Abstract
The most frequent cause of familial clear cell renal cell carcinoma (RCC) is von Hippel–Lindau disease and the VHL tumor suppressor gene (TSG) is inactivated in most sporadic clear cell RCC. Although there is relatively little information on the mechanisms of tumorigenesis of clear cell RCC without VHL inactivation, a subset of familial cases harbors a balanced constitutional chromosome 3 translocation. To date nine different chromosome 3 translocations have been associated with familial or multicentric clear cell RCC; and in three cases chromosome 6 was also involved. To identify candidate genes for renal tumorigenesis we characterized a constitutional translocation, t(3;6)(q22;q16.1) associated with multicentric RCC without evidence of VHL target gene dysregulation. Analysis of breakpoint sequences revealed a 1.3-kb deletion on chromosome 6 within the intron of a 2 exon predicted gene (NT_007299.434). However, RT-PCR analysis failed to detect the expression of this gene in lymphoblast, fibroblast, or kidney tumor cell lines. No known genes were disrupted by the translocation breakpoints but several candidate TSGs (e.g., EPHB1, EPHA7, PPP2R3A RNF184, and STAG1) map within close proximity to the breakpoints.
INTRODUCTION
Although only 2% of renal cell carcinoma (RCC) cases are familiar, the identification of familial RCC genes (e.g., the VHL gene) has provided important insights into the pathogenesis of sporadic RCC. Thus, somatic inactivation of the VHL tumor suppressor gene (TSG) is found in ~60% of sporadic clear cell RCC (Kim and Kaelin, 2004; Maher, 2004). VHL inactivation leads to stabilization of the HIF-1 and HIF-2 transcription factors and overexpression of downstream target genes, including VEGF, cyclin D1, and carbonic anhydrase IX (CA-IX) (Maxwell et al., 1999; Zatyka et al., 2002; Staller et al., 2003). VHL mutations are rare in nonclear cell RCC, but there is also evidence for a nonVHL dependent pathway of renal tumorigenesis in clear cell RCC. Thus, a subset of sporadic cases do not have VHL inactivation and linkage to VHL has been excluded in kindreds with familial nonVHL clear cell RCC (Teh et al., 1997; Woodward et al., 2000).
A subgroup of patients with familial clear cell RCC harbors a constitutional chromosome 3 translocation. Nine different chromosome 3 translocations have been associated with RCC susceptibility. In seven cases the translocations t(1;3)(q32; q13), t(2;3)(q33;q21), t(2;3)(q35;q21), t(3;4)(p13; p16), t(3;6)(q12;q15), t(3;8)(p13;q24), and t(3;8) (p14.1; q24.23) (see Melendez et al., 2003 and references within) were associated with familial clear cell RCC and in two cases, individuals with a familial t(3;6) (p13;q25.1) (Kovacs et al., 1989) and t(3;6)(q23; q16.2) (Subramonian et al., 1998), developed multicentric clear cell RCC. Cloning of the translocation breakpoint has, in some cases, led to the identification of novel RCC susceptibility genes. Thus characterization of the t(3;8)(p14;q24) breakpoints revealed that the FHIT and TRC8 genes were disrupted (Ohta et al., 1996; Gemmill et al., 1998). Subsequently, loss of FHIT expression has been shown to occur in a variety of human cancers, including RCC, and FHIT has been demonstrated to have TSG activity (Pekarsky et al., 2002). TRC8 has also been proposed as a candidate TSG (Gemmill et al., 2002). In another family, a t(1;3)(q32;q13) associated with familial RCC was characterized and two candidate TSGs, NORE1A, and LSAMP, were disrupted (Chen et al., 2003). Both genes are silenced by promoter methylation in a subset of sporadic RCC (Chen et al., 2003; Morris et al., 2003). However, for many RCC-associated constitutional chromosome 3 translocations, characterization of the translocation breakpoints has led to the identification of genes that were not implicated in the pathogenesis of RCC or the breakpoints were found not to directly disrupt known genes (Druck et al., 2001; Bodmer et al., 2002, 2003). In such cases susceptibility to RCC may result from chromosome 3 instability (predisposing to loss of 3p which contains several TSGs) or because the translocations have long-range effects on gene expression. Thus, in disorders such as aniridia and campomelic dysplasia, translocations many hundreds of kilobases away from PAX6 or SOX9, respectively, may alter gene expression and cause disease (Crolla and Van Heyningen, 2002). Here we report the molecular characterization of a t(3;6)(q22;q16.2) associated with multicentric RCC (Subramonian et al., 1998).
MATERIALS AND METHODS
Cell Line
An EBV-transformed lymphoblastoid cell line was established from the proband. Cells were maintained at 37°C under 5% CO2 and cultured in Dulbecco's Modified Eagle Medium supplemented with 10% FCS, 0.1 mM nonessential amino acids, 1% glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Genomic DNA was prepared using a Nucleon Kit (Amersham, Little Chalfont, Bucking-hamshire, UK). SKRC39 cells were also cultured under the same conditions.
Chromosome Preparation and Flow Sorting
Derivative chromosomes, der(3) and der(6) were flow sorted from the patient's cell line and used as templates for whole genome amplification (Genomiphi), following the manufacturer's instructions (Amersham) (Fiegler et al., 2003). The amplified DNAs were used in array painting experiments and as templates for further PCRs.
CGH Array
Array CGH was carried out as described previously (Gribble et al., 2005). About 450 ng of genomic DNA from the cell line containing the translocation was labeled and hybridized against 450 ng pooled male DNA (DNA from 20 individuals).
Array Painting
The experimental procedures used for array painting were essentially as described (Fiegler et al., 2003). Briefly, the two derivative chromosomes were separated by flow sorting, amplified, and 450 ng of each were differentially labeled and hybridized onto DNA arrays.
Construction of BAC Contigs and Fluorescent In Situ Hybridization Analyses
Nine 3q22.2-3q22.3 and twelve 6q16.1 BAC clones were obtained from the BACPAC Resource Center (Children's Hospital, Oakland Research Institute, Oakland, CA). The clones were selected based on information in the BAC clone mapping databases and the human genome sequence draft database. BAC DNA was prepared using Phase-Prep BAC DNA Kit (Sigma, Gillingham, UK) and then labeled directly by nick translation according to the manufacturer's instructions (Vysis), with Spectrum Green or Spectrum Red-dUTPs (Vysis, Downers Grove, IL). Probes were blocked with Cot-1 DNA to suppress repetitive sequences. Metaphase spreads obtained from the patient's cell line and two normal controls were hybridized overnight at 37°C with labeled probes. Chromosome 3 and 6-specific centromeric probes were included in hybridization mixture (Vysis). After posthybridization washes the chromosomes were counterstained with DAPI in Vectashield antifade solution (Vector Laboratories, Peterborough, UK).
BAC Clones used for the Construction of BAC Contigs and FISH Analyses
The 9 BAC clones from 3q22.2-3q22.3 are RP11-192L3, RP11-1079L18, RP11-73J21, RP11-354M3, RP11-767H24, RP11-452N24, RP11-219P9, RP11-1096B15, and RP11-93K21. The 12 BAC clones from 6q16.1 are RP11-777M22, RP11-461I4, RP11-244K16, RP11-960B10, RP11-119L14, RP11-25909, RP11-419I4, RP11-630M20, RP11-654K3, RP11-44P5, RP11-939I18, and RP11-243P16.
STS-PCR and Amplification of Breakpoint Sequences
Sequence information derived from the two breakpoint spanning BACs was used to design primers to amplify regions along the sequences of interest. Flow sorted der(3) and der(6) DNA, as well as genomic DNA were used as templates. This approach enabled the region containing the breakpoints to be narrowed until it was possible to PCR across the breakpoint. Breakpoint fragments generated using pairs of primers from the different chromosomes were then sequenced using an ABI BigDye Cycle Sequencing Kit (Perkin–Elmer, Rotkreuz, Switzerland) and analyzed using an ABI Prism377 DNA sequencer (Perkin–Elmer).
RESULTS
Clinical Case Report
A 49-years-old man presented with hematuria and loin pain. Renal ultrasound and CT scans demonstrated bilateral multicentric RCC (Subramonian et al., 1998). A left partial nephrectomy was performed and histopathological examination revealed a well-differentiated clear cell RCC. Two months later, a radical right nephrectomy was performed for the two large (4.5 cm) tumors in the right kidney. Histopathological examination demonstrated clear cell RCC with occasional areas of papillary cancer. Subsequently, he developed three new RCCs in his residual left kidney and a left nephrectomy was performed 6 years after the first operation. At Clinical Genetics assessment there was no evidence of VHL disease or tuberose sclerosis, and no family history of cancer. VHL gene analysis demonstrated no abnormality, but cytogenetic analysis revealed a constitutional t(3;6)(q22;q16.2). The translocation was not present in his father and two siblings (his mother had died and was not available for analysis).
Mapping and Characterization of Translocation Breakpoints
Mapping of translocation breakpoints by Array Painting
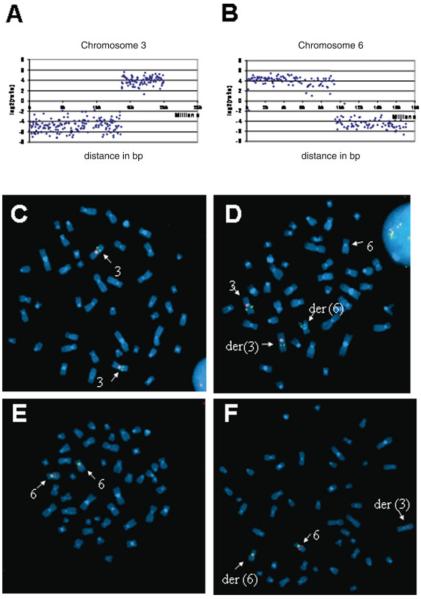
Array Painting using a 1-Mb resolution BAC array was performed on derivative chromosomes flow sorted from a lymphoblastoid cell line. DNA from the two derivative chromosomes was differentially labeled and cohybridized. The ratios of fluorescence intensity were plotted against the distance of the clones along the chromosome from the p termini. Fluorescence ratio profiles are shown in Figure 1. Sharp transitions in ratio values define the breakpoint region. Thus Array Painting mapped the breakpoints to an approximately 0.9-Mb interval between BAC clones RP11-91O5 and RP11-311K2 on chromosome 3, and an approximately 1.6-Mb region between clones RP11-21G12 and RP11-58A9 on chromosome 6. Array CGH analysis was also performed and did not detect any genomic imbalances.
Figure 1.
Panels A and B: Array Painting analysis demonstrating changes in hybridization signals at translocation breakpoints on chromosomes 3 and 6. Panels C–F: metaphase spreads prepared from lymphoblast cell line established from normal control (C and E) and patient with t(3;6) translocation (D and F). C and D show chromosome 3 identified using chromosome 3 specific centromeric probe, labeled with spectrum-orange. Normal and t(3;6) spreads were hybridized with a spectrum-green labeled chromosome 3 BAC probe, RP11-354M3, that contained the breakpoint (as shown by the split signal). E and F show corresponding images for chromosome 6. The breakpoint mapped within RP11-44P5 as shown by the split signal. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Refining translocation breakpoints by FISH analysis
Human genome sequence and BAC mapping data were used to construct in silico (a) a contig of 9 BAC clones (See Materials and Methods section) spanning the 0.9-Mb region containing the t(3) breakpoint and, (b) a contig of 12 BAC clones covering the t(6) breakpoint region. BAC clones were hybridized to metaphase spreads from patient and normal lymphoblast cells. The presence of split signals within the patient metaphase spreads identified two breakpoint-spanning BACs, RP11-354M3 on chromosome 3 and RP11-44P5 on chromosome 6 (Fig. 1).
Characterization and sequencing of translocation breakpoints
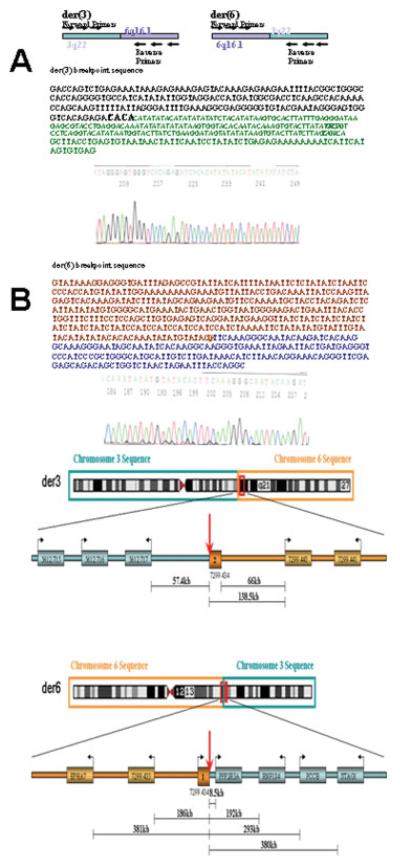
To map more precisely the t(3;6) breakpoints, primers were designed for STSs within the two breakpoint spanning BACs. PCR amplification was then performed using amplified DNA from each derivative chromosome and this approach enabled the region containing the breakpoints to be narrowed to within ~300 bp on chromosome 3, and to within ~1.7 kb on chromosome 6. We then attempted to PCR across the breakpoints (Fig. 2) and generated fragments of ~1.1 kb for der(3) and ~0.5 kb for der(6). Sequencing of these fragments revealed a 1.3-kb deletion on chromosome 6, a repeat motif comprising tandem tetranucleotide repeats (CTAT)n1(CCAT)n2, in which n1 was reduced, as compared with the available genome sequence, from 13 to 7 and n2 was reduced from 7 to 5, and an 8 bp loss at the immediate breakpoint site. The breakpoints did not directly disrupt any known genes, although the chromosome 6 breakpoint was located within the intron of a 3 exon predicted gene (NT_007299.434). RT-PCR analysis, however, failed to detect expression of this gene in lymphoblast, fibroblast or kidney tumor cell lines.
Figure 2.
Panels A and B show breakpoint region sequences for derivative chromosome 3 (Panel A) and derivative chromosome 6 (Panel B) (see text for details). Panel C illustrates the location of genes in relation to translocation breakpoints. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
DISCUSSION
We have characterized the breakpoints of a 3;6 translocation associated with multiple RCC. Multicentric RCC is rare, occurring in ~2% of all cases (Blute et al., 2003). Although bilateral RCC is characteristic of inherited RCC susceptibility, it may occur sporadically in older individuals (mean 65 years) so the early age at diagnosis and multiple tumors (n = 6) in our patient, coupled with the known association between constitutional chromosome 3 translocations and RCC, is consistent with a causal association between the t(3;6) and multicentric RCC. It is intriguing that chromosome 6 is the most commonly involved partner in chromosome 3 translocations associated with RCC, although there is no evidence of common breakpoints: t(3;6)(q12;q15) (Eleveld et al., 2001), t(3;6)(p13;q25.1) (Kovacs et al., 1989), and t(3;6)(q23;q16.2) (Subramonian et al., 1998), perhaps suggesting that the translocation might predispose to renal neoplasia by instability of the derivative chromosomes and/or long-range effects on gene expression and not by direct disruption of specific genes. Clinically our case resembles the German family with a t(3;6)(p13;q25.1) in three consecutive generations in which the only translocation carrier aged >50 years of age developed multiple bilateral renal-cell carcinomas (Kovacs et al., 1989).
Characterization of RCC-associated translocations has led to the identification of several genes that are disrupted by the breakpoints (e.g., FHIT, TRC8, DIRC1, DIRC2, DIRC3, LSAMP, and NORE1 (Gemmill et al., 1998; Druck et al., 2001; Bodmer et al., 2002, 2003; Chen et al., 2003). However, the role of DIRC1, DIRC2, and DIRC3 in renal tumorigenesis requires confirmation and, as in our case, there are several cases where no transcribed genes appeared to be disrupted by the breakpoints. Thus, analysis of a t(3;8)(p14.2;q24.2) revealed a 5-kb microdeletion at the chromosome 3 breakpoint but no genes were disrupted by either breakpoint (Rodriguez-Perales et al., 2004). Evidence that RCC-associated translocations might cause instability of the translocated chromosome is provided by the observation that in some tumors associated with a constitutional chromosome 3 translocation, the derivative 3 was lost in the tumor and a somatic VHL mutation was also detected (such that the tumor was homozygous for VHL mutation/loss, a 3-hit model of tumorigenesis) (Schmidt et al., 1995; Bodmer et al., 1998; Kanayama et al., 2001). However, in other cases tumors may not show VHL inactivation, implicating a VHL-independent pathway of tumorigenesis (Valle et al., 2005). In our case molecular genetic analysis of tumor material for VHL mutations failed because of the poor quality DNA but immunostaining for CA-IX expression (Mandriota et al., 2002) showed no evidence of increased expression in the two RCC studied, providing indirect evidence for a possible VHL-independent mechanism for tumorigenesis.
Balanced translocations have been reported to produce long range (e.g., up to 1.3 Mb) “position effects” on gene expression (Crolla and Van Heyningen, 2002; Velagaleti et al., 2005). Hence genes in the vicinity of the translocation might be implicated in the pathogenesis of RCC. The known genes closest to the chromosome 3 and 6 breakpoints are PPP2R3A, PCCB, STAG1, RNF184, and EPHB1 and EPHA7, respectively. PCCB encodes the beta subunit of propionyl-CoA carboxylase (germline PCCB mutations cause propionicacidemia) and we could find little evidence to implicate PCCB in tumorigenesis. PPP2R3A is a regulatory subunit of protein phosphatase 2A (PP2A), a heterotrimeric serine/threonine phosphatase (Hendrix et al., 1993). PP2A is a candidate TSG and inactivation of the regulatory PR65/A subunit occurs frequently in human tumors (Janssens et al., 2005). Hence, it can be hypothesized that the t(3;6) might lead to altered PPP2R3A expression and disturbances in PP2A function. Ephrins are the ligands for the Ephrin receptors that form a large family of receptor tyrosine kinases and play a key role in cell–cell interactions and cell migration. Ephrins and their receptors are frequently overexpressed in human cancers (Surawska et al., 2004). Interestingly, EPHA7 was the closest known gene to the chromosome 6 breakpoint, and EPHB1 mapped ~500 kb from the chromosome 3 breakpoint. The EPHB1 ephrin receptor interacts with the adaptor molecule Grb7 and regulates integrin-mediated attachment and migration (Han et al., 2002; Huynh-Do et al., 2002). EPHA7 is highly expressed in the kidney vasculature and plays a key role in normal brain development by regulating apoptosis of neural progenitor cells (Hafner et al., 2004; Depaepe et al., 2005). These observations suggest that a possible relationship between RCC and EPHB1 and EPHA7 would merit further investigation. While our experimental studies were in progress, RNF184 (ring finger protein 184) was listed as an unannotated gene (FLJ10546); however, after these studies were completed it was attributed a putative role in ubiquitin ligase function. In view of the role of VHL in an E3 ubiquitin ligase complex, the possible role of RNF184 in renal tumorigenesis may also merit further investigation. Finally, the STAG1 gene product (Stromal antigen-1, also known as SA-1), associates with the cohesin proteins, SMC1, SMC3 (CSPG6), and SCC1 (RAD21) to form one of the two characterized cohesin complexes in mammalian somatic cells. The cohesin complex has a key role in chromatid segregation and mitotic DNA replication (Uhlmann, 2004) and the STAG component of cohesin complexes may be implicated in transcriptional regulation (Lara-Pezzi et al., 2004). Subunits of the cohesin complexes have been linked to cancer susceptibility. Thus SMC1 is an effector of the ataxia telangiectasia mutated/Nijmegen breakage syndrome pathway and forms part of the DNA damage response network (Kim et al., 2002; Yazdi et al., 2002) and RAD21 expression is induced by BRCA1 (Atalay et al., 2002) and mutations in human RAD21 gene have been related with the radiosensitivity in some cancer patients (Severin and Leong, 2001). Thus, we suggest that further studies are indicated to evaluate the role of genes close to the translocation breakpoints (e.g., PPP2R3A, PCCB, STAG1, RNF184, EPHB1, and EPHA7) in the pathogenesis of familial and sporadic RCC and, possibly, other tumor types (Kemp et al., 2006).
Acknowledgments
Supported by: Cancer Research, UK, and the Wellcome Trust (EP, SG, BN, and NPC).
REFERENCES
- Atalay A, Crook T, Ozturk M, Yulug IG. Identification of genes induced by BRCA1 in breast cancer cells. Biochem Biophys Res Commun. 2002;299:839–846. doi: 10.1016/s0006-291x(02)02751-1. [DOI] [PubMed] [Google Scholar]
- Blute ML, Itano NB, Cheville JC, Weaver AL, Lohse CM, Zincke H. The effect of bilaterality, pathological features and surgical outcome in nonhereditary renal cell carcinoma. J Urol. 2003;169:1276–1281. doi: 10.1097/01.ju.0000051883.41237.43. [DOI] [PubMed] [Google Scholar]
- Bodmer D, Eleveld M, Kater-Baats E, Janssen I, Janssen B, Weterman M, Schoenmakers E, Nickerson M, Linehan M, Zbar B, van Kessel GA. Disruption of a novel MFS transporter gene, DIRC2, by a familial renal cell carcinoma-associated t(2;3)(q35;q21) Hum Mol Genet. 2002;11:641–649. doi: 10.1093/hmg/11.6.641. [DOI] [PubMed] [Google Scholar]
- Bodmer D, Eleveld MJ, Ligtenberg MJ, Weterman MA, Janssen BA, Smeets DF, de Wit PE, Van den Berg A, Van den Berg E, Koolen MI, van Kessell GA. An alternative route for multistep tumorigenesis in a novel case of hereditary renal cell cancer and a t(2;3)(q35;q21) chromosome translocation. Am J Hum Genet. 1998;62:1475–1483. doi: 10.1086/301888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer D, Schepens M, Eleveld MJ, Schoenmakers EF, van Kessel GA. Disruption of a novel gene, DIRC3, and expression of DIRC3-HSPBAP1 fusion transcripts in a case of familial renal cell cancer and t(2;3)(q35;q21) Genes Chromosomes Cancer. 2003;38:107–116. doi: 10.1002/gcc.10243. [DOI] [PubMed] [Google Scholar]
- Chen J, Lui WO, Vos MD, Clark GJ, Takahashi M, Schoumans J, Khoo SK, Petillo D, Lavery T, Sugimura J, Astuti D, Zhang C, Kagawa S, Maher ER, Larsson C, Alberts AS, Kanayama HO, Teh BT. The t(1;3) breakpoint-spanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell. 2003;4:405–413. doi: 10.1016/s1535-6108(03)00269-1. [DOI] [PubMed] [Google Scholar]
- Crolla JA, Van Heyningen V. Frequent chromosome aberrations revealed by molecular cytogenetic studies in patients with aniridia. Am J Hum Genet. 2002;71:1138–1149. doi: 10.1086/344396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaepe V, Suarez-Gonzalez N, Dufour A, Passante L, Gorski JA, Jones KR, Ledent C, Vanderhaeghen P. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435:1244–1250. doi: 10.1038/nature03651. [DOI] [PubMed] [Google Scholar]
- Druck T, Podolski J, Byrski T, Wyrwicz L, Zajaczek S, Kata G, Borowka A, Lubinski J, Huebner K. The DIRC1 gene at chromosome 2q33 spans a familial RCC-associated t(2;3)(q33;q21) chromosome translocation. J Hum Genet. 2001;46:583–589. doi: 10.1007/s100380170025. [DOI] [PubMed] [Google Scholar]
- Eleveld MJ, Bodmer D, Merkx G, Siepman A, Sprenger SH, Weterman MA, Ligtenberg MJ, Kamp J, Stapper W, Jeuken JW, Smeets D, Smits A, Van Kessel AG. Molecular analysis of a familial case of renal cell cancer and a t(3;6)(q12;q15) Genes Chromosomes Cancer. 2001;31:23–32. doi: 10.1002/gcc.1114. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Gribble S, Burford D, Carr P, Prigmore E, Porter K, Clegg S, Crolla J, Dennis N, Jacobs P, Carter NP. Array painting: A method for the rapid analysis of aberrant chromosomes using DNA microarrays. J Med Genet. 2003;40:664–670. doi: 10.1136/jmg.40.9.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill RM, Bemis LT, Lee JP, Sozen MA, Baron A, Zeng C, Erickson PF, Hooper JE, Drabkin HA. The TRC8 hereditary kidney cancer gene suppresses growth and functions with VHL in a common pathway. Oncogene. 2002;21:3507–3516. doi: 10.1038/sj.onc.1205437. [DOI] [PubMed] [Google Scholar]
- Gemmill RM, West JD, Boldog F, Tanaka N, Robinson LJ, Smith DI, Li F, Drabkin HA. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc Natl Acad Sci USA. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SM, Prigmore E, Burford DC, Porter KM, Ng BL, Douglas EJ, Fiegler H, Carr P, Kalaitzopoulos D, Clegg S, Sandstrom R, Temple IK, Youings SA, Thomas NS, Dennis NR, Jacobs PA, Crolla JA, Carter NP. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet. 2005;42:8–16. doi: 10.1136/jmg.2004.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner C, Schmitz G, Meyer S, Bataille F, Hau P, Langmann T, Dietmaier W, Landthaler M, Vogt T. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50:490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- Han DC, Shen TL, Miao H, Wang B, Guan JL. EphB1 associates with Grb7 and regulates cell migration. J Biol Chem. 2002;277:45655–45661. doi: 10.1074/jbc.M203165200. [DOI] [PubMed] [Google Scholar]
- Hendrix P, Mayer-Jaekel RE, Cron P, Goris J, Hofsteenge J, Merlevede W, Hemmings BA. Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A: Evidence for different size forms produced by alternative splicing. J Biol Chem. 1993;268:15267–15276. [PubMed] [Google Scholar]
- Huynh-Do U, Vindis C, Liu H, Cerretti DP, McGrew JT, Enriquez M, Chen J, Daniel TO. Ephrin-B1 transduces signals to activate integrin-mediated migration, attachment and angiogenesis. J Cell Sci. 2002;115:3073–3081. doi: 10.1242/jcs.115.15.3073. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J, Van Hoof C. PP2A: The expected tumor suppressor. Curr Opin Genet Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Kanayama H, Lui WO, Takahashi M, Naroda T, Kedra D, Wong FK, Kuroki Y, Nakahori Y, Larsson C, Kagawa S, Teh BT. Association of a novel constitutional translocation t(1q;3q) with familial renal cell carcinoma. J Med Genet. 2001;38:165–170. doi: 10.1136/jmg.38.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp Z, Carvajal-Carmona L, Spain S, Barclay E, Gorman M, Martin L, Jaeger E, Brooks N, Bishop DT, Thomas H, Tomlinson I, Papaemmanuil E, Webb E, Sellick GS, Wood W, Evans G, Lucassen A, Maher ER, Houlston RS. Evidence for a colorectal cancer susceptibility locus on chromosome 3q21-q24 from a high-density SNP genome-wide linkage scan. Hum Mol Genet. 2006;15:2903–2910. doi: 10.1093/hmg/ddl231. [DOI] [PubMed] [Google Scholar]
- Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- Kovacs G, Brusa P, De Riese W. Tissue-specific expression of a constitutional 3;6 translocation: Development of multiple bilateral renal-cell carcinomas. Int J Cancer. 1989;43:422–427. doi: 10.1002/ijc.2910430313. [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E, Pezzi N, Prieto I, Barthelemy I, Carreiro C, Martinez A, Maldonado-Rodriguez A, Lopez-Cabrera M, Barbero JL. Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J Biol Chem. 2004;279:6553–6559. doi: 10.1074/jbc.M307663200. [DOI] [PubMed] [Google Scholar]
- Maher ER. Von Hippel-Lindau disease. Curr Mol Med. 2004;4:833–842. doi: 10.2174/1566524043359827. [DOI] [PubMed] [Google Scholar]
- Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe PJ. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GTW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliff PJ. The tumor suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Melendez B, Rodriguez-Perales S, Martinez-Delgado B, Otero I, Robledo M, Martinez-Ramirez A, Ruiz-Llorente S, Urioste M, Cigudosa J, Benitez J. Molecular study of a new family with hereditary renal cell carcinoma and a translocation t(3;8)(p13;q24.1) Hum Genet. 2003;112:178–185. doi: 10.1007/s00439-002-0848-6. [DOI] [PubMed] [Google Scholar]
- Morris MR, Hesson LB, Wagner KJ, Morgan NV, Astuti D, Lees RD, Cooper WN, Lee J, Gentle D, Macdonald F. Multigene methylation analysis of Wilms' tumour and adult renal cell carcinoma. Oncogene. 2003;22:6794–6801. doi: 10.1038/sj.onc.1206914. [DOI] [PubMed] [Google Scholar]
- Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Palamarchuk A, Huebner K, Croce CM. FHIT as tumor suppressor: Mechanisms and therapeutic opportunities. Cancer Biol Ther. 2002;1:232–236. doi: 10.4161/cbt.73. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Perales S, Melendez B, Gribble SM, Valle L, Carter NP, Santamaria I, Conde L, Urioste M, Benitez J, Cigudosa JC. Cloning of a new familial t(3;8) translocation associated with conventional renal cell carcinoma reveals a 5 kb microdeletion and no gene involved in the rearrangement. Hum Mol Genet. 2004;13:983–990. doi: 10.1093/hmg/ddh111. [DOI] [PubMed] [Google Scholar]
- Schmidt L, Li F, Brown RS, Berg S, Chen F, Wei MH, Tory K, Lerman MI, Zbar B. Mechanism of tumorigenesis of renal carcinomas associated with the constitutional chromosome 3;8 translocation. Cancer J Sci Am. 1995;1:191. [PubMed] [Google Scholar]
- Severin MD, Leong T. Novel DNA sequence variants in the hHR21 DNA repair gene in radiosensitive cancer patients. Int J Radiat Oncol Biol Phys. 2001;50:1323–1331. doi: 10.1016/s0360-3016(01)01608-x. [DOI] [PubMed] [Google Scholar]
- Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- Subramonian K, Weston PM, Curley P. Multifocal renal cancer associated with renal artery aneurysm and a unique genetic change. Br J Urol. 1998;82:761–762. doi: 10.1046/j.1464-410x.1998.00832.x. [DOI] [PubMed] [Google Scholar]
- Surawska H, Ma PC, Salgia R. The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev. 2004;15:419–433. doi: 10.1016/j.cytogfr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Teh BT, Giraud S, Sari NF, Hii SI, Bergerat JP, Larsson C, Limacher JM, Nicol D. Familial non-VHL non-papillary clear-cell renal cancer. Lancet. 1997;349:848–849. doi: 10.1016/S0140-6736(05)61751-5. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. The mechanism of sister chromatid cohesion. Exp Cell Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Valle L, Cascon A, Melchor L, Otero I, Rodriguez-Perales S, Sanchez L, Cruz Cigudosa J, Robledo M, Weber B, Urioste M. About the origin and development of hereditary conventional renal cell carcinoma in a four-generation t(3;8)(p14.1;q24.23) family. Eur J Hum Genet. 2005;13:570–578. doi: 10.1038/sj.ejhg.5201371. [DOI] [PubMed] [Google Scholar]
- Velagaleti GV, Bien-Willner GA, Northup JK, Lockhart LH, Hawkins JC, Jalal SM, Withers M, Lupski JR, Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward ER, Clifford SC, Astuti D, Affara NA, Maher ER. Familial clear cell renal cell carcinoma (FCRC): Clinical features and mutation analysis of the VHL, MET, and CUL2 candidate genes. J Med Genet. 2000;37:348–353. doi: 10.1136/jmg.37.5.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatyka M, da Silva NF, Clifford SC, Morris MR, Wiesener MS, Eckardt KU, Houlston RS, Richards FM, Latif F, Maher ER. Identification of cyclin D1 (CCND1) and other novel targets for the VHL tumour suppressor gene by expression array analysis and investigation of CCND1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 2002;62:3803–3811. [PubMed] [Google Scholar]