Summary
The advent of genome-wide association studies has allowed considerable progress in the identification and robust replication of common gene variants that confer susceptibility to common diseases and other phenotypes of interest. These genetic effect sizes are almost invariably moderate-to-small in magnitude and single studies, even if large, are underpowered to detect them with confidence. Meta-analysis of many genome-wide association studies improves the power to detect more associations, and to investigate the consistency or heterogeneity of these associations across diverse datasets and study populations. In this review, we discuss the key methodological issues in the set-up, information gathering and processing, and analysis of meta-analyses of genome-wide association datasets. We illustrate, as an example, the application of meta-analysis methods in the elucidation of common genetic variants associated with type 2 diabetes. Finally, we discuss the prospects and caveats for future application of meta-analysis methods in the genome-wide setting.
Keywords: meta-analysis, genome-wide association studies, replication, heterogeneity, methods
Impetus for meta-analysis of genome-wide association (GWA) studies
Until recently, the field of complex disease genetics had been plagued by irreproducibility of published results [1,2]. In retrospect, studies with small sample sizes given what are now known to be small effects [3], limited coverage of the genetic variability [4] and liberal use of statistical significance thresholds for claiming discovery were probably responsible for this poor replication record. The last few years have witnessed changes leading to the feasibility of genome-wide association (GWA) scans and better study designs. Larger samples have become available and researchers have recognised the value of collaborating to combine resources [5]. Advances in genotyping technologies have enabled high-throughput pipelines and accurate, reproducible genotyping. Finally, efforts like the International HapMap project [4], have improved our understanding of human sequence variation both by providing a more complete catalogue of common SNPs and by helping to characterise linkage disequilibrium (LD) patterns across the genome. Several large-scale GWA scans for common complex diseases have been carried out in the last couple of years, taking advantage of these advances [6].
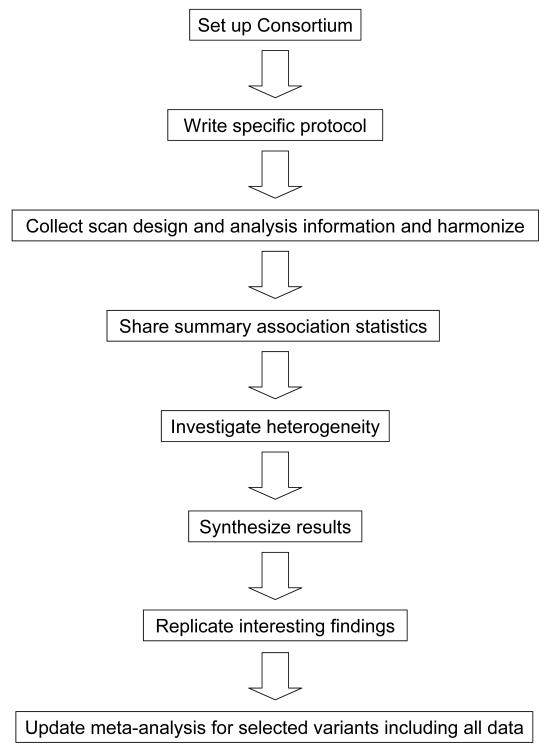
Optimal power is very important in finding new disease genes. Increased power may be achieved by combining datasets. Meta-analysis is a set of methods that allows the quantitative combination of data from multiple studies. These methods also allow the quantitative evaluation of the consistency or inconsistency/heterogeneity of the results across multiple datasets. Meta-analysis methods have been applied for several decades in a large variety of scientific fields and there are already several textbooks and handbooks thereof [7,8], some of which also cover genetic epidemiology [8]. However, the combination of large-scale data from GWA studies offers a new challenge for quantitative synthesis. In this review, we will focus on the peculiarities and specific issues that arise in setting up, gathering and processing information, and analysing data in meta-analyses of GWA studies (Figure 1).
Figure 1.
Typical work flow for conducting a meta-analysis of GWA datasets
Basic principles
Meta-analysis of GWA datasets can increase the power to detect association signals by increasing sample size and by examining more variants throughout the genome than each dataset alone. Different datasets may have used different platforms and may have thus genotyped different variants. However, current approaches [9] allow imputing genotypes at untyped variants using a reference such as HapMap. Directly-typed or imputed genotype information can currently be combined across datasets on up to several millions of common variants. Meta-analysis can be conducted in a sequential, cumulative manner; as more datasets become available, these may be included in the calculations resulting in more discoveries of previously unrecognized variants. Cumulative meta-analysis thus potentially represents a “replication ad infinitum” process.
Set-up issues: consortia and selection biases
Ideally, all GWAS research in a specific field should be conducted with the upfront aim that the data generated from different teams will be combined prospectively as they accumulate. The development of consortia of multiple teams can facilitate this process [10]. Consortia have been developed in many disease fields and they may cover a large number of teams [11]. In some diseases, several non-overlapping consortia exist, each one comprised of multiple teams of investigators, while other teams may not participate in any consortium, but may nevertheless conduct their own GWA and/or replication studies. Moreover, most GWA datasets are generated in epidemiological studies that have been set up in the past, and many important aspects of their design, conduct, data and sample collection can no longer be altered. Therefore, certain limitations that arise from retrospective features are almost unavoidable, even for the most inclusive, international, prospective effort.
A key challenge for any meta-analysis is to avoid selection biases. Biases arise if only some data are available for inclusion in the meta-analysis calculations and availability is dependent on the nature of the results. Publication bias, selective outcome and analysis reporting bias are extensively discussed in the traditional meta-analysis literature [12], but the ways that they may operate in GWA studies and whether GWA studies may have some particular immunity to them is poorly understood. In theory, a major advantage of GWA studies is their agnostic approach which allows comprehensive coverage of the genome [13]. However, if one were to simply increase the number of analyses, but still focus on making available only the most favourable results, selection bias could be detrimental. Conversely, if GWA studies are coupled with full availability of all the produced data and accompanying analyses, selection biases would be minimized. Consortia may have full availability of the entire datasets across all their participating teams and can also benefit from the implementation of common, standardized (or at least harmonized) approaches to data collection, definitions, measurements, and analysis [10]. Public repositories are also developed for GWA datasets, in particular those funded by public and not-for-profit funds [14,15]. However, public data availability poses several challenges, e.g. assurance of anonymity and proper use of the deposited information for appropriate scientific purposes.
Besides performing analyses for millions of genetic variants across the genome, one may also analyze thousands of phenotypes of interest for association with these variants. “Phenome scans” superimposed on GWA scans can yield interesting new discoveries, but they also add an extra dimension of multiplicity of comparisons. One should clarify whether one is searching agnostically a large number of phenotypes or is focusing on a specific type of phenotype(s). For phenome scan-derived associations, typical levels of “genome-wide statistical significance” used for single-phenotype analyses [16] may not be sufficient. If data are selectively available only for the “winner” phenotypes, even a meticulous meta-analysis can yield misleading conclusions. Finally, sometimes there may exist multiple analytical options for the same data (e.g. different genetic models, use or not of different adjustments, and so forth) [17].
As in any meta-analysis in any field, a GWA meta-analysis requires a robust protocol that should be set up before any quantitative analyses are undertaken. The protocol should specify the eligibility criteria for the combined datasets, the eligibility criteria for the specific genotype and phenotype data to be used, steps taken to ensure that the information is not selectively available or selectively analyzed, and the explicit analytical plans for the combined data.
Information for a GWA meta-analysis
Several pieces of information are required before embarking on a meta-analysis of GWA datasets (Table 1). Eventually, for each dataset, summary association statistics for each variant and phenotype of interest are generated from the individual-level information. This is done either locally at each participating team or centrally by the coordinating meta-analysis team, if individual-level data are shared. The ability to reproduce the analyses both locally and centrally offers an additional safeguard against errors and inconsistencies or even simple misunderstandings about some aspects of the data. However, such double analysis is often not possible if individual-level data have not been shared. The derived summary statistics may be odds ratios, standardized effect sizes, or other metrics along with their uncertainty (e.g. variance or 95% confidence interval) and/or the accompanying p-values. However, for each dataset, there are also many other items of information that are crucial to deal with. These include:
Table 1.
Informational aspects essential to deal with in a meta-analysis of GWA datasets and selected examples
| Epidemiological design features underlying each study and dataset |
| Design features, with emphasis on peculiarities and potential sources of bias |
| Quality checks including |
| Evaluation of Hardy-Weinberg equilibrium |
| Missing rate |
| Imputation accuracy scores |
| Analytical methods, definitions and adjustments used in each dataset |
| Central versus local analysis |
| Harmonized analysis of imputed genotypes |
| Standardization, harmonization, consistency in outcome definition |
| Standardization, harmonization, consistency in adjusting variables |
| Independence of the samples |
| Adjusting for cryptic or overt relatedness |
| Adjustment for population stratification |
| Handling overlapping samples |
| Strand and build of the human genome on which individual study results are provided |
| Handling of inconsistencies |
| Direct genotyping versus imputation |
| Relative availability of directly genotype vs. imputed data |
| Methods of imputation |
Epidemiological design features underlying each study and dataset
A checklist for improved reporting of the epidemiological design of genetic association studies (STREGA) has recently been developed [18]. Essential features of epidemiological design are often missing in reported genetic association studies [19]. When the pursued associations represent odds ratios of 1.1-1.3, knowing about design features that can spuriously generate observed odds ratios of at least that large magnitude on their own is essential.
Quality checks
These include, but are not limited to [5], evaluation of Hardy-Weinberg equilibrium, missing rate, and, in the case of imputed variants, imputation accuracy scores. Rules should be set upfront on what thresholds are not acceptable for each such quality check (e.g. p<0.0001 for Hardy-Weinberg violation, call rate <95%, or imputation accuracy <90%). Such quality checks may help reduce the impact of genotyping and other errors.
Analytical methods, definitions and adjustments used in each dataset
Ideally, all datasets should use the same definitions and adjustments (e.g. age, gender, body mass index and so forth) and there should be consensus on what adjustments will be used upfront. This is easier if a central facility analyzes all the data, but as discussed above, this may often not be feasible. The definitions of adjusting variables also should ideally be agreed upfront; since some information may have already been collected in the past, it is important to ensure sufficient consistency in these definitions. The same applies to the definitions of main phenotypes/outcomes of interest. Some fields have an extreme variability of options on how to define outcomes, e.g. there were almost 500 different outcomes and analyses reported on an evaluation of asthma pharmacogenetics [20]. Clearly, some consensus is needed in such fields. In other cases, if the differences are subtle, it may be reasonable to accept them, e.g. at least 3 different sets of criteria are commonly used to define Parkinson’s disease worldwide and their concordance is high. For most teams in the field, it is extremely difficult to go back and re-define cases and controls based on different criteria. Conversely, with stark differences in definitions, an effort at harmonization is essential. Exploratory analyses may be performed with different definitions and/or adjustments. However, picking the best results with particular definitions/adjustments post hoc may generate spuriously inflated signals of significance [17].
Independence of the samples
It is important to be aware of any relatedness among individuals within a study. Appropriate methods should be used to account for such relatedness. Through the use of unrelated markers, one can estimate how many times the chi-squared statistic for the association is inflated, and thus correct the chi-squared for the inflation factor λ. For a test of association with one degree of freedom (e.g. a 2×2 table), this is equivalent to inflating the standard deviation of the natural logarithm of the odds ratio by the square root of λ. In a similar vein, it is important to know whether/how population stratification was accounted for in each study. Population stratification can pose a serious threat for subtle effects. Ideally one should adjust for any evidence for population stratification [21-23] at the level of the individual study before carrying out the meta-analysis, and then after having combined studies also. Finally, one should examine whether any samples overlap across studies. If overlap is unavoidable, the overlap/covariance can be accounted for. The variance of each dataset increases (and thus its weight decreases), when the between-dataset correlation is included in the calculations.
Strand and build of the human genome on which individual study results are provided
Differences in strand need to be corrected before any data can be combined. Data generated with reference to a different build may show discrepancies. Ideally, all data should refer to the same build for full correspondence.
Direct genotyping versus imputation
Due to the limited overlap between different genotyping platforms, variants are likely to be imputed in some studies and directly-typed in others. Analysis of directly-typed and imputed variants is carried out in each scan separately, and controlled for population stratification separately, before summary statistics are combined. Within-study analysis of imputed data should conform to a predefined analysis plan and account for the uncertainty of genotype assignments.
In all, the importance of rigorous quality control and careful attention to detail at all steps and for all the issues listed above, cannot be overstated. The combination of insufficiently cleaned-up data may yield spurious associations.
Heterogeneity
There are over a dozen different statistics or metrics that can be used to evaluate heterogeneity [7,24,25]. Some are more popular in the literature, without necessarily being more useful than others.
A typical used measure of heterogeneity is Cochran’s Q, which tries to answer the question on whether there is statistically significant heterogeneity or not. It is calculated as the weighted sum of squared differences between individual study effects and the summary effect across studies. Q is distributed as a chi-square statistic with k-1 (k=number of studies) degrees of freedom. When the number of studies combined is small, the test has low power to detect heterogeneity if it is present. Conversely, if the number of studies is large, the test is likely to detect significant heterogeneity, even if the absolute magnitude of the variability is unimportant. As in any underpowered statistical test, a positive result does not ensure that heterogeneity is present indeed, and a negative result does not really exclude the possibility that heterogeneity exists. Acknowledging these caveats, the traditional threshold of claiming significant heterogeneity based on Q is p<0.10. It is not appropriate to adjust this for the number of SNPs tested as is the case for association statistics. With such improper adjustment, the power to detect significant heterogeneity vanishes.
The I2 statistic [25,26] describes the percentage of variation across studies that seems not to be due to chance. It is occasionally proposed as a measure of inconsistency, as opposed to heterogeneity, but the distinction of these terms is subtle and most authors still use them interchangeably. I2 is calculated as (Q-number of degrees of freedom)/Q. As shown by the formula, I2 differs from Q in that it also accounts for the number of studies. I2 has the advantage that it does not depend on the number of combined studies. It usually has large uncertainty and its 95% confidence intervals can be readily calculated [27].
The between-study variance, τ2, is the most direct measure of the magnitude of heterogeneity, but is rarely reported. When seen in isolation, it is difficult to interpret and it cannot be compared across different meta-analyses. If we want to see the magnitude of the heterogeneity juxtaposed to the magnitude of the effect size, a more useful metric may be h, which is defined as the ratio of τ over the effect size, i.e. it says how big the between-study standard deviation is compared with the summary effect size. A large h may be seen with large between-study variance or a small effect size [28].
There are many reasons that can underlie heterogeneity [29]. Besides heterogeneity due to chance, we still have limited insight into how much heterogeneity may be due to errors and biases differently affecting the results of different datasets, or to what extent heterogeneity may represent genuine differences in genetic effects across different populations and different biological setting, i.e. truly informative heterogeneity [30,31]. Informative heterogeneity may reveal interesting facts about biology, e.g. the mechanism through which the variant is acting on disease risk. One has to be cautious to avoid discarding such associations as replication failures. Heterogeneity could also result from the presence of variable LD between the typed marker and the causal variant.
However, if heterogeneity exists, then this association may not be possible to extend equally well to diverse populations. In the presence of considerable heterogeneity, if random effects calculations are used, no meta-analysis, no matter how large would have enough power to detect an association at genome-wide significance [28]. Moreover, the discovery process in the GWA setting is such that for discovered associations, on average, the observed heterogeneity may be less than the true heterogeneity: those associations that happen to have more observed heterogeneity in the data get more inflated confidence intervals by random effects calculations and are less likely to pass the threshold of genome-wide significance; while those that by chance have no or less heterogeneity than their true heterogeneity are more likely to be considered replicated. This makes interpretation of homogeneity particularly difficult.
Summarizing results
There are several ways to combine datasets in a meta-analysis framework (Figure 2). Meta-analyses can combine p-values or effect sizes. P-value meta-analysis methods have a long history of applications in the social sciences [7], but they became unpopular and had been practically abandoned in the biomedical sciences, until some investigators started using them again in the GWA era. Limitations of p-value meta-analyses (difficulties in interpretation of the combined estimate, inability to provide effect sizes, difficulties in addressing heterogeneity, differences in p-values obtained with parametric and non-parametric methods, handling of p-values with p>0.5, among others) have been recognized for decades, and our recommendation is that effect sizes should be combined whenever the data are available, even if there is still debate about the ability of meta-analysis of GWA to yield summary effect sizes that can be readily differentiated in magnitude among themselves (see below).
Figure 2.
Common meta-analysis methods
Studies or datasets combined in a meta-analysis are given different weights. The general principle is that studies with more precision should weigh more in the calculations than studies with lower precision. Two commonly used approaches are fixed and random effects models [7,8,24].
Methods of fixed effect meta-analysis are based on the assumption that a single common (or ‘fixed’) effect underlies every study in the meta-analysis, i.e. if every study were infinitely large, the results of every study would be identical because there is no between-study heterogeneity. Fixed effects analysis provides for testing of the null hypothesis of no association in any of the study populations being analyzed. This is useful if we simply aim to optimize power for detecting association. However, if heterogeneity exists, then this association may not be possible to extend equally well to diverse populations.
A random effects analysis makes the assumption that individual studies are estimating different effects. We assume that the different effects have a distribution with some mean value and some degree of variability dictated by the between-study variance. The idea behind a random effects meta-analysis is to learn about this distribution of effects across different studies.
In the absence of between-study heterogeneity, fixed and random effects calculations yield identical point estimates and confidence intervals. With increasing between-study heterogeneity, the random effects summary estimates have larger variance (wider confidence intervals) and usually, but not always, they have less prominent statistical significance.
Most meta-analysts in different fields would typically run both models, but prefer placing emphasis on random effects. For many GWA studies, however, only fixed effects are reported. Statistically significant associations in fixed or random effects calculations certainly need further documentation and replication. Random effects may sometimes yield spuriously large summary effects when selection biases operate differently in small versus larger studies. If a meta-analysis of GWA studies can ensure that there are no such biases within its confines (e.g. including all data from a well-circumscribed consortium), then this is no longer an issue. Conversely, when small studies have smaller effects than larger studies and results are heterogeneous, random effects calculations may give stronger overall statistical significance for the summary effect.
For GWA meta-analyses, discriminating different magnitudes of effect size can sometimes be problematic. The detected associations are usually (but not always) so modest in terms of effect size that it is difficult to differentiate and rank them with certainty. Effect sizes would be important to know with precision, if this information were to be used successfully for predictive risk modelling [32].
It is also anticipated that discovered associations that pass a threshold of required genome-wide statistical significance are likely to have observed effect sizes that are inflated compared with the true effect size [33]. Moreover, it is usually impossible to tell upfront whether a newly discovered genetic variant is the direct culprit or simply linked to the (yet unknown) causal variant. The analytical methods used, e.g. genetic model specification (or misspecification), may also affect the magnitude of the effect size. Finally, discovery of a genetic locus has important implications on its own, regardless of the observed effect size, e.g. it may highlight some interesting biological pathway and may give some insights into developing new therapeutics.
Cumulative, Bayesian, and sequential meta-analysis issues
The concept of cumulative meta-analysis [34], where many studies or datasets are sequentially incorporated in the calculations, as they become available, fits very nicely to a Bayesian framework. Previous studies form the prior belief. Estimates are updated with each new study to generate a posterior belief. Besides this simple concept, there is also a wide literature of formal Bayesian methods that can be applied in meta-analysis [35-37]. These methods require that specific priors be applied to the uncertainty parameters and one can examine the robustness of conclusions based on these priors, i.e. whether different priors may change the results perceptibly. While we need more empirical data on how these methods would perform on the GWA setting, Bayesian meta-analysis is gradually gaining ground in other biomedical fields, since it allows for a more general view of meta-analysis, of which the typically used fixed and random effects are only a special case. Bayesian estimates of summary effects usually have increased uncertainty [37], especially if more uncertainty is introduced in the prior assumptions of between-study variance, effect size, genetic model, etc. However, there is also the possibility that, in some circumstances, borrowing strength from external prior evidence may increase certainty.
Another approach to cumulative incorporation of more datasets stems from the frequentist methods that try to correct levels of statistical significance for sequential designs, e.g. alpha-spending functions. These approaches are routine in large clinical trials, where it is widely appreciated that the required p-value threshold should depend on how many times and when the data are analyzed, as they are sequentially assembled. In the GWA setting, one would similarly argue that if several updates in the meta-analysis are performed, more stringent p-values should be required to claim genome-wide significance as new data are introduced. While the concept is valid and methods exist for such adjustments [38], they have not yet been applied to GWA meta-analyses.
In all, besides the statistical significance of the summary estimate, it is useful to interpret summary estimates in terms of the Bayes factors and credibility that they confer to the postulated association. Several straightforward Bayesian approaches are readily available [39-41] and allow estimating the credibility of the meta-analysis results under different assumptions.
Replication of genome-wide meta-analysis findings
Following a GWA meta-analysis, researchers should prioritize interesting signals (whether they reach genome-wide significance or not) for follow-up and further replication. Follow-up sample sets should be adequately powered to detect the diminishingly low effect sizes observed at newly identified variants. Issues of set-up, information aggregation, estimation of heterogeneity and summary effects in such extended replication efforts are similar to those described above for GWA meta-analysis.
An example: Application of meta-analysis to T2D GWA data
We describe here in brief the application of meta-analysis to GWA studies of T2D [42]. Three T2D GWA scans (WTCCC, DGI, FUSION) [43-46] were combined in a meta-analysis framework. Details for the design of these studies can be found in their original publications [43-46]. Genotypes at untyped SNPs were imputed across the HapMap in each of the 3 individual studies. Stringent quality control was carried out for directly typed and imputed variants, and individual studies were corrected for population stratification before combining summary results across a total of 10,128 samples. Approximately 2.2 million SNPs survived quality control. To combine the data, both fixed effects OR-based and p value-based meta-analysis approaches were used.
In combining the 3 GWA scans, several challenges were encountered and analytically overcome. The 3 studies had been carried out on different genotyping platforms. To overcome this, SNPs were imputed across the HapMap, and this allowed maximized use of the information available. The scans had different ascertainment schemes, for example in terms of matching cases and controls for BMI, a known T2D risk factor. To address this, both fixed- and random effects meta-analyses were carried out and evidence for informative heterogeneity was examined. One prime example of informative heterogeneity is the FTO locus, the first robustly replicating obesity susceptibility locus [47], which was associated with T2D in some, but not other scans. In particular, this locus did not achieve significant evidence for association with T2D in scans that had matched cases and controls for BMI.
Different imputation methods had been used to infer genotypes at untyped variants across the 3 studies. A stringent approach to quality control of the imputed genotypes was therefore taken. The robustness of results was ensured by directly genotyping in the original samples any SNPs that gave significant signals based on imputed genotype data.
Finally, population stratification was dealt with in multiple ways, to ensure that the rising signals were not spurious associations. Among the different approaches taken, directly typed and imputed data were adjusted for genomic control in each scan separately, and then adjusted for genomic control in the combined meta-analysis dataset.
A 3-stage approach was pursued (Table 2). In the first stage, data on the 2.2 million SNPs were combined across 10,128 samples in a meta-analysis framework. In the second stage, 69 SNPs were selected and genotyped in 22,426 additional independent samples, all of European descent. In the third stage, 11 of these promising signals were taken forward to a further independent set of samples of European descent, consisting of 57,366 T2D cases and controls.
Table 2.
Type 2 diabetes study design
| Stage | Description | Number of samples |
Number of evaluated SNPs |
Main findings |
|---|---|---|---|---|
| 1 | 3-way GWA scan meta-analysis | 10,128 | 2,202,892 | Clear deviation from the null hypothesis of no association |
| 2 | Follow-up of interesting signals in independent samples |
22,426 | 69 | Excess of T2D-associated loci in the same direction as stage 1 |
| 3 | Further follow-up of promising signals in independent samples |
57,366 | 11 | Six novel loci surpassing specified genome- wide significance levels |
A large excess of associated SNPs in stages 2 and 3 was observed. In stage 2, out of 65 independent signals, 10 achieved a p-value<0.05 for association with T2D, whereas only 1.6 were expected to achieve this by chance. In stage 3, out of 10 independent signals, 7 reached a p-value<0.05 with T2D, as opposed to an expected 0.25 under the null. It is noteworthy that the associated SNPs all had small observed effect sizes, with the largest allelic odds ratio being only 1.15. This underlines the need for very large-scale meta-analysis replication efforts.
Table 3 summarizes the genetic loci that have substantial statistical support for association with T2D as of the summer of 2008. As shown, some of these loci were identified through non-GWA approaches. Others emerged out of single GWA studies combined with data from several replication datasets; while a number of them have emerged only recently as the result of meta-analysis of several GWAs and replication datasets, as described above. A common feature is that rarely has a single study been able to reach genome-wide significance for specific loci. This has required the combination of data from several studies, GWA scans and focused replication efforts.
Table 3.
Gene loci with common variants associated with type 2 diabetes and their method of identification.
| Representative SNP | Chr | Position | Region / gene | Identification |
|---|---|---|---|---|
| rs10010131 | 4 | 6343816 | WFS1 | Non-GWA |
| rs1801282 | 3 | 12368125 | PPARG | Non-GWA |
| rs757210 | 17 | 33170628 | TCF2 | Non-GWA |
| rs5219 | 11 | 17365206 | KCNJ11 | Non-GWA |
| rs7901695 | 10 | 114744078 | TCF7L2 | Non-GWA |
| rs10811661 | 9 | 22124094 | CDKN2A/B | GWA |
| rs10946398 | 6 | 20769013 | CDKAL1 | GWA |
| rs13266634 | 8 | 118253964 | SLC30A8 | GWA |
| rs4402960 | 3 | 186994389 | IGF2BP2 | GWA |
| rs5015480 | 10 | 94455539 | HHEX/IDE | GWA |
| rs8050136 | 16 | 52373776 | FTO * | GWA |
| rs2237895 | 11 | 2813770 | KCNQ1 | GWA |
| rs10923931 | 1 | 120230001 | NOTCH2 | GWA meta-analysis |
| rs12779790 | 10 | 12368016 | CDC123/CAMK1D | GWA meta-analysis |
| rs4607103 | 3 | 64686944 | ADAMTS9 | GWA meta-analysis |
| rs7578597 | 2 | 43644474 | THADA | GWA meta-analysis |
| rs7961581 | 12 | 69949369 | TSPAN8/LGR5 | GWA meta-analysis |
| rs864745 | 7 | 27953796 | JAZF1 | GWA meta-analysis |
reflecting an association with body mass index and obesity risk rather than type 2 diabetes per se
Moreover, the causal variants at these loci have not yet been identified. Even very large-scale GWA meta-analyses would require extensive fine-mapping and targeted resequencing experiments before the truly causal variants can be confidently identified [5]. Finally, evaluation of the extent of generalizability of these associations would need even further replication studies in different settings and types of participants (e.g. different racial groups, or different risk populations [48]).
Future perspective
GWA studies and their meta-analyses have already resulted in an unprecedented number of genetic associations with strong statistical support [6]. At the same time, they have convincingly shown that effect sizes are almost always moderate or small and that with current platforms and sample sizes we are still unable to explain the large majority of genetic risk for most common diseases. We still don’t know whether what we have found with current resources is the tip of the iceberg or the bottom of the barrel [49]. This means that larger and larger studies and meta-analyses will probably continue to be performed in an effort to unearth more genetic risk variants of interest, if the resources can be met. The large majority of research to-date has been performed with case-control samples from pre-existing studies and in populations of Caucasian descent. There will be a challenge to extend the observations in new prospective cohorts and biobanks [50] and across populations with different ancestries. Moreover, incorporating environmental and lifestyle components into meta-analytic evaluations of large-scale evidence will also be a challenge.
There is already a huge interest and even commercial pressure for marketing prognostic services based on common genetic variants [51]. However, this would require explicit knowledge of the exact identity and magnitude of the effect sizes associated with causal variants, an expansion of the list of associated variants and even more extensive replication across diverse populations and settings [52], with accumulation of evidence that can be confidently graded as having high credibility [53]. The extent to which genetic risk may lie in rare variants and may need to be approached and/or complemented with other high-throughput platforms (e.g. copy number variation) remains unquantified. Meta-analysis methods for such variants are likely to pose new challenges and would require careful interpretation. At the same time, the blossoming of human genome epidemiology research means that large numbers of teams may have access to high throughput facilities and that many GWA teams, consortia, replication studies, and isolated efforts may continue to perform research in parallel. The synthesis and synopsis [54] of all these data in what might be called conglomerate meta-analyses and the updating of the total evidence-base on genetic associations is an evolving challenge. Large-scale, better-designed efforts hold the promise of further advancing our understanding of the genetic landscape of complex disorders.
Executive summary
Genetic effects of common variants associated with major diseases are mostly small and require very large sample sizes to identify with any certainty.
Meta-analysis of genome-wide association data can improve the power for detecting and validating such associations.
Meta-analysis of genome-wide data from studies using different platforms is enhanced by the use of imputed genotype data.
Selection biases need to be considered as in any meta-analysis
Careful collection and quality-checking of information is essential to avoid errors given the massive scale of analyses.
Meta-analysis allows the exploration of the extent and reasons for heterogeneity of the genetic effects across datasets, besides providing summary effects.
A wide array of methods may be used, including fixed effects, random effects, and Bayesian meta-analysis and they have particular advantages and disadvantages.
Application of the meta-analysis methodology in genome-wide data has been particularly successful in identifying more disease-related genes for some conditions such as type 2 diabetes.
References
- 1.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet. Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Burton PR, Hansell AL, Fortier I, et al. Size matters: just how big is BIG?: Quantifying realistic sample size requirements for human genome epidemiology. Int. J. Epidemiol. 2008 Aug 25; doi: 10.1093/ije/dyn147. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9(5):356–69. doi: 10.1038/nrg2344. *An extensive review of current status and challenges on genome-wide association studies
- 6.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118(5):1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutton AJ, Abrams KR, Jones DR, Sheldon TA, Song F. Methods for meta-analysis in medical research. Wiley; Chichester: 2000. [Google Scholar]
- 8.Higgins JP, Little J, Ioannidis JP, et al. Turning the pump handle: evolving methods for integrating the evidence on gene–disease association. Am. J. Epidemiol. 2007;166:863–866. doi: 10.1093/aje/kwm248. [DOI] [PubMed] [Google Scholar]
- 9.Anderson CA, Pettersson FH, Barrett JC, et al. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am. J. Hum. Genet. 2008;83(1):112–9. doi: 10.1016/j.ajhg.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seminara D, Khoury MJ, O’Brien TR, et al. The emergence of networks in human genome epidemiology: challenges and opportunities. Epidemiology. 2007;18:1–8. doi: 10.1097/01.ede.0000249540.17855.b7. [DOI] [PubMed] [Google Scholar]
- 11.Ioannidis JP, Bernstein J, Boffetta P, et al. A network of investigator networks in human genome epidemiology. Am. J. Epidemiol. 2005;162:302–304. doi: 10.1093/aje/kwi201. [DOI] [PubMed] [Google Scholar]
- 12.Rothstein HR, Sutton AJ, Borestein M, editors. Publication bias in meta-analysis–prevention, assessment and adjustments. Wiley; Chichester: 2005. [Google Scholar]
- 13.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat. Genet. 2006;38(6):659–62. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 14.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39(10):1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GAIN Collaborative Research Group. Manolio TA, Rodriguez LL, Brooks L, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat. Genet. 2007;39(9):1045–51. doi: 10.1038/ng2127. *Presentation of the GAIN initiative for collaboration in genome-wide association studies
- 16.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet. Epidemiol. 2008;32(2):179–85. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19(5):640–8. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 18.Little J, et al. Strengthening the reporting of genetic association studies. Ann. Intern. Med. doi: 10.7326/0003-4819-150-3-200902030-00011. (in press) [DOI] [PubMed] [Google Scholar]
- 19.Yesupriya A, Evangelou E, Kavvoura FK, et al. Reporting of human genome epidemiology (HuGE) association studies: an empirical assessment. BMC Med. Res. Methodol. 2008;8:31. doi: 10.1186/1471-2288-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contopoulos-Ioannidis DG, Alexiou GA, Gouvias TC, Ioannidis JP. An empirical evaluation of multifarious outcomes in pharmacogenetics: beta-2 adrenoceptor gene polymorphisms in asthma treatment. Pharmacogenet. Genomics. 2006;16:705–711. doi: 10.1097/01.fpc.0000236332.11304.8f. [DOI] [PubMed] [Google Scholar]
- 21.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum. Genet. 2008;123(1):1–14. doi: 10.1007/s00439-007-0445-9. *Short overview of quantitative methods for meta-analysis in genetic epidemiology
- 25.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335:914–916. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP. Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc. Natl. Acad. Sci. U S A. 2008;105(2):617–22. doi: 10.1073/pnas.0705554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum. Hered. 2007;64:203–13. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- 30.Evangelou E, Maraganore DM, Ioannidis JP. Meta-analysis in genome-wide association datasets: strategies and application in Parkinson disease. PLoS ONE. 2007;2:e196. doi: 10.1371/journal.pone.0000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS ONE. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle--will we get our wish? N. Engl. J. Med. 2008;358(2):105–7. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 33.Zollner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am. J. Hum. Genet. 2007;80(4):605–15. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau J, Schmid CH, Chalmers TC. Cumulative meta-analysis of clinical trials builds evidence for exemplary medical care. J. Clin. Epidemiol. 1995;48:45–57. doi: 10.1016/0895-4356(94)00106-z. (discussion 59-60) [DOI] [PubMed] [Google Scholar]
- 35.Spiegelhalter DJ, Thomas A, Best NG, Lunn D. WinBUGS version 1.4 users manual. 2003.
- 36.Spiegelhalter DJ, Abrams KR, Myles PJ. Evidence synthesis. Wiley; Chichester: 2004. [Google Scholar]
- 37.Salanti G, Higgins JP, Trikalinos TA, Ioannidis JP. Bayesian meta-analysis and meta-regression for gene-disease associations and deviations from Hardy–Weinberg equilibrium. Stat. Med. 2007;26:553–567. doi: 10.1002/sim.2575. [DOI] [PubMed] [Google Scholar]
- 38.Thorlund K, Devereaux PJ, Wetterslev J, et al. Can trial sequential monitoring boundaries reduce spurious inferences from meta-analyses? Int. J. Epidemiol. doi: 10.1093/ije/dyn179. (in press) [DOI] [PubMed] [Google Scholar]
- 39.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ioannidis JP. Calibration of credibility of agnostic genome-wide associations. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B(6):964–972. doi: 10.1002/ajmg.b.30721. [DOI] [PubMed] [Google Scholar]
- 42.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40(5):638–45. doi: 10.1038/ng.120. *Large-scale application of meta-analysis of genome-wide association datasets in type 2 diabetes
- 43.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for Type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 44.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of Type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in U.K. samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore AF, Jablonski KA, McAteer JB, et al. Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes. 2008;57(9):2503–10. doi: 10.2337/db08-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iles MM. What can genome-wide association studies tell us about the genetics of common disease? PLoS Genet. 2008;4(2):e33. doi: 10.1371/journal.pgen.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott P, Peakman TC. UK Biobank: The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int. J. Epidemiol. 2008;37(2):234–44. doi: 10.1093/ije/dym276. [DOI] [PubMed] [Google Scholar]
- 51.Janssens AC, Gwinn M, Bradley LA, et al. A critical appraisal of the scientific basis of commercial genomic profiles used to assess health risks and personalize health interventions. Am. J. Hum. Genet. 2008;82(3):593–9. doi: 10.1016/j.ajhg.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 53.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int. J. Epidemiol. 2008;37(1):120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 54.Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40(7):827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]