
2.3. Primers
Primers are listed in Table I and those specific for MDV-1 genes were selected on the basis of MDV-1 Md5 sequence [38].
Table I.
Primers used in this study.
| Primer | Sequence 5′-3′ | |
|---|---|---|
| Bac17-F | GGATCCATGGAGGCGCATATAGAAAGCGAA | Underlined, BamHI site |
| Bac17-R | GCGGCCGCTCACATATACACCTCTGAAACGTA | Underlined, NotI site |
| pF-Forw | GATATCCGCCACCATGTTTGCAGTGAGCGC | Underlined, EcoRV site |
| pF-Rev | AGATCTTACACAGCTGTCTGAGACATCG | Underlined, BglII site |
| 2×-HA-Fw | GAATTCTATCCGTATGATGTTCCTGATTATGCTAGCCTCTATCCGTATGATGTTCCTGATTATGCTAGCCTCGTGTAGGCTGGAGCTGCTTC | Bold, EcoRI site; underlined, 2×HA tag sequence |
| 2×-HA-Rev | GAATTCCATATGAATATCCTCCTTAG | Bold, EcoRI |
| UL17-Fw | AAGATGCAACAGGTGCTACGACCTTACGTTTCAGAGGTGTATATGTATCCGTATGATGTTCCTGATTAT | Underlined, sequences annealing to template plasmid pSU311-2×HA |
| UL17-Rev | TCTTCAGTCTCTGCGTAGTCATAATTCAATACCTGTGTTGAGGATTGGCATATGAATATCCTCCTTAG | |
| UL49-Fw | CGTCGCCATCATTCATCCGCCCGCGGATATCGTAGCAGTGATAGCGAATATCCGTATGATGTTCCTGATTAT | |
| UL49-Rev | TAGATAAGATATATATGTACTAGTTTTTAATTCGGATGTCTATAAAAGCATATGAATATCCTCCTTAG | |
| UL48-Fw | AATTCAATTTCCCCTGGTGATCCTGTTGCCACTACCATCAGTACTTTATATCCGTATGATGTTCCTGATTAT | |
| UL48-Rev | CGAATACGACAAATTCGTTTATTAAACGTCACATTTACGTATAATATCCATATGAATATCCTCCTTAG | |
| PX1 | GCGGCAGCGTGAGGGGATCTTGAAG | Specific of KanR gene |
| PX2 | CCCCAGCTTCAAAAGCGCTCTGA | Specific of KanR gene |
| T497 | GCATCTAGCACTTGGAGA | Specific of UL49 gene |
| T500 | CCCTGCCCATGCCTCCCA | Specific of UL48 gene |
| DelTag17 | CAGGTGGCTGTGTACGTTC | Specific of UL17 gene |
| UL17delFw | GGTATCTTCAGTCTCTGCGTAGTCATAATTCAATACCTGTGTTGAGGACATATGAATATCCTCCTTAG | Deletion of UL17 |
| UL17delRev | GACCTTAGTCACAACATTGGTGCACGTAGGCAACACGACGGATAACAGCGGTGTAGGCTGGAGCTGCTTCG | |
| UL17delRev2 | GAAACTATATATTCATTATCTAACTTTGATCGGGGGGCGTAGCGACATCTGTTAACACATAGGATGACGACGATAAGTAGGG | Mutation of the 2d start codon of UL17. Bold, complementary sequences; grey box, mutation; underlined, sequences annealing to template plasmid pEPkan-S |
| UL17delFw2 | CATTCATCAGGTATTATTATATGTGTTAACAGATGTCGCTACGCCCCCCGATCAAAGTTACAACCAATTAACCAATTCTGATTAG | |
| UL17ResA | CTATGCACGGTGTCGTCGGT | |
| UL17ResB | TCCTGCATAAACGAATCCGG | |
| UL17seqA | CCCGTAGCATTCTGTCGTATTTGAACG | |
| UL17RFP1 | gtatcttcagtctctgcgtagtcataattcaatacctgtgttgaggattggTCACAAGGCGCCGGTGGAGTGG | Uppercase, sequences annealing to template plasmid pEPmRFPin |
| UL17RFP2 | accaacaagatgcaacaggtgctacgaccttacgtttcagaggtgtatatgATGGCCTCCTCCGAGGACGTCATC | |
| B17F | cccgGGATCCatggaggcgcatatagaaag | Underlined, BamHI site |
| Xh17r | agatCTCGAGctacacgaggctagcataatc | Underlined, XhoI site |
2.4. Deletion of UL17 and generation of rescued viruses
A first mutant (pRB-1BΔ17) was generated in pRB-1B, in which the entire UL17 gene was replaced by a KanR cassette obtained by PCR amplification with pKD4 [8] as a template plasmid and primers UL17delFw and UL17delRev (Tab. I). Selection for recombinant BAC and subsequent analyses were performed as described [29]. A second mutant, pRB-1B17stop, was constructed by mutating the second start codon from ATG to TAG. Mutant pRB-1B17stop was obtained by using the two-step red mutagenesis method [35] with primers UL17delFw2 and UL17delRev2. Rescuant viruses were obtained by co-transfection of mutagenized BAC with a complementing plasmid harbouring UL17 gene and 500 bp sequences immediately up and downstream of the gene. The region encompassing UL17 and additional sequences was PCR amplified from pRB-1B using primers UL17ResA and UL17ResB (Tab. I), and cloned in pGEM-T Easy, yielding pGEM-T17R. Rescued viruses were obtained as described [14], by co-transfection of either pRB-1BΔ17 or pRB-1B17stop DNA with pGEM-T17R, and named rRB-1BΔ17R or rRB-1B17stopR.
2.5. Epitope tagging of MDV genes
2.5.1. HA epitope tagging at the 3′ end of genes
Epitope tagging of targeted MDV genes was based on the method described by Uzzau et al. [39]. A template plasmid containing a repeat of a HA epitope and the kanamycin resistance gene (KanR) flanked by FRT sites and was constructed by cloning the PCR fragment obtained with pKD4 plasmid as a template [8] and primers 2×-HA-Fw and 2×-HA-Rev (Tab. I). The forward primer contained a 33 nt extension corresponding to the sequence of the HA tag [23]. The PCR fragment was cloned in pGEM-T Easy, sequenced, released by EcoRI digestion and ligated into EcoRI digested pSU311 plasmid to obtain template plasmid pSU311-2×-HAKanR. To insert the HA epitope at the end of a gene, a PCR amplification was performed with pSU311-2×-HAKanR as a template and primers carrying extensions homologous to the end of the gene (forward primer) and to the genome region immediately downstream of it (reverse primer). Primers UL17-Fw and -Rev were used to tag UL17, UL49-Fw and -Rev for UL49 and UL48-Fw and -Rev for UL48 (Tab. I). PCR fragments were inserted in pRB-1B as described [14, 20, 29]. Insertion of the tag cassette was checked by PCR, Southern blotting and sequencing. PCR and sequencing were performed by using primers PX1, PX2, T497, T500 and Deltag17 (Tab. I).
2.5.2. Insertion of the mRFP at the end of UL17
Fusion of the mRFP to the C-terminus of pUL17 was performed in pRB-1B and Bac20 using primers UL17RFP1 and UL17RFP2 (Tab. I), as described [35].
2.6. DNA analyses
BAC modifications were characterized by RFLP and Southern blot analysis, by using a KanR specific probe and a chemoluminescence detection kit (ECLTM, Amersham-Biosciences, Little Chalfont, UK) [14]. Primers PX1 and PX2 (Tab. I) were used to sequence verify the correct insertion of the cassette and in pRB-1B17stop the mutation was confirmed by sequencing with primer UL17seqA (Tab. I).
2.7. MDV-1 reconstitution and propagation
Transfection of CESC by BAC DNA, subsequent viral passages and growth kinetic studies of pRB-1B and pRB-1Btag17 derived viruses were performed as described [14, 27].
2.8. Indirect immunofluorescence
CESC were seeded on round coverslips (14 mm in diameter), in 24-well plates, at 1 × 105 cells per well. One-day old CESC were either transfected with BAC DNA or infected with viruses. Cells were then incubated at 41 °C for 2 to 4 days, fixed, permeabilised and probed with antibodies. Anti-HA antibodies used were either a mouse Mab HA-7 (Sigma-Aldrich, St. Quentin-Fallavier, France) at 1:10 000 or a polyclonal rabbit anti-HA (Sigma H6908) at 1:4 000. Monoclonal antibodies to tegument or capsid proteins have been described [13, 14, 34]. Anti-mouse IgG Alexa-Fluor conjugates (Invitrogen) were used to label bound antibodies. Double immunostaining was carried out by using anti-VP22 or anti-VP5 Mabs conjugated to Oregon Green (FluoReporter labelling kit, Invitrogen). Cell nuclei were stained with Hoechst 33342 dye and coverslips were mounted on a drop of Vectashield medium (Vector lab. ABCYS, Paris, France). A minimum of 50 infected cells were examined for the nuclear or cytoplasmic localization of viral proteins with a Zeiss Axiovert 200 microscope equipped with the Apotome imaging system. Images were generated and analyzed by using the Axiovision Software (Carl Zeiss SA, Le Pecq, France).
2.9. Immunoblot analyses
Cell extracts were prepared from infected or mock-infected CESC at 4 days post infection. Cells were trypsinized, washed in PBS and centrifuged at 200 × g for 10 min at 4 °C. Cell pellets were then incubated in RIPA buffer (50 mM Tris-HCL pH 8, 150 mM NaCl, 1 mM MgCl2, 1% NP40), containing protease inhibitors (Complete Mini EDTA free, Roche, Meylan, France), 0.5 mM PMSF (Sigma-Aldrich), 1 mM DTT and benzonase (25 U/μL) for 90 min at 4 °C. After centrifugation at 7 800 × g for 30 min at 4 °C, cell pellets were resuspended in sample buffer [19] and heated in boiling water for 2 min. Proteins were separated on SDS-polyacrylamide gels, electroblotted to nitrocellulose membranes (Qbiogene, Strasbourg, France) [13]. Protein extracts corresponding to 2 × 106 cells were loaded in each lane. Immunodetection was done as described [13], and tagged proteins were detected by using the anti-HA7 Mab diluted 1:5 000 and an anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich). Bound antibodies were detected by using the NBT-BCIP substrate (Zymed, Invitrogen).
2.10. Cloning of MDV-1 genes in expression vectors
MDV-1 UL17 gene was PCR amplified from pRB-1B with primers Bac17F and Bac17R. The fragment was cloned in pGEM-T Easy, sequenced, released by BamHI – NotI digestion and cloned in the same sites of pFastBac1 vector (Invitrogen). The UL17 gene was then released from pFastBac1-UL17 by BamHI – HindIII and cloned into the BglII HindIII sites of pEGFP-C1, yielding pEGFP-UL17. The MDV-UL17 tagged gene was PCR amplified from pRB-1Btag17 by using primers B17F and Xh17R (Tab. I) and cloned in BamHI-XhoI sites of pCDNA3 vector yielding pCDNA3-UL17HA. The MDV-1 UL14 gene was PCR-amplified with primers pF-Forw and pF-Rev and the 700 bp fragment was cloned in pGEM-T, sequenced and transferred to pFuseFc plasmid (InvivoGen – CAYLA InvivoGen) by using EcoRV and BglII sites. The recombinant pFuse-UL14 plasmid encoded the UL14 gene fused to the Fc portion of a mouse IgG2a.
3. Results
3.1. pUL17 is essential for replication of MDV in cultured cells
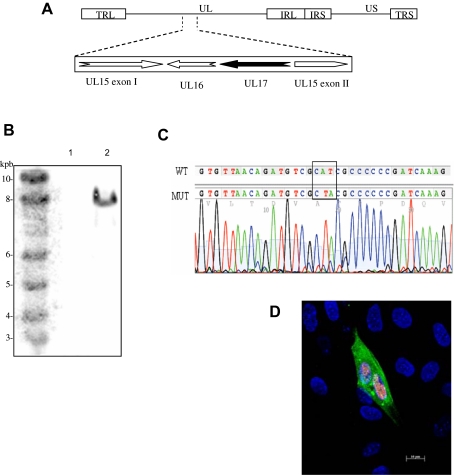
Insertion of the KanR cassette at the UL17 locus (Fig. 1A) in pRB-1BΔ17 was confirmed by RFLP and Southern blot analyses (Fig. 1B). Sequencing confirmed the correct insertion of the cassette (data not shown). pRB-1BΔ17 DNA was transfected into CESC to analyze the effect of UL17 open reading frame (ORF) deletion on viral growth. At 3 days post-transfection the presence of isolated cells expressing the major tegument and capsid protein VP22 and VP5 could be detected; but no cell to cell transmission of the virus was observed. After four serial blind passages on CESC no viral replication could be detected in three independent transfections. Parallel transfection of the parental pRB-1B DNA consistently yielded viable virus. As co-transfection of pRB-1BΔ17 with rescue plasmid pGEM-T17R yielded a viable rescued virus, rRB-1BΔ17R, we concluded that the UL17 knock-outed virus was not able to undergo a complete replication cycle that would result in the production of a progeny. However the localization of UL17 in the intron of another putatively essential gene, UL15, prompted us to verify that the phenotype observed was directly related to the absence of UL17 and not to a negative effect of the deletion on the expression of neighbouring genes. Site specific mutagenesis of the second start codon to a stop codon (ATG > TAG) was performed. After verification of the sequence (Fig. 1C), the resulting BAC DNA (pRB-1B17Stop) was transfected in CESC. As described for the UL17 deletion mutant, no viral progeny could be obtained from 3 independent transfection. Major capsid and tegument proteins could be detected in single cells in the same conditions as above (Fig. 1D). A rescued virus (rRB-1B17Stop) was obtained by co-transfection of pRB-1B17stop DNA and pGEM-T17R. As both rRB-1B17stop and rRB-1BΔ17 showed no obvious differences in replication when compared with vRB-1B virus originating from the parental BAC pRB-1B (data not shown), we concluded that MDV-1 UL17 gene was essential for viral growth in vitro.
Figure 1.
Inactivation of UL17 gene is lethal for MDV replication. (A) Schematic representation of the genomic localization of the MDV UL17 gene. The unique long (UL), unique short (US), and repeated (IR, TR) sequences are indicated. (B) Southern blot analysis of HindIII digests of pRB-1B (lane 1) and pRB-1B∆17 (lane 2). Hybridization with a KanR specific-probe reveals the insertion of the cassette in the UL17 locus. (C) Sequence analysis of UL17 gene confirming the alteration of codon 21 from ATG (CAT in the complementary strand) in pRB-1B (WT) to TAG in the pRB-1B17stop mutant (MUT). (D) vRB-1B17Stop mutant viruses does not spread to neighbouring cells: CESC transfected with pRB-1B17stop DNA were fixed at 120 h p.i. Co-staining of VP22 (green) and VP5 (red) shows replication of mutant virus limited to the transfected cell.
3.2. Epitope tagging of MDV-1 UL17 gene
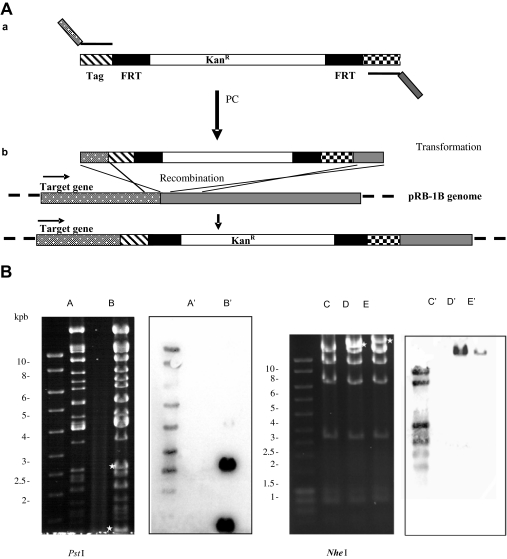
As no antibody against pUL17 was available, we constructed a recombinant MDV-1 expressing a pUL17 with a repetition of the HA epitope, fused to its C terminal end. The insertion of a tag sequence at the 3′ end of a gene was performed using Red recombination in EL250 [20] containing pRB-1B, with a PCR fragment obtained by using the primers described and pSU311-2×-HA-KanR as a template (Fig. 2A). We obtained pRB-1Btag17, a mutated BAC carrying a tagged UL17 gene. Two others tagged mutants were constructed as controls, pRB-1Btag49 and pRB-1Btag48 in which genes UL49 and 48 were similarly modified to encode tagged VP22 and VP16 respectively. After an initial screening based on PCR, the genotypes of the resulting BAC were confirmed by RFLP and Southern blotting (Fig. 2B). Digestion with PstI generated 2 additional fragments (2.7 and 1.3 kB) in pRB-1B17tag DNA (Fig. 2B). Similarly, two new fragments of approximately 13 and 12 kbp were generated by NheI digestion of pRB-1Btag49 or -48 (Fig. 2B). Southern blotting analysis confirmed the presence of the KanR gene in the tagged mutants (Fig. 2B). Sequencing of the recombination site with primers PX1 and PX2 confirmed the insertion of the fragment in the desired genome loci (data not shown).
Figure 2.
Tagging of viral genes. (A) Schematic representation of the tagging strategy: (a) A DNA fragment beginning with the HA-encoding sequence (hatched box) and including a kanamycin-resistance cassette (KanR) flanked by the FRT sites (filled box) was amplified with primers carrying an extension of 45-48 nt homologous to the region immediately upstream of the translation stop signal of the target gene (dotted box) and to a region downstream from it (shaded box). (b) The modified sequence was incorporated by homologous recombination in pRB-1B. (B) Restriction enzyme digestion and Southern blot analysis of tagged pRB-1B clones. pRB-1B (A) and pRB-1Btag17 (B) DNA were digested with PstI, while pRB-1Btag49 (D) and pRB-1Btag48 (E) and, as a control, pRB-1B (C) DNA were digested with NheI. Asteriks indicate new fragments in tagged BAC DNA. Southern blot analysis revealed the presence of the cassette in the newly generated fragments (lane B′, pRB-1Btag17; lanes D′ & E′, pRB-1B tag49 and 48 respectively).
3.3. Reconstitution of MDV-1 expressing pUL17 tagged protein
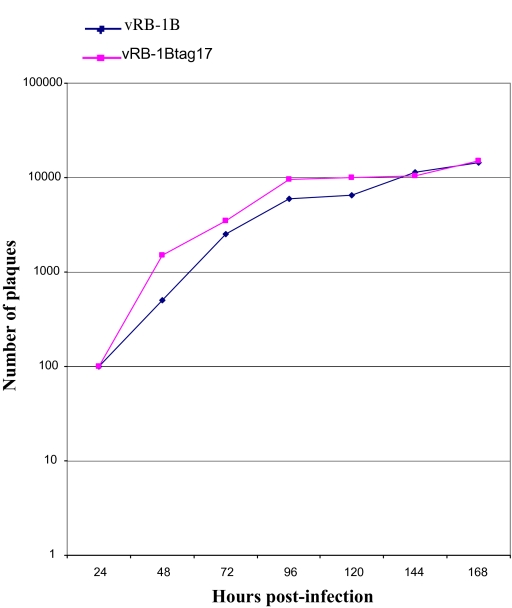
The DNA from pRB-1Btag17 or pRB-1B were transfected into CESC to obtain tagged (vRB-1Btag17) or parental (vRB-1B) viruses. Typical MDV-1 plaques were observed after transfection of both parental and tagged mutant. The growth kinetics of the mutant and parental virus were similar as no significant differences could be observed (Simstat statistical analyses on two independent experiments) (Fig. 3).
Figure 3.
Growth curves of vRB-1B and vRB-1Btag17 viruses. CESC were infected with 100 PFU of virus, and titres were determined at the indicated time p.i.
3.4. Characterization of pUL17 in MDV-infected cells
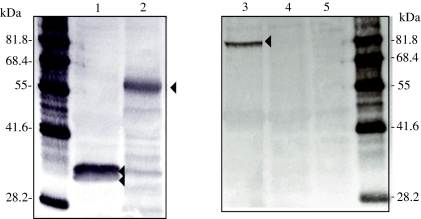
The expression of pUL17 was monitored by western blotting analyses in infected CESC lysates. In NP40 insoluble extracts of CESC infected with vRB-1Btag17, Mab HA-7 reacted with a protein of approximately 82 kDa (Fig. 4, lane 3), which is in good agreement with the predicted molecular mass of pUL17 (81 kDa). Control experiments showed no back ground staining in lysates from CESC or vRB-1B infected CESC (Fig. 4, lanes 4 & 5), whereas MabHA-7 clearly delineated a doublet of bands (33 & 30 kDa) corresponding to VP22 in vRB-1Btag49 lysates and a unique protein migrating at 55 kDa (VP16) in vRB-1Btag48 infected CESC (Fig. 4, lanes 1 & 2). The tagged VP22 was also detected by a monoclonal antibody L13a (data not shown). Our data show that HA tagging of pUL17 allowed its specific identification in CESC infected with vRB-1BTag17 and allowed further analyses of its expression.
Figure 4.
pUL17 expression in CESC. Proteins in NP40 insoluble fraction of CESC infected with vRB-1Btag49 (1) vRB-1BTag48 (2) vRB-1Btag17 (3) vRB-1B (4) or mock-infected (5) were separated by PAGE, blotted and probed with MAb HA-7. Arrowheads point to the respective positions of tagged proteins. Molecular mass markers are on the left and right (Chemicon).
3.5. MDV-1 pUL17 is a nuclear protein
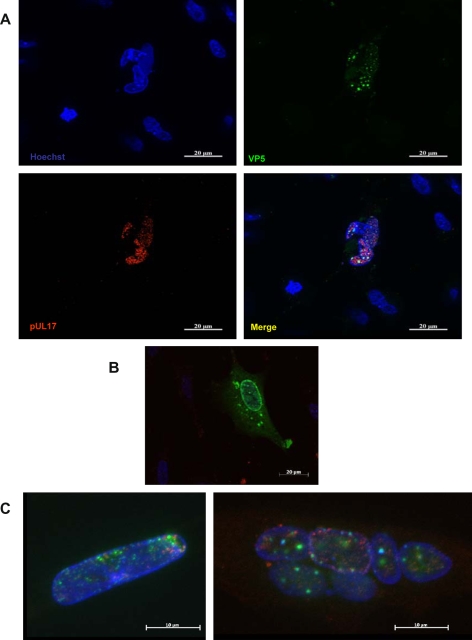
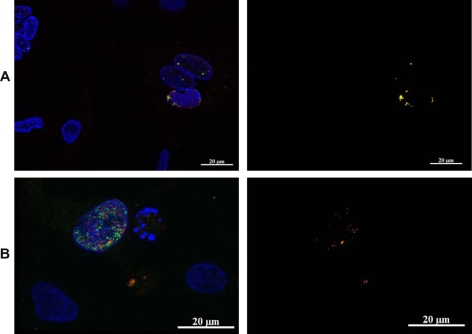
Intracellular localization of pUL17 was analyzed in vRB-1Btag17 infected cells, and its colocalization with other viral proteins was established by double immunostaining. When stained with anti-HA antibodies, the tagged pUL17 associated signal appeared as a punctuate intranuclear signal, partly associated with the nuclear membrane (Fig. 5, panel A). The pUL17 staining was observed only in vRB-1Btag17 infected CESC, in association with two other intranuclear viral proteins, the MCP VP5 (Fig. 5, panel A) and the major tegument protein VP13/14 (see below). The intranuclear localisation of pUL17 associated fluorescence was not an artefact related to its tagging with HA peptide as a similar location was observed when pUL17 was fused in frame with the mRFP, in vRB-1BmRFPUL17 and vBAC20mRFPUL17 (Fig. 5, panel C). A very limited colocalization of pUL17 with MCP VP5 or VP13/14 could also be observed in the nuclei, close to or in association with the nuclear membrane (Fig. 6, panel A & B). By using a tagging strategy, we show that pUL17 localizes in the nuclei of infected CESC, in association with the major capsid protein VP5 and with the tegument protein VP13/14.
Figure 5.
Expression and subcellular location of MDV-1 pUL17. CESC were infected with vRB-1Btag17 (A) or vRB-1B (B) and stained for pUL17 (anti-HA tag-Red) and either VP5 (A) or VP22 (B) (green). The localisation of pUL17 is unaffected by the size of the tag sequence (C): CESC were infected with vRB-1B-UL17mRFP (left) or vBac20UL17mRFP (right). The mRFP tagged protein displays the red signal in nuclei where the presence of VP5 is also detected (green).
Figure 6.
pUL17 colocalizes partially with the major capsid protein VP5 (A) and the tegument protein VP13/14 (B) in the nucleus of vRB-1B-tag17 infected CESC. Anti-HA rabbit antibody (red) reveals the pUL17 tagged protein and VP5 or VP13/14 (green) were identified using specific monoclonal antibodies H18 and L13b. The colocalized signals were extracted from a single slice and presented in the left panel of A and B.
3.6. The discrete nuclear localization of pUL17 is not an intrinsic property
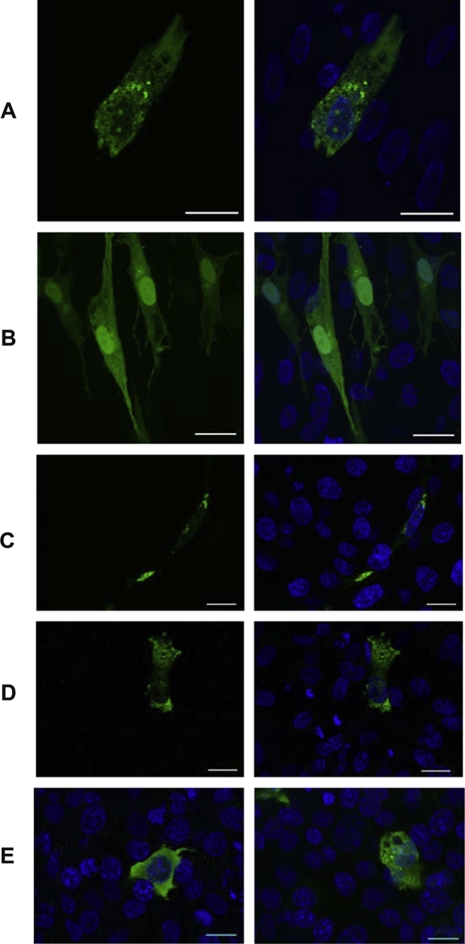
As no canonical NLS could be predicted from the amino-acid sequence of pUL17, we checked whether the protein would display a similar location when expressed under the PCMV IE promoter. A first green fluorescent protein (GFP) fusion was constructed in pEGFPC1 in which the GFP was fused to the N term of pUL17. In CESC the fusion protein localized predominantly in the cytoplasm, forming brightly fluorescent aggregates although a lower amount of the protein could be detected in the nucleus (Fig. 7A). In the same cells eGFP was evenly distributed in the nucleus and cytoplasm of the cells (Fig. 7B). These aggregates appeared early after transfection, in primary CESC as well as in DF1 cells (Fig. 7C). To ascertain that the tagging strategy or the expression under a strong promoter were not inducing an artifactual expression of pUL17, we cloned the tagged UL17-HA from pRB-1Btag17 in pCDNA3 and compared the subcellular localizations of the HA tagged protein with the GFP-UL17, in chicken DF1 and in mammalian COS-7 cells. The localization of UL17-HA expressed from pCDNA3-UL17-HA, was predominantly cytoplasmic, although again a faint intranuclear signal could be observed (Fig. 7D). The C-terminal HA tagged protein was considerably less prompt to aggregation than the GFP fusion protein, in the chicken cells (Fig. 7C & D). In mammalian cells (COS-7) the UL17-HA protein showed a localization similar to what was already observed in DF1 (Fig. 7E – left panel), but GFP-UL17 did not form large cytoplasmic aggregates and showed a pattern of localization much similar to the one of UL17-HA (Fig. 7E – right panel). From these experiment we concluded that the localization of pUL17 in transfected cells was predominantly cytoplasmic and that this distribution was representative of the expression of pUL17 as a transgene in avian and mammalian cells. As this subcellular localization differed considerably from what was reported in infected cells, we wondered whether any tegument protein could retain or relocate pUL17 to the nucleus. However none of the tegument protein tested (pUL11/12; pUL13/14; VP22, and pUL14) were able to retain or relocate pUL17 in the nucleus (Data not shown).
Figure 7.
The discrete pUL17 nuclear localisation is not an intrinsic property. When expressed in fusion with eGFP, pUL17 localizes mainly in the cytoplasm in transfected CESC (A) whereas eGFP alone is randomly distributed in the cell (B). A similar localization is observed for the GFP-UL17 in DF1 cells (C). The localization of UL17-HA, expressed from pCDNA3-UL17-HA, was also predominantly cytoplasmic however the recombinant protein did not form large aggregates in the cytoplasm (D). When expressed in COS-7 cells, the HA tagged (E left panel) or the GFP fusion protein (E right panel) showed similar fluorescence patterns. The HA tag was revealed by staining with anti-HA7 monoclonal antibody and an anti-mouse antibody conjugated to Alexa 488. The nuclei were counterstained with Hoechst 33342 dye.
4. Discussion
We aimed at characterizing pUL17 of MDV-1 by generating deletion and tagged mutants in pRB-1B. By deleting (pRB-1BΔ17) or mutating (pRB-1B17stop) the UL17 gene we showed that this ORF was essential for MDV-1 growth in cell culture, and that this growth defect could be cured by restoring the UL17 locus as previously shown for UL17 gene orthologs in HSV-1 and PrV [18, 26]. To further characterize the protein coded by UL17 in MDV-1, we developed a tagging strategy enabling the fusion of the HA epitope [23], at the C-terminus of the protein. We designed the method to (i) generate recombinant viruses in a one-step Red recombination procedure; (ii) avoid the isolation of false-positive double resistant colonies by using a π dependent plasmid [39]. We also show that we could generate tagged mutants without introducing deletions in downstream intergenic sequences. We constructed a recombinant pRB-1B molecule encoding a C-terminally tagged UL17 protein and used it to characterize the expression of pUL17 in MDV-1. In pRB-1Btag17, as well as in other tagged recombinants, we found that the insertion had no obvious impact on virus viability, protein expression level and location1. We demonstrate that the use of commercially available anti-HA antibodies enabled us to detect the tagged pUL17, as well as another weakly expressed MDV-1 tegument protein, VP16, in infected cell-lysates. This improved detection of viral proteins is in good agreement with the recent report by Adler et al. [1] who described a higher sensitivity of anti-HA antibody compared to a monospecific antiserum for the detection of HCMV pUL131A. MDV-1 pUL17, a 82 kDa protein, was associated with the NP40 (and Triton X100) insoluble fraction and this precluded any attempt to probe it for interaction with other proteins by co-immunoprecipitation. The cellular location of pUL17 in rRB-1Btag17 infected cells was nuclear, as reported for other alphaherpesviruses. Protein pUL17 colocalized partially with VP5, probably delineating the replication compartment(s) [15, 18]. A discrete and very limited co-localization was also observed with VP13/14, an observation that had not been reported earlier. Whether this co-localization corresponds to the delineation of the same replication compartment remains to be established, but VP13/14 was also reported to localize to replication compartments in HSV-1 infected cells [11, 12]. In PrV it was hypothesized that the nuclear localization of pUL17 was mediated by a NLS [18], but no such signal could be predicted from the amino-acid sequence of MDV-1 pUL17. The absence of a strong NLS was confirmed as pUL17 was observed mainly in the cytoplasm of the cells transfected with plasmids expressing either HA tagged or GFP fusion protein. The presence of minute amounts of nuclear pUL17 was recorded in almost all instances, although no amino acid sequence could be predicted as mediating this relocation. Whether the viral pUL17 is retained to the nucleus or relocated through its interaction with one or several proteins remains to be established, but our results strongly suggest that, for MDV-1, this (these) protein(s) are either of viral origin or induced by viral infection. Further identification of MDV pUL17 ligands was rendered difficult due to the insolubility of the protein, limited virus titres and high cell-association of MDV-1. No information on pUL17 ligands could be obtained from literature [41]. In HSV-2, pUL14 was reported to influence the intracellular location of a number of proteins including VP26, pUL33 and pUL17 [43]. However none of the tegument proteins tested so far, including MDV-1 pUL14, were able to retain or relocate pUL17 to the nucleus. The interaction of pUL17 with capsid proteins has not been extensively studied but recent publications on the capsid structure showed that the pUL17-pUL25 complex is located at the penton base [37]. If pUL17 is relocated to the nucleus, it could be through a direct or indirect interaction with the triplex protein VP19c as it is known to relocate both VP5 and VP23 to the nucleus [25], and has recently shown to be in a good conformation to interact with the pUL17-pUL25 complex [37].
Acknowledgments
The Ph.D. thesis of N. Chbab was supported by a fellowship from the French ministry of Research and by a grant from the Animal Health Department of INRA. The laboratory of J.F. Vautherot is a member of IFR 136. Nikolaus Osterrieder was supported by National Research Initiative grants 2003-02234 and 2005-01806, awarded through the Cooperative State, Research and Extension Service of the USDA. The pRB-1B BAC was a kind gift of Dr V. Nair (IAH – Compton, United Kingdom).
Footnotes
Chbab N., Réplication et morphogénèse du virus MDV-1 : caractérisation d’une protéine de tégument produit du gène UL17 essentiel à la réplication virale, Thèse d’Université, Université François Rabelais, Tours, France, 2006.
References
- 1.Adler B., Scrivano L., Ruzcics Z., Rupp B., Sinzger C., Koszinowski U.. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 2006;87:2451–2460. doi: 10.1099/vir.0.81921-0. [DOI] [PubMed] [Google Scholar]
- 2.Buckmaster A.E., Scott S.D., Sanderson M.J., Boursnell M.E., Ross N.L., Binns M.M.. Gene sequence and mapping data from Marek’s disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J. Gen. Virol. 1988;69:2033–2042. doi: 10.1099/0022-1317-69-8-2033. [DOI] [PubMed] [Google Scholar]
- 3.Calnek B.W., Adldinger H.K., Kahn D.E.. Feather follicle epithelium: a source of enveloped and infectious cell-free herpesvirus from Marek’s disease. Avian Dis. 1970;14:219–233. [PubMed] [Google Scholar]
- 4.Churchill A.E., Biggs P.M.. Agent of Marek’s disease in tissue culture. Nature. 1967;215:528–530. doi: 10.1038/215528a0. [DOI] [PubMed] [Google Scholar]
- 5.Churchill A.E.. Herpes-type virus isolated in cell culture from tumors of chickens with Marek’s disease, I. Studies in cell culture. J. Natl. Cancer Inst. 1968;41:939–950. [PubMed] [Google Scholar]
- 6.Churchill A.E., Biggs P.M.. Herpes-type virus isolated in cell culture from tumors of chickens with Marek’s disease. II. Studies in vivo. J. Natl. Cancer Inst. 1968;41:951–956. [PubMed] [Google Scholar]
- 7.Cohen J.I., Seidel K.. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 1994;68:7850–7858. doi: 10.1128/jvi.68.12.7850-7858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison A.J.. Evolution of the herpesviruses. Vet. Microbiol. 2002;86:69–88. doi: 10.1016/s0378-1135(01)00492-8. [DOI] [PubMed] [Google Scholar]
- 10.Denesvre C., Blondeau C., Lemesle M., Le Vern Y., Vautherot D., Roingeard P., Vautherot J.F.. Morphogenesis of a highly replicative EGFPVP22 recombinant Marek’s disease virus in cell culture. J. Virol. 2007;81:12348–12359. doi: 10.1128/JVI.01177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly M., Elliott G.. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 2001;75:2575–2583. doi: 10.1128/JVI.75.6.2575-2583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnelly M., Elliott G.. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 2001;75:2566–2574. doi: 10.1128/JVI.75.6.2566-2574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorange F., El Mehdaoui S., Pichon C., Coursaget P., Vautherot J.F.. Marek’s disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 2000;81:2219–2230. doi: 10.1099/0022-1317-81-9-2219. [DOI] [PubMed] [Google Scholar]
- 14.Dorange F., Tischer B.K., Vautherot J.F., Osterrieder N.. Characterization of Marek’s disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 2002;76:1959–1970. doi: 10.1128/JVI.76.4.1959-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goshima F., Watanabe D., Takakuwa H., Wada K., Daikoku T., Yamada M., Nishiyama Y.. Herpes simplex virus UL17 protein is associated with B capsids and colocalizes with ICP35 and VP5 in infected cells. Arch. Virol. 2000;145:417–426. doi: 10.1007/s007050050033. [DOI] [PubMed] [Google Scholar]
- 16.Himly M., Foster D.N., Bottoli I., Iacovoni J.S., Vogt P.K.. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 17.Johannsen E., Luftig M., Chase M.R., Weicksel S., Cahir-McFarland E., Illanes D.. et al. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp B.G., Granzow H., Karger A., Mettenleiter T.C.. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J. Virol. 2005;79:13442–13453. doi: 10.1128/JVI.79.21.13442-13453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U.K.. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L.. et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 21.Lee L.F., Wu P., Sui D., Ren D., Kamil J., Kung H.J., Witter R.L.. The complete unique long sequence and the overall genomic organization of the GA strain of Marek’s disease virus. Proc. Natl. Acad. Sci. USA. 2000;97:6091–6096. doi: 10.1073/pnas.97.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossman K.L., Sherburne R., Lavery C., Duncan J., Smiley J.R.. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 2000;74:6287–6299. doi: 10.1128/jvi.74.14.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niman H.L., Houghten R.A., Walker L.E., Reisfeld R.A., Wilson I.A., Hogle J.M., Lerner R.A.. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petherbridge L., Brown A.C., Baigent S.J., Howes K., Sacco M.A., Osterrieder N., Nair V.K.. Oncogenicity of virulent Marek’s disease virus cloned as bacterial artificial chromosomes. J. Virol. 2004;78:13376–13380. doi: 10.1128/JVI.78.23.13376-13380.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rixon F.J., Addison C., McGregor A., Macnab S.J., Nicholson P., Preston V.G., Tatman J.D.. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 26.Salmon B., Cunningham C., Davison A.J., Harris W.J., Baines J.D.. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher D., Tischer B.K., Fuchs W., Osterrieder N.. Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 2000;74:11088–11098. doi: 10.1128/jvi.74.23.11088-11098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumacher D., Tischer B.K., Reddy S.M., Osterrieder N.. Glycoproteins E and I of Marek’s disease virus serotype 1 are essential for virus growth in cultured cells. J. Virol. 2001;75:11307–11318. doi: 10.1128/JVI.75.23.11307-11318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher D., Tischer B.K., Trapp S., Osterrieder N.. The protein encoded by the US3 orthologue of Marek’s disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown. J. Virol. 2005;79:3987–3997. doi: 10.1128/JVI.79.7.3987-3997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taus N.S., Salmon B., Baines J.D.. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology. 1998;252:115–125. doi: 10.1006/viro.1998.9439. [DOI] [PubMed] [Google Scholar]
- 31.Thurlow J.K., Rixon F.J., Murphy M., Targett-Adams P., Hughes M., Preston V.G.. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 2005;79:150–158. doi: 10.1128/JVI.79.1.150-158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurlow J.K., Murphy M., Stow N.D., Preston V.G.. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 2006;80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tischer B.K., Schumacher D., Messerle M., Wagner M., Osterrieder N.. The products of the UL10 (gM) and the UL49.5 genes of Marek’s disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 2002;83:997–1003. doi: 10.1099/0022-1317-83-5-997. [DOI] [PubMed] [Google Scholar]
- 34.Tischer B.K., Schumacher D., Chabanne-Vautherot D., Zelnik V., Vautherot J.F., Osterrieder N.. High-level expression of Marek’s disease virus glycoprotein C is detrimental to virus growth in vitro. J. Virol. 2005;79:5889–5899. doi: 10.1128/JVI.79.10.5889-5899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tischer B.K., von Einem J., Kaufer B., Osterrieder N.. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 36.Tischer B.K., Kaufer B.B., Sommer M., Wussow F., Arvin A.M., Osterrieder N.. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J. Virol. 2007;81:13200–13208. doi: 10.1128/JVI.01148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trus B.L., Newcomb W.W., Cheng N., Cardone G., Marekov L., Homa F.L.. et al. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol. Cell. 2007;26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tulman E.R., Afonso C.L., Lu Z., Zsak L., Rock D.L., Kutish G.F.. The genome of a very virulent Marek’s disease virus. J. Virol. 2000;74:7980–7988. doi: 10.1128/jvi.74.17.7980-7988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L.. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varnum S.M., Streblow D.N., Monroe M.E., Smith P., Auberry K.J., Pasa-Tolic L.. et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vittone V., Diefenbach E., Triffett D., Douglas M.W., Cunningham A.L., Diefenbach R.J.. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 2005;79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinheimer S.P., Boyd B.A., Durham S.K., Resnick J.L., O’Boyle D.R. 2nd. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamauchi Y., Wada K., Goshima F., Takakuwa H., Daikoku T., Yamada M., Nishiyama Y.. The UL14 protein of herpes simplex virus type 2 translocates the minor capsid protein VP26 and the DNA cleavage and packaging UL33 protein into the nucleus of coexpressing cells. J. Gen. Virol. 2001;82:321–330. doi: 10.1099/0022-1317-82-2-321. [DOI] [PubMed] [Google Scholar]