Abstract
Deletions within the neurexin 1 gene (NRXN1; 2p16.3) are associated with autism and have also been reported in two families with schizophrenia. We examined NRXN1, and the closely related NRXN2 and NRXN3 genes, for copy number variants (CNVs) in 2977 schizophrenia patients and 33 746 controls from seven European populations (Iceland, Finland, Norway, Germany, The Netherlands, Italy and UK) using microarray data. We found 66 deletions and 5 duplications in NRXN1, including a de novo deletion: 12 deletions and 2 duplications occurred in schizophrenia cases (0.47%) compared to 49 and 3 (0.15%) in controls. There was no common breakpoint and the CNVs varied from 18 to 420 kb. No CNVs were found in NRXN2 or NRXN3. We performed a Cochran–Mantel–Haenszel exact test to estimate association between all CNVs and schizophrenia (P = 0.13; OR = 1.73; 95% CI 0.81–3.50). Because the penetrance of NRXN1 CNVs may vary according to the level of functional impact on the gene, we next restricted the association analysis to CNVs that disrupt exons (0.24% of cases and 0.015% of controls). These were significantly associated with a high odds ratio (P = 0.0027; OR 8.97, 95% CI 1.8–51.9). We conclude that NRXN1 deletions affecting exons confer risk of schizophrenia.
INTRODUCTION
Copy number variants (CNVs) are emerging as an important genomic cause of disease, particularly autism where large de novo CNVs may account for up to 10% of cases (1-3). These CNVs appear to be highly deleterious and may account for the dramatically reduced fecundity in autism; in line with autistic individuals, schizophrenics have fewer children on average than the general population and risk increases with paternal age (4,5). It also appears that the same CNVs can increase susceptibility to more than one psychiatric phenotype (6). Because of this, CNVs are also strong candidates for involvement in the aetiology of schizophrenia.
The role of CNVs in schizophrenia is less well documented, but there are classical examples such as the translocation disrupting the DISC1 gene in a large Scottish pedigree (7) and velo-cardio-facial syndrome (VCFS), caused by a large deletion at 22q11 which is associated with psychosis in ~30% of cases (8). Several recent papers have also indicated that there might be further schizophrenia associated CNVs, especially de novo deletions and duplications (9-14).
Both Kirov et al. (9) and Walsh et al. (11) investigated individuals with schizophrenia in order to identify microdeletions and microduplications >100 kb potentially associated with the disease. Each study found a deletion disrupting the neurexin 1 gene (NRXN1) at 2p16.3 in one family, in a mother and two affected siblings, and identical twins concordant for child-onset schizophrenia, respectively. However, neither was able to determine the prevalence, penetrance or odds ratio of these CNVs because of their rarity and the small population size examined. CNVs in NRXN1 have also previously been implicated in autism and mental retardation (6,15-19). In the present study, we assessed the association of NRXN1 CNVs with schizophrenia as part of a genome-wide scan in a sample of 2977 schizophrenia patients and 33 746 controls from seven European populations (Iceland, Finland, Norway, Germany, The Netherlands, Italy and UK) using microarray data.
RESULTS
For the NRXN1 locus on 2p16.3, we limited our search to include the region between Mb 49.90–51.50 (Human Genome build36), encompassing all exons of NRXN1 and several hundred kilobases of the upstream sequence. For comparison, the deletions reported in schizophrenic patients by Kirov et al. (11) spans Mb 51.04–51.29 (Genome build36, 250 Kb) and the deletion reported by Walsh et al. (13) Mb 50.023–50.137 (Genome build 36, 115 kb). Similarly, we examined the interval for deletions or duplications and tested for their enrichment in schizophrenia (see Materials and Methods section, CNV detection subsection).
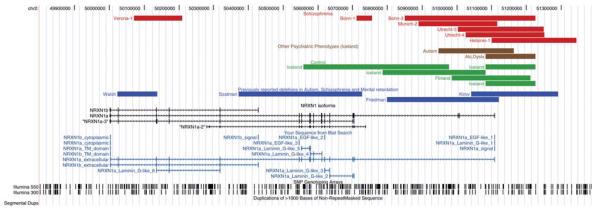
We examined the region in 2977 schizophrenia patients and 33 746 controls from seven European populations (Iceland, Finland, Norway, Germany, The Netherlands, Italy and UK) (Supplementary Material, Table S1). We found 61 NRXN1 deletions, including a de novo deletion, and 5 duplications in the combined sample (Fig. 1). The CNVs were found throughout the locus, and varied, plus a further five deletions in subjects excluded from the controls (a subject with autism, three subject with alcohol dependence and one first degree relative of a schizophrenia subject) in size from 18 to 420 kb, with no common breakpoint, although some single breakpoints may be shared. We found no obvious sequence elements such as low copy repeats (LCRs) in the region that could mediate the rearrangements. In contrast, there were no deletions or duplications found at the NRXN2 (11p13.1) or NRXN3 (14q31.1) loci, which were searched for using an identical strategy.
Figure 1.
UCSC browser output showing the positions of the 2p16.3 CNVs from the Kirov et al. (2008), Walsh et al. (2008), Friedman et al. (2008) and Szatzmar et al. (2008) studies (blue lines) relative to the NRXN1 gene, and the CNVs discovered in the present study (schizophrenia, red lines; other psychiatric diagnoses, brown lines; controls, green lines). The majority of the discovered CNVs are deletions, asterisks indicate duplications. Markers from the Illumina 300 K and 550 K arrays, segmental duplications of >1000 bp as well as LD structure of the Hapmap CEU sample (r2) is also shown.
We found 12 deletions and 2 duplications of NRXN1 in cases (0.47%) compared to 49 and 3 (0.15%) in the controls, all of which encompassed part of the NRNX1 gene or its promoter (Fig. 1). The most common deletion was found in 27 samples including one homozygote. One of these was schizophrenic, and two had a diagnosis of alcoholism; the remainders were controls (Supplementary Material, Table S2). Haplotype background suggests that all carriers for the common deletion derive from a single founder. A further six deletions and one duplication were seen in two, three and four samples, respectively, while 24 deletions and one duplication were only observed once.
We performed Cochran–Mantel–Haenszel exact test to estimate association between all deletions and duplications and schizophrenia. This analysis gave suggestive results, P = 0.13 (OR = 1.73; 95% CI 0.81–3.50) for all NRXN1 CNVs in schizophrenia versus controls. Because it is likely that the pathogenicity and penetrance of CNVs at this locus vary, depending on the functional effect they have on the gene, we next restricted the association analysis to those CNVs which disrupt exons (Fig. 2). We found seven in cases (0.24%) and five in controls (0.015%); these were more strongly associated with schizophrenia and had a higher odds ratio (P = 0.0027; OR 8.97; 95% CI 1.8–51.9).
Figure 2.
UCSC browser output showing the positions of exon-disrupting CNVs discovered in the study relative to the 2p16.3 CNVs from the Kirov et al. (2008), Walsh et al. (2008), Friedman et al. (2008) and Szatzmar et al. (2008) studies and known (schizophrenia, red lines; other psychiatric diagnoses, brown lines; controls, green lines; previously described CNVs, blue lines). The four putative Neurexin isoforms are shown below the deletions, along with protein domains aligned to genomic sequence.
Because almost half of the cases (but not controls) with CNVs were from Germany (6/14), we analysed haplotypes in the 5′ region of the NRXN1 gene region to see if we could detect evidence of a common ancestor for these six CNVs; however, there was no evidence for haplotype sharing in the region, unlike the haplotype sharing observed for the common small deletion, supporting the notion that the majority of CNVs at this locus are not derived from a founder.
There was no sex difference among carrier subjects; CNVs were found in eight female and six male schizophrenia subjects, 31 female and 21 male controls. CNV type, geographic origin, SNP boundaries, position (Human genome build 36) size, phenotype and sex are shown in Supplementary Material, Table S2. In addition to the deletions in patients with schizophrenia and control subjects with no known mental illness, there were five further deletions found in the Icelandic population, in a subject with autism, three subjects with alcohol dependence and one first degree relative of a schizophrenia subject who did not participate in this study. These subjects are shown in Figure 1, but were not included in the statistical analysis.
The clinical characteristics of the deletion subjects are shown in Table 1; there seems to be nothing remarkable about these cases. We also tested allelic association for all SNP markers in the 2p16.3 region studied here but no marker remained significantly associated at the 5% level after correction for number of performed tests (data not shown).
Table 1.
Phenotypic features of schizophrenia cases with 2p16.3 deletions
| Centre | Diagnosis | Family history of schizophrenia | CNV boundaries | Age at onset |
Sex | Other |
|---|---|---|---|---|---|---|
| Munich 1 | DSMIV:295.3 | No | chr2:50,856,110-50,900,862 | 27 | F | |
| Munich 2a | DSMIV:295.3 | No | chr2:50,890,216-51,116,653 | 25 | M | Social contact problems in childhood |
| Munich 3 | DSMIV:295.3 | No | chr2:51,147,600-51,225,851 | 31 | F | |
| Veronaa,b | ICD10: F20.3 | Brother with mental illness (not specified) | chr2:50,071,499-50,208,992 | 36 | F | Chronic, positive symptoms, low IQ (82), low educational level |
| Verona 2 | ICD10: F20.3 | Yes, other psychiatric disorder | chr2:50,822,312-50,948,557 | 14 | F | Chronic, negative symptoms, episode of aggression |
| Helsinki 1a | DSMIV:295.3 | Yes, other psychiatric disorder | chr2:51,101,161-51,344,213 | 37 | M | Alcoholism |
| Utrecht 1 | DSMIV:295.3 | Probably psychosis in first or second degree relative | chr2:50,735,657-50,800,548 | 29 | F | Not mentioned |
| Utrecht 2b | DSMIV:295.9 | Not examined | chr2:50,786,446-50,900,862 | 19 | M | Not mentioned |
| Utrecht 3a | DSMIV:295.3 | chr2:51,002,576-51,250,922 | M | |||
| Utrecht 4a | DSMIV:295.9 | None | chr2:51,0249,62-51,251,873 | Unknown | F | Episodic with partial remission and negative symptoms |
| Reykjavik 1 | RDC:126.4 | None | chr2:51,211,406-51,299,436 | 26 | M | Not mentioned |
| Bonn 1a | DSMIV:295.3 | None | chr2:50,711,199-50,756,435 | 18 | M | Myoclonic seizures in the right shoulder |
| Bonn 2 | DSMIV:295.3 | None | chr2:50,836,690-50,936,258 | 21 | F | Resistancy in left lower abdomen with otherwise unremarkable finding |
| Bonn 3a | DSMIV:295.3 | Yes | chr2:50,850,456-51,225,851 | 27 | F | Mental retardation (mild), Tardive dyskinesia |
Exon disrupting.
Duplication.
To address the issue of the role of splicing in NRXN1 deletions, we reviewed all available genomic data across the NRXN1 locus for evidence of alternative NRXN1 isoforms and identified EST and cDNA transcript evidence to support two additional isoforms, which share many exons with NRXN1a, which we term NRXN1a-2 and NRXN1 a-3 (Fig. 3). The NRXN1a-2 isoform shares exons 6–14 with NRXN1a, but employs alternative first and last exons. These alternative exons allow for distinct expression analysis of the NRXN1a-2 isoform, using probe 1558708_at on the Affymetrix HG-U133_Plus_2.0 GeneChip (Supplementary Material, Fig. S1c). This shows very low expression of NRXN1a-2, predominantly in the brain, compared to much higher brain-specific expression of NRXN1a and NRXN1b (Supplementary Material, Figs S1a and S1b). The NRXN1a-2 isoform has no signal peptide and no transmembrane domain; however, it does contain Laminin G domains 2–4 and EGF domains 2 and 3 (Fig. 3). Evidence for the NRXN1a-3 isoform is much less conclusive as it shares all exons with exons 4–18 of NRXN1a.
Figure 3.
Protein domain organization of Neurexin 1 isoforms. NRXN1a contains an N-terminal signal peptide, six laminin G (LamG) domains, three epidermal growth factor-like (EGF) domains, a transmembrane domain and a short cytoplasmic domain. NRXN1b contains a different signal peptide, but shares the last LamG domain, transmembrane domain and cytoplasmic domain with NRXN1a. NRXN1a-2 shares the 2nd, 3rd, 4th and 5th LamG domains and the 2nd and 3rd EGF domains with NRXN1a, but does not contain a signal peptide or transmembrane domain. NRXN1a-3 has no signal peptide and has a truncated version of the 2nd LamG domain but shares all remaining domains with NRXN1a. The five regions in the NRXN1 gene where alternative splicing occurs, leading to insertion or deletion of amino acids is indicated by arrows (SS no. 1–5). Protein domain annotation was generated using SMART (http://smart.embl-heidelberg.de/), using swissprot accessions Q9ULB1 (NRXN1a) and P58400 (NRXN1b) and translations from the genbank transcript AK093260 (NRXN1a-2) and Ensembl transcript ENST00000331040 (NRXN1a-3).
DISCUSSION
In the present study, we used 2977 schizophrenia patients and 33 746 controls, from seven European populations, to examine CNVs at the neurexin 1 locus on 2p16.3. Deletions at this locus have previously been associated with autism and mental retardation (6,15-19), and deletions were also found in two families with schizophrenia. In addition, we examined the other two known neurexin genes, NRXN2 (11p13.1) and NRXN3 (14q31.1) to see if they also harbour potentially pathogenic deletions. Kirov et al. (9) screened 93 cases, and found a deletion at 2p16.3 in one affected proband, as well as the unaffected mother and an affected sibling. Walsh et al. (11) found a different deletion disrupting NRXN1 in identical twins concordant for childhood-onset schizophrenia. However, because of the small sample size employed, it is not possible to determine extent of association and the prevalence and odds ratio for disease risk from their data. In our study population, we found an excess of CNVs in the NRXN1 gene in schizophrenia, 0.47% of cases compared to 0.15% of controls, which did not reach statistical significance. Conditioning on exon disrupting deletions or duplications (i.e. those more likely to be pathogenic), the association became significant (P = 0.0027; OR 8.97, 95% CI 1.8-51.9) indicating that CNVs in the region may have different pathogenicity or penetrance. There were no CNVs detected in the NRXN2 or NRXN3 loci, indicating that deletion or duplication of either of these genes is not a significant factor in disease.
De novo versus segregating deletions
One of the 2p16.3 deletions reported here (autistic subject, Supplementary Material, Table S2), was also found de novo in the Icelandic population (12), and the remaining rare deletions probably occur de novo because they do not show any evidence of a founder effect and the boundaries differ between individuals. However, we cannot be sure of this as in the present study, the majority of cases with deletions do not have parents and other relatives available for analysis. In contrast, the smallest common deletion appears to be a segregating polymorphism with a single founder. In the Kirov et al. (9) study, the NRXN1 deletion they found was scored as a segregating polymorphism as it was present in a mother and son; in the Walsh et al. (11) study, it was found in a pair of MZ twins, with the parents not tested, so it is unclear if it arose by a de novo event. These deletions may still be predominantly de novo as in the cases found by Kirov et al. (9) the deletion in the mother could have occurred de novo in a transmission from a grandparent and subsequently been transmitted to her offspring; strong negative selection pressure (the reduced fecundity in male schizophrenics) would then mean that this deletion is quickly extinguished from the population. Reduced fecundity is well established in schizophrenia, and earlier onset (such as appears to occur in many cases with large CNVs) is also associated with lower fecundity. Although this is seen in both males and females, it is more pronounced in males (20-22). As Bassett et al. (20) suggest, these epidemiological observations are consistent with ‘a high mutation rate, consistent with unstable mutations, to explain the high prevalence of schizophrenia’.
Heterogeneity of deletions and potential impact on the complex splicing of neurexin
The heterogeneity of the deletions we have identified at the NRXN1 locus also presents difficulties when it comes to assessing association with schizophrenia. Caution is needed in interpreting these results as the association we found was not significant when all CNVs were examined collectively, and required conditioning on exon-disrupting deletions. It is likely that pathogenicity of the deletions and duplications at this locus will vary, depending on the functional importance of the DNA deleted. Conditioning on exons is the simplest method of addressing this, as protein structure and splicing will be affected, and under this test the association between exonic deletions and schizophrenia in our sample is highly significant.
The complexity of neurexin regulation at the level of alternative splicing has already been suggested, with six splice sites with perhaps 1000 isoforms (23) and the potential to theoretically generate over 2000 isoforms encoding variant extracellular domains (24). Although the exact impact of deletions of the first exons of NRXN1a on alternative splicing is unclear, it is possible that the expression of alternative NRXN1 isoforms may compensate for the loss of the NRXN1a transcript. It is interesting that the first exon of NRXN1a-2 seems to be deleted in the small founder deletion observed in 24 subjects most of whom were controls. Considering the very low levels of expression of NRXN1a-2, and its frequent deletion in controls, we therefore suggest that NRXN1a-2 may have a very limited biological role in Neurexin signalling. However, activity of the NRXN1a-2 isoform may become significant against a background of deletion of the other Neurexin isoforms. In such circumstances, even low-level dominant negative activity may become biologically relevant. As most known Neurexin interactions are extracellular it is unclear if NRXN1a-2 has any function, although if it is secreted it may exert a dominant negative effect by competitive binding with NRXN1a interactors; alternatively improper cellular localization of NRXN1a-2 may also be deleterious.
Evidence for the NRXN1a-3 isoform is much less conclusive than other isoforms as it shares all exons with exons 4–18 of NRXN1a. This excludes the possibility of differentiation of NRXN1a and NRXN1a-3 by the expression analysis. There is a high level of mammalian conservation before the first exon of NRXN1a-3 and there is also EST evidence to support the existence of this isoform although, these ESTs cannot be clearly differentiated from NRXN1a transcripts. The putative initiation methionine of the NRXN1a-3 isoform correlates with NRXN1a Met321 roughly a third of the way into Laminin G domain 2 of the protein (see Fig. 3).
The truncated domain of NRXN1a-3 may not be functional as it has lost a critical Ca2+ binding residue Asp306, which also mediates interactions with Neurexophilins (25,26). Loss of Neurexophilin interactions may be particularly relevant to schizophrenia pathology, as Neurexophilin-3 knockout mice show no anatomical defects, but remarkable functional abnormalities in sensory information processing and motor coordination, evident by increased startle response, reduced prepulse inhibition and poor Rotarod performance (27). It is tempting to speculate that the disruption of NRXN1/Neurexophilin interactions that are likely to be seen in all our observed exonic deletions may explain some of the pathology of schizophrenia. More detailed bioinformatic and functional biology may be able to shed further light on the genotype–phenotype and functional consequences of various deletions at this locus.
Haploinsufficieny as a mechanism of pathogenicity
In addition to the disruption of splicing and the effects of this on the interaction of neurexin proteins with protein partners, the association between NRXN1 deletions and schizophrenia could also result from haploinsufficiency of the gene. Given that we see a strong association when conditioning on disruption of an exon in the two known RefSeq sequences for NRXN1, and the most common and smallest deletion of intron-only sequence [although the first exon of the novel splice variant NRXN1a-2 (Fig. 3) appears to be deleted] shows a founder effect and is thus not likely to be under strong negative selection pressure, it is not likely that all the deletions we found at the NRXN1 locus have the same biological effect on disrupting the function of NRXN1, and some will probably be neutral variants. Intron-only deletions could of course disrupt splicing or remove essential regulatory elements such as splicing enhancers and/or silencers, and the neurexin 1 gene is extensively spliced, with more than 2000 potential isoforms (23). In order to provide further information on the effects of these deletions, it will be necessary to analyse mRNA and protein expression, for example, using relative allelic expression of non-deleted exons in NRXN1 mRNA to demonstrate which deletions result in haploinsufficiency.
Founder effects and the distribution of neurexin 1 deletions in Europe
There is also a question of how to treat the sample with regard to geographical origin, as most of the deletions in cases were from the German population (6/14; 43%) although, of course, as 25% of the examined patients were German in origin, this over-representation could have occurred by chance. If these deletions are truly recurrent, and depend on a random mechanism involving DNA sequences or recombination rates in the region, one would not expect significant geographic clustering. Clustering may indicate deletions which are segregating in the population, perhaps from an ancestral founder, or a pre-mutation or polymorphic DNA sequence which increases the probability of a de novo event and is more prevalent in one population than others. A haplotype analysis that we performed on the German samples with deletions showed no excess sharing of alleles around the NRXN1 5′-region, which, when combined with the observed heterogeneity of the deletions, strongly opposes a founder effect. The most common deletion, which occurred in 29 subjects, was also the smallest and showed evidence of a single founder. This may be a neutral segregating polymorphism which does not significantly affect NRXN1 function and is not relevant to risk of schizophrenia.
Penetrance and frequency of neurexin deletions
Rare NRXN1 deletions have also been reported in the normal population. Pinto et al. (28) found an intronic 71 kb deletion in a sample of 506 unrelated healthy individuals from Northern Germany and 270 HapMap individuals (chr2:50,826,443-50, 897,556) which overlaps with some of the intronic deletions seen in the present study, and both Redon et al. (29) (chr2:50,643,118-50,686,339 Build 36) and Wong et al. (30) found similar sized deletions (chr2:50,651,965-50,714,553) in single control subjects, one of which removes four exons of the gene. These subjects have no known neuropsychiatric phenotype, consistent with the notion that although deletions of NRXN1 are strongly associated with mental illness, they are not necessarily fully penetrant, as seen in the data from the present study, and some individuals may be able to compensate for dosage loss, for example, through a high expressing residual copy of the gene, another modifying locus, or an environmental event which results in a gene-by-environment interaction pathway to disease.
The 2p16.3 deletions we found are moderately rare, occurring at a rate of about 4.7 per 1000 cases, and, from the control population in the present study, at ~0.15% of live births. This is substantially higher than the VCFS deletion, found in ~0.01% of live births (31), making it one of the most common gene deletions known. However, if we exclude the deletions in NRXN1 which do not disrupt exons and are less likely to be pathogenic, the deletion rate is 2.4 per 1000 (0.24%) in the cases compared to 0.015% in the controls, a rate very comparable to VCFS deletions.
The structural mutations showing association to schizophrenia disrupt the NRXN1 gene, rather than deleting or duplicating the entire gene. A generalized study of structural mutations by Lupski and Stankiewicz (32) showed that such mutations are much more likely to have biological consequences, in our case deletion of exons from the most prevalent NRXN1a isoform may favour the expression of the rarer NRXN1a-2 and NRXN1a-3 isoforms, which may possibly exert a dominant negative effect on Neurexin signalling, particularly through Neurexophilins.
The odds ratio (~10-fold increase in risk) is less than that associated with the VCFS deletion, where ~25% of subjects develop psychosis and the odds ratio for the VCFS deletion is about 30, assuming a population rate of non-VCFS deletion schizophrenia of ~1% (33). It is possible that there are many more CNVs in the human genome which can cause schizophrenia, and these could make up a significant proportion of genetic risk for the disease which could prove clinically useful, for example, through genetic testing.
Potential deletion mechanisms at the NRXN1 locus
The nature of the loci studied here raises questions about the mechanisms underlying CNV events. Many chromosomal rearrangements are not random events, but the result of genome architecture that may create inherent instability (34), such as the long LCRs typical of many other regions where CNVs have been reported. These result in non-allelic homologous recombination (NAHR) between the LCRs on opposite flanks of the deleted/duplicated segment. However, several genomic disorders are associated with rearrangements whose breakpoints do not cluster within LCRs, as in the case of NRXN1 deletions. There we see no evidence of LCRs flanking the breakpoints, nor do we find common breakpoints or evidence of sequence duplications in the region. The LD pattern is particularly unstructured in the 5′-end of the gene and the upstream region where we observe the deletions. This may suggest increased recombination rate in the region that may be related to the aggregation of deletions at the site, through an unknown mechanism. There are suggestions that CNV events are also associated with the presence of processed pseudogenes, the presence of microsatellites and nonhomologous end joining mediated by micro-homologies (<25 bp homology), especially in subtelomeric regions, or DNA replication-based mechanisms, rather than a NAHR mechanism mediated by larger matching repeats (32,35,36). Similar to NRXN1, CNVs in the PLP1 gene do not show common breakpoints. Lee et al. (36) examined this locus and uncovered evidence for sequence complexity at some junctions, consistent with a DNA replication-based mechanism of CNV formation (replication Fork Stalling and Template Switching, FoSTeS). This mechanism might explain the non-recurrent apparently de novo deletions with multiple breakpoints seen in NRXN1. To examine this further, we performed long range phasing analysis (37) of the single case of autism known to have a de novo NRXN1 deletion in the Icelandic population. This showed that no recombinations had occurred in the region around the de novo deletion (data not shown), providing further evidence against a role for an NAHR mechanism in this case.
Schizophrenia and autism: aetiological overlap?
The data from our sample of cases and controls provide support for a role for deletions within the NRXN1 gene in schizophrenia, and raise the concept that this disorder has shared genetic risk factors with autism. If this is indeed the case, it would change the way the distinctions between the diseases is formulated, and therefore has profound clinical implications. In support of this are previous observations such as the VCFS deletions at 22q11.2, which appear to increase the risk of both autism (38) and schizophrenia (33,39), and data from the present and other studies indicating that neurexin deletions are seen in both disorders as well as other behavioural syndromes (9,11,15-19). On the other hand, the concept of diagnostic overlap between the two disorders is controversial: while Rzhetsky et al. (40) provide some theoretical support for aetiological overlap, evidence from clinical and epidemiological study is less supportive (41,42). In general, there have been few genetic and epidemio-logical studies of potential overlap between the disorders, and the present and other studies point the way towards more comprehensive evaluation of potential diagnostic overlap between these and indeed other psychiatric disorders in the future.
MATERIALS AND METHODS
Samples
A total of 2977 affected and 33 746 controls from six European populations were successfully examined for CNVs at the NRXN1, NRXN2 and NRXN3 loci studied here (Supplementary Material, Table S1); 1435 schizophrenia patients and 28 554 control individuals from the Iceland, Scotland, Germany, England, Italy and Finland (The SGENE sample; http://www.SGENE.eu), 491 affected and 881 controls from Bonn, Germany, 806 cases and 4039 controls from the Netherlands and 245 cases and 272 controls from Norway. For a full description of samples, see Supplementary Information. Ethical approval was obtained from the local Ethics Committees. All participants gave written informed consent.
The SGENE samples were typed on the HumanHap300 BeadArray™ (Illumina, San Diego, CA, USA) at deCODE genetics. The samples from Bonn were typed at Bonn University on HumanHap550v3 BeadArray™ (Illumina, San Diego, CA, USA). The Dutch samples from Utrecht University were genotyped at the University of California, Los Angeles, on HumanHap550v3 BeadArray™. The remaining Dutch samples were genotyped at deCODE genetics on HumanHap300 BeadArray™. The Norwegian samples were genotyped on Affymetrix GeneChip GenomeWide SNP 6.0 array and analysed using the Affymetrix Power Tools 1.8.0. Samples with Contrast QC below 0.4 were excluded as recommended by the manufacturer.
CNV detection
DosageMiner software, developed at deCODE genetics, and QuantiSNP (43; see below for description) were used to identify deletions and duplications around the regions reported by Kirov et al. (9) and Walsh et al. (11) in all samples except the Norwegian sample, where only QuantiSNP was used. DosageMiner, described in detail elsewhere (12), uses the intensities from SNP probes on the Illumina microarrays to estimate copy number of genomic regions, and models factors such as SNP effect, sample effect and GC-content the in neighbouring region to normalize the intensities. The software then automatically registers SNP loci where intensities fall above or below an empirical threshold. We conditioned our search to include only CNVs with 10 or more contiguous markers registered as duplicated or deleted by DosageMiner, in order to reduce the level of false positive CNVs identified by the software. We then inspected the intensities by eye for all samples with a CNV identified by the software. We used SNP genotypes for further confirmation in case of deletions by conditioning on homozygosity for all markers within the boundaries of the suspected CNV. In some instances, the actual CNV may be shorter than 10 contiguous markers, because the boundaries called by DosageMiner may not always be entirely accurate.
The QuantiSNP software, developed at the Wellcome Trust Centre for Human Genetics and at the University of Oxford (http://www.well.ox.ac.uk/QuantiSNP/), relies on an Objective Bayes Hidden–Markov model to estimate copy number variations (43). In this model, the hidden states denote the unknown copy number at the inspected SNPs. Genotype data were used to compute different states. Based on the ratio of fluorescent dye ratios (logR) and stretches of homozygosity, the algorithm computes a Bayes factor that is used to calibrate the model to a fixed type I (false-positive) error rate. A Bayes factor threshold of 10 is considered as a promising value for the possible presence of a CNV. Usually, such values occur when 5–10 consecutive SNPs are deleted/duplicated. Differences in GC base pairs may result in biased hybridization behaviour of SNP probes bearing the risk of miscalling genotypes. To normalize for this, QuantiSNP assigns a locus-specific GC value to each probe. Again, all potential CNVs detected were subsequently visually inspected and confirmed using the Bead-Studio software (Illumina, San Diego, CA, USA). We used the Cochran–Mantel–Haenszel exact test for the association analysis, which includes a correction for population of origin in the analysis. In order to condition on exons, we scored a CNV as exon-disruption if it overlapped any known exonic sequence in NRXN1, based on reviewed sequence from Human mRNA from RefSeq (NM_004801 for NRXN1-alpha). Long-range phasing analysis to detect recombination events in the Icelandic case of autism with a de novo NRXN1 deletion was performed using the method of Kong et al. (37).
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank the subjects and their relatives and staff at the recruitment centres.
FUNDING
This work was sponsored by EU grant LSHM-CT-2006-037761 (Project SGENE).
NOTE ADDED IN PROOF
Some of the data published in this paper (submitted May 16 2008; accepted October 22 2008) has also appeared in a parallel publication taking a discovery approach rather than a candidate gene approach (Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E; Genetic Risk and Outcome in Psychosis (GROUP) Consortium, Sabatti C, Geurts van Kessel A, Brunner HG, Ophoff RA, Veltman JA. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008 Oct;83(4):504–510; submitted July 2nd 2008; accepted October 20 2008). Specifically, the 54+752 cases tested Vrijenhoek et al. are the same 806 cases from The Netherlands reported in the present paper, and the Utrecht controls (n=706) of our Dutch controls in the present study are also the same. For the purposes of clarification and to assist meta analysis, we have calculated the results without the Dutch sample as follows. For exon-disrupting cnvs without the Dutch sample, we found the following (Country, aff-carriers/aff-non-carriers, ctrl-carriers/ctrl-non-carriers) Germany 3/684, 0/1078; Iceland 0/648, 4/27747; Italy 1/85, 0/91; Finland 1/62, 1/149. The P-value = 0.04, and the alternative hypothesis: true common odds ratio is not equal to 1, 95 percent confidence interval: 0.853866-38.921971. Common odds ratio: 5.4. This should be borne in mind for combined or meta analysis of data.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao XA, Leotta V, Kustanovich C, Lajonchere DH, Geschwind K, Law P, Law S, Qiu C, Lord J, Sebat J, et al. A unified genetic theory for sporadic and inherited autism. Proc. Natl Acad. Sci. USA. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 4.Reichenberg A, Gross R, Weiser M, Bresnahan M, Silverman J, Harlap S, Rabinowitz J, Shulman C, Malaspina D, Lubin G, et al. Advancing paternal age and autism. Arch. Gen. Psychiatry. 2006;63:1026–1032. doi: 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- 5.Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D, Susser ES. Advancing paternal age and the risk of schizophrenia. Arch. Gen. Psychiatry. 2001;58:361–367. doi: 10.1001/archpsyc.58.4.361. [DOI] [PubMed] [Google Scholar]
- 6.Bish JP, Bearden CE, Ding L, Ferrante S, Nguyen V, Gee JC, McDonald-McGinn DM, Zackai EH, Emanuel BS. A multilevel analysis of cognitive dysfunction and psychopathology associated with chromosome 22q11.2 deletion syndrome in children. Dev. Psychopathol. 2005;17:753–784. doi: 10.1017/S0954579405050364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 8.Murphy KC. Schizophrenia and velo-cardio-facial syndrome. Lancet. 2002;359:426–430. doi: 10.1016/S0140-6736(02)07604-3. [DOI] [PubMed] [Google Scholar]
- 9.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 10.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 11.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 12.Stefansson H, Rujescu D, Cichon S, Pietiläinen OPH, Ingason A, Fossdal R, Steinberg S, Sigurdsson E, Sigmundsson T, Hansen T, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuguchi T, Hashimoto R, Itokawa M, Sano A, Shimokawa O, Yoshimura Y, Harada N, Miyake N, Nishimura A, Saitsu H, et al. Microarray comparative genomic hybridization analysis of 59 patients with schizophrenia. J. Hum. Genet. 2008;53:914–919. doi: 10.1007/s10038-008-0327-6. [DOI] [PubMed] [Google Scholar]
- 15.Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH, Jr, Skinner C, Schwartz CE, Sommer SS. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci. Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T, Marra MM, Friedman JM. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1{alpha} J. Med. Genet. 2007;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 19.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassett AS, Chow EW, Weksberg R, Brzustowicz L. Schizophrenia and genetics: new insights. Curr. Psychiatry Rep. 2002;4:307–314. doi: 10.1007/s11920-996-0051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haukka J, Suvisaari J, Lönnqvist J. Fertility of patients with schizophrenia, their siblings, and the general population: a cohort study from 1950 to 1959 in Finland. Am. J. Psychiatry. 2003;160:460–463. doi: 10.1176/appi.ajp.160.3.460. [DOI] [PubMed] [Google Scholar]
- 22.Terzian AC, Andreoli SB, Razzouk D, Chaves AC, Mari Jde J. Fertility and fecundity of an outpatient sample with schizophrenia. Rev. Bras. Psiquiatr. 2006;28:305–307. [PubMed] [Google Scholar]
- 23.Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 24.Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, Graveley BR. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- 25.Sheckler LR, Henry L, Sugita S, Südhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+ binding and the effects of alternative splicing. J. Biol. Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missler M, Hammer RE, Südhof TC. Neurexophilin binding to alpha-neurexins. A single LNS domain functions as an independently folding ligand-binding unit. J. Biol. Chem. 1998;273:34716–34723. doi: 10.1074/jbc.273.52.34716. [DOI] [PubMed] [Google Scholar]
- 27.Beglopoulos V, Montag-Sallaz M, Rohlmann A, Piechotta K, Ahmad M, Montag D, Missler M. Neurexophilin 3 is highly localized in cortical and cerebellar regions and is functionally important for sensorimotor gating and motor coordination. Mol. Cell. Biol. 2005;25:7278–7288. doi: 10.1128/MCB.25.16.7278-7288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum. Mol. Genet. 2007;16(Spec no. 2):R168–R173. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- 29.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tezenas Du Montcel S, Mendizabai H, Ayme S, Levy A, Philip N. Prevalence of 22q11 microdeletion. J. Med. Genet. 1996;33:719. doi: 10.1136/jmg.33.8.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 34.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. 2004;13(Spec no. 1):R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 35.Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 37.Kong A, Masson G, Frigge ML, Gylfason A, Zusmanovich P, Thorleifsson G, Olason PI, Ingason A, Steinberg S, Rafnar T, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antshel KM, Aneja A, Strunge L, Peebles J, Fremont WP, Stallone K, Abdulsabur N, Higgins AM, Shprintzen RJ, Kates WR. Autistic spectrum disorders in velo-cardio facial syndrome (22q11.2 deletion) J. Autism Dev. Disord. 2007;37:1776–1786. doi: 10.1007/s10803-006-0308-6. [DOI] [PubMed] [Google Scholar]
- 39.Chow EW, Bassett AS, Weksberg R. Velo-cardio-facial syndrome and psychotic disorders: implications for psychiatric genetics. Am. J. Med. Genet. 1994;54:107–112. doi: 10.1002/ajmg.1320540205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc. Natl Acad. Sci. USA. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkmar FR, Cohen DJ. Comorbid association of autism and schizophrenia. Am. J. Psychiatry. 1991;148:1705–1707. doi: 10.1176/ajp.148.12.1705. [DOI] [PubMed] [Google Scholar]
- 42.Esterberg ML, Trotman HD, Brasfield JL, Compton MT, Walker EF. Childhood and current autistic features in adolescents with schizotypal personality disorder. Schizophr. Res. 2008;104:265–273. doi: 10.1016/j.schres.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.