Abstract
Small interfering RNAs (siRNAs) and genome-encoded microRNAs (miRNAs) silence genes via complementary interactions with mRNAs. With thousands of miRNA genes identified and genome sequences of diverse eukaryotes available for comparison, the opportunity emerges for insights into origin and evolution of RNA interference (RNAi). The miRNA repertoires of plants and animals appear to have evolved independently. However, conservation of the key proteins involved in RNAi suggests that the last common ancestor of modern eukaryotes possessed siRNA-based mechanisms. Prokaryotes have a RNAi-like defense system that is functionally analogous but not homologous to eukaryotic RNAi. The protein machinery of eukaryotic RNAi seems to have been pieced together from ancestral proteins of archaeal, bacterial and phage origins that are involved in DNA repair and RNA-processing pathways.
The miRNA and siRNA machinery
Recent transcriptome analyses have shown that most of the eukaryotic genome is transcribed [1, 2], and the genomes of all cellular life forms, in addition to protein-coding genes, contain varying numbers of non-protein-coding RNA [3, 4]. MicroRNAs (miRNAs) are an abundant class of small (21-22 nucleotides) non-protein-coding RNAs that regulate translation in eukaryotes [5]. MiRNAs are key components of a major, evolutionarily conserved system of gene regulation in plants and animals that typically post-transcriptionally down-regulates gene expression either by inducing degradation of the target mRNAs, or by blocking their translation [6, 7]. MiRNA-mediated pathways belong to a vast network of regulatory systems known as RNA interference (RNAi) [4, 8]. RNAi consists of three major branches. These branches are small interfering (si)RNA-mediated pathways, miRNA-based pathways that are involved, respectively, in defense against viruses and transposable elements and in regulation of eukaryotic gene expression, and piwi-interacting RNA (piRNA) pathway that appears to be mechanistically distinct from the other two pathways [9].
There are two key differences between miRNA and siRNA-mediated systems:
miRNAs are endogenous non-protein-coding RNA molecules that are encoded by their own, distinct genes; by contrast, there are no dedicated genes for siRNAs. Instead, siRNAs are either generated by degradation of exogenous (e.g., viral) dsRNAs or transcribed from transposable elements integrated in the genome, or from other types of inverted repeats;
siRNAs are fully complementary to their targets, whereas miRNAs, at least in animals, show limited complementarity to their recognition sites.
Although the structures and mechanisms of animal and plant miRNAs differ substantially, the same or homologous key proteins are involved both in miRNA biogenesis and in the siRNA pathways, suggesting that animal and plant miRNAs derive from the same, ancestral proto-RNAi system, but subsequently evolved along widely different trajectories so that the extant repertoires of miRNAs are unrelated. Prokaryotes have no RNAi systems homologous to the eukaryotic ones but seem to possess an independently evolved, analogous defense mechanism. However, apparent ancestors of the key protein components of eukaryotic RNAi can be identified among prokaryotic proteins involved in other processes.
Here we discuss the origin and evolution of RNAi systems and miRNAs, with an emphasis on their deep eukaryotic roots and prokaryotic connections. We review evidence on conservation of the key proteins involved in RNAi suggesting that the last common ancestor of modern eukaryotes (LECA) already possessed siRNA-based mechanisms, and implying that miRNA-like regulation in plants and metazoans evolved before the divergence of these two distinct types of multicellular life.
Origin and evolution of eukaryotic RNA interference systems
The staggering complexity of RNAi phenomena notwithstanding, there are only three key proteins involved in RNAi, namely Ago-Piwi, Dicer-like protein that typically consists of RNAseIII and helicase domains, and RNA-dependent RNA polymerase (RdRP). The diversity of RNAi-related complexes and pathways is created by multiple paralogous forms of these proteins that emerged via numerous duplications at different stages of eukaryotic evolution, along with several accessory proteins (Table 1, Fig. 1 and [8]). These three key components are present in at least some members of 4 of the 5 supergroups of eukaryotes. Notably, excavates either lack dicer homologs or possess dicer-like proteins that lack either the helicase component or the tandem RNase III portion (Table 1 and [10]). The existence of RNAi in most of the excavates remains an open question [11], but considering the “star phylogeny” of the 5 supergroups that is the current best approximation of the early eukaryotic evolution [12], it appears most likely that a functional RNAi system containing, at least, these three proteins antedates the last common ancestor of the extant eukaryotes (Last Eukaryotic Common Ancestor, or LECA [13]). Moreover, members of the ancient paralogous pair Ago-Piwi show a scattered distribution in 4 eukaryotic supergroups (Table 1, [8, 14]) suggesting that, multiple losses during subsequent evolution notwithstanding, this duplication antedates the radiation of the supergroups. Thus, LECA might have had the capacity for small-RNA-mediated gene silencing both at the level of translation (which involves Ago) and at the level of transcription (which involves Piwi).
Table 1.
Distribution of RNAi machinery components in the 5 supergroups of eukaryotesa
| Species | Argonaute-PIWI-like | Dicer-like | RdRP | miRNAs (Rfam v10) | |
|---|---|---|---|---|---|
| Argonaute | PIWI | ||||
| Excavata | |||||
| Giardia intestinalis | 0 | 1 | (1) no helicase domain; might not be dicer ortholog | 1 | - |
| Trypanosoma brucei | 1 | 1 | (1)dicer-like helicase only | - | - |
| Trypanosoma cruzi | - | - | - | - | - |
| Leishmania major | - | - | - | - | - |
| Leishmania braziliensis | - | 1 | - | - | - |
| Chromalveolata | |||||
| Paramecium tetraurelia | - | 4 | 2 (Dcr1p without helicase domain) | 2 | - |
| Tetrahymena thermophila | - | 4 | 2 (Dcr1p without helicase domain) | 1 | - |
| Plasmodium falciparum | - | - | - | - | - |
| Phytophthora infestans | 1 | - | ? | ? | - |
| Thalassiosira pseudonana | - | - | - | - | - |
| Archaeplastida | |||||
| Cyanidioschyzon merolae | - | - | - | - | |
| Chlamydomonas reinhardtii | 2 | - | 3 | - | + |
| Arabidopsis thaliana | 10 | - | 4 | 6 | + |
| Oryza sativa (japonica) | 18 | - | 5 | 5 | + |
| Unikonta | |||||
| Dictyostelium discoideum | - | 4 | 2 (no helicase domain) | 3 (2 fused to Dicer-like helicase) | |
| Entamoeba histolytica | - | 3 | - | 1 | - |
| Saccharomyces cerevisiae | - | - | - | - | - |
| Schizosaccharomyces pombe | 1 | - | 1 | 1 | - |
| Neurospora crassa | 1 | - | 1 | 3 | - |
| Aspergillus nidulans | 1 | - | 1 | 2 | - |
| Caenorhabditis elegans b | 5 | 3 | 2 (Dicer + Drosha) | 4 | + |
| Drosophila melanogaster | 2 | 3 | 3 (2 Dicers + Drosha) | - | + |
| Strongylocentrotus purpuratus | 1 | 1 | 2 (Dicer + Drosha) | - | - |
| Danio rerio | 4 | 4 | 2 (Dicer + Drosha) | - | + |
| Homo sapiens | 4 | 4 | 2 (Dicer + Drosha) | - | + |
The number of detectable paralogs in each genome is indicated; the data are from references [8] and [14](Argonaute-PIWI) and additional BLASTP searches (for species that encode a single version of the respective protein or no more than 4 paralogs, all sequences were used as queries to search the NCBI non-redundant protein sequence database; for species with more than four paralogs, 2-3 representative sequences were used).
In addition, the nematode genome encodes a large family of highly derived argonaute proteins, the Argonaute group 3 [58].
Figure 1.
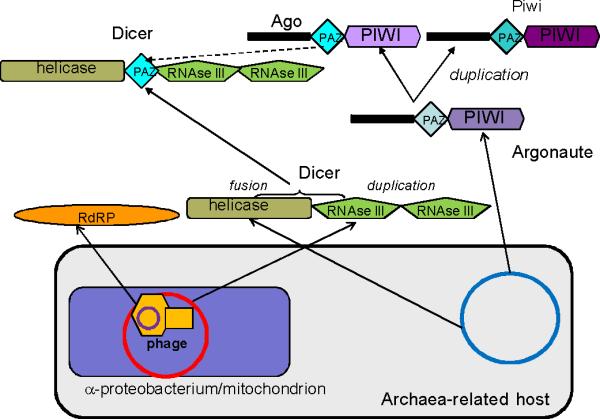
A scenario for the origin and earliest stages of evolution of the eukaryotic RNAi protein machinery. The scenario is based on the symbiotic model of the eukaryotic cell origin [73]. Domains are depicted by unique shapes; proteins are not shown to scale. Arrows denote inferred derivation of the coding regions for proteins or (in the case of Dicer) individual domains from homologous sequences residing in the genomes of the postulated archaea-related host, the protomitochondrial (α-proteobacterial) endosymbiont and a bacteriophage. Considering the spread of protein involved in RNAi among the eukaryotic supergroups (Table 2), the assembly of Dicer from domains of distinct origins and the duplication of Argonaute that produced Ago and Piwi is mapped to the “stem” phase of eukaryotic evolution (between the symbiosis event that is assumed here to have triggered eukaryogenesis and the radiation of the supergroups that is shown as a multifurcation in the upper part of the figure as per [12]). The broken arrow shows the likely derivation of the PAZ domain of Dicer subsequent to the argonaute duplication after [8]. Evolution of the eukaryotic supergroups involved multiple duplications and losses of the key components of RNAi that are not elaborated on the figure (see Table 1 and [8, 13, 14])
Considering the major difference in the miRNA structures in plants and animals (Box 1), it seems likely that the ancestral RNAi system functioned, primarily, in defense against viruses (via cytoplasmic, Ago-centered pathways) and transposons, (via nuclear, Piwi-based pathways) [15-18]. The notion that the ancestral RNAi primarily had a defense function is further supported by the inference that LECA possessed RdRP, because in extant eukaryotes the RdRP is primarily involved in siRNA amplification but not in miRNA pathways [19, 20]. The defense role of RNAi is widely conserved among eukaryotes but, in addition, distinct miRNA pathways appear to have evolved in animals and plants, and perhaps in fungi as well. An intriguing question remains as to whether or not LECA also possessed proto-miRNAs, and if it did, what the structure and mechanism of these was.
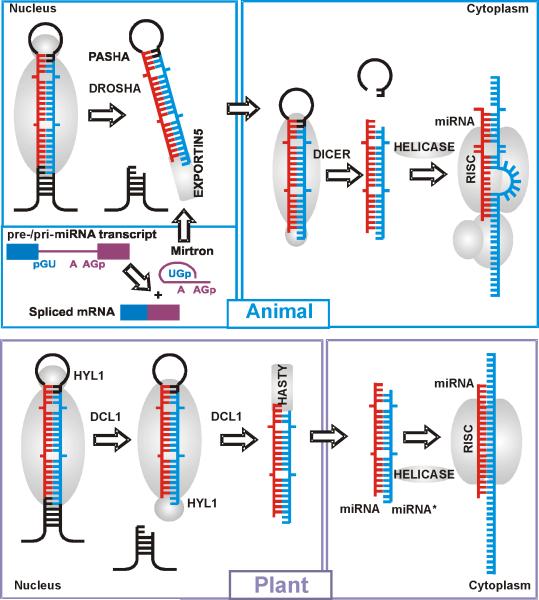
Box 1. Plant and animal miRNAs.
In animals, miRNA genes mostly reside within introns of annotated genes and often form clusters that can be transcribed as a single primary transcript [60]. The majority of plant miRNAs are produced from individual transcription units that are located in intergenic regions [61]. In both animals and plants, the mechanism through which miRNAs are generated is dictated by the highly conserved secondary structures of their transcripts that form stable imperfectly paired hairpins. These hairpins are processed by RNAi machinery into miRNA precursor duplexes [6, 62]. The miRNA duplex has essentially the same structure as a double-stranded siRNA [63-66] except that the mature miRNA strand is only partially paired to the complementary miRNA* strand.
Processing of miRNA precursors is mediated by proteins of the Dicer family that contain a helicase domain and two tandem RNAse III domains [67]. miRNA processing pathways vary in plants and animals in multiple aspects (see Table I and Figure I). Plants lack an ortholog of Drosha that is predominantly localized in the nucleus in animals (see Figure I). Instead, it is thought that the function of Drosha is carried out by one or more of the specialized Dicer paralogs found in plants.
However, Drosha-mediated cleavage is not the only way to produce canonical pre-miRNA hairpins in animals. An alternative pathway utilizes splicing of suitable RNAs transcribed from introns (mirtrons) that mimic the structural features of pre-miRNAs and enter the miRNA-processing pathway without Drosha-mediated cleavage (Figure Ia). This pathway has been identified in flies, worms [68, 69], and mammals [70, 71]. The evolutionary conservation of mammalian mirtrons suggests that mirtrons were incorporated into endogenous regulatory pathways at a very early stage. Thus, there seems to be a direct link between the evolution of introns and the evolution of miRNAs. Given that the positions of numerous introns have been conserved throughout the evolution of eukaryotes [72] and massive intron invasion is thought to be one of the pivotal events of eukaryote genesis [73], it seems plausible that mirtrons were already operational in LECA.
The miRNA/miRNA* duplex is further processed in the cytosol by the RNA-induced silencing complex (RISC). The RISC directs gene silencing in both miRNA and siRNA pathways in plants and animals, although the subunit compositions of the miRNA-specific and siRNA-specific RISCs differ (Fig I). The common features of miRNAs and siRNAs in plants suggest that complex RNA-silencing systems evolved before multicellularity and were a feature of primitive eukaryotic cells [74].
Plant miRNAs are almost perfectly complementary to their mRNA targets. In contrast, the complementarity between animal miRNAs and their targets is usually much lower and restricted to the 5′ core miRNA region and, in some cases, an additional region in the 3′ part of the miRNA that compensates for imperfect core pairing [75]. The degree of complementarity between miRNAs and their targets determines, at least in part, the regulatory mechanism (cleavage versus translation repression).
Several lineages of unicellular eukaryotes from different supergroups, namely Saccharomyces cerevisiae (Unikonta), Trypanosoma cruzi and Leishmania major (Excavata), Cyanidioschyzon merolae (Archaeplastida), and Plasmodium falciparum (Chromalveolata), appear to have lost the RNAi machinery independently and completely. This supports the notion that, however prominent as a defense and regulatory mechanism, RNAi is non-essential in eukaryotes, specifically, in unicellular forms [8]. In many multicellular eukaryotic lineages, RNAi seems to have become essential (as evidenced by the embryonic lethality of Dicer mutants) due to the involvement of miRNAs in developmental gene regulation [21].
Origin of the key proteins Ago-Piwi, Dicer, and RdRP
There seems to be no prokaryotic ancestor of the eukaryotic RNAi as a functional system. However, prokaryotic homologs of the three key proteins of eukaryotic RNAi have been identified. These proteins show a peculiar gamut of archaeal and bacterial connections (Table 2). The two enzymatic domains of Dicer, namely, RNAse III and Superfamily II RNA helicase, are common in prokaryotes but their combination in a single protein is a eukaryotic signature that is shared by all eukaryotic supergroups. The helicase domain of Dicer is specifically related to the archaeal family of Superfamily II helicases (Hef proteins) [22]. Archaeal Hef proteins have been shown to play an important role in DNA replication [23, 24]. By contrast, RNAse III domain, the nuclease domain of Dicer, is mostly of bacterial provenance. In bacteria, RNAse III is involved in essential reactions of rRNA processing as well as mRNA degradation [25]. Conceivably, fusion of the helicase and RNAse III domains from different sources was one of the pivotal, early events that led to the consolidation of the eukaryotic RNAi system.
Table 2.
The prokaryotic connections of the key components of the eukaryotic RNAi machinery
| Protein | Taxonomic range of homologs in prokaryotes | Closest archaeal homolog (E-value, % identity) | Closest bacterial homolog (E-value, % identity) | Functions of prokaryotic homologs (reference) |
|---|---|---|---|---|
| Dicer-helicasea | All archaea; no bacteria (only distantly related helicases with statistically insignificant similarity) | ERCC4-like helicase (Hef), uncultured crenarchaeote31-F-01 (5e-19; 24%) | No significant similarity | Resolution of stalled replication forks[23] |
| Dicer-RNaseIIIa | Several mesophilic archaea; all bacteria | RNaseIII, Methanococcus maripaludis S2 (9e-14; 25%) | RNase III, Mannheimia haemolytica PHL213 (1e-14; 27%) | rRNA and mRNA processing [25] |
| Argonauteb | Scattered distribution among archaea and bacteria (mostly, in Cyanobacteria) | No significant similarity | No significant similarity | No direct evidence; prokaryotic PIWI-domain proteins are DNA-guided RNA endonucleases, so thought to participate in chromatin remodeling [27, 59] |
| PIWIc | Homolog of the eukaryotic argonaute protein, implicated in translation or RNA processing, Methanopyrus kandleri AV19, (0.14, 24%); limited sequence conservation only in PIWI domain | No significant similarity | ||
| RdRPd | Bacteriophages and prophages from diverse bacteria; uncharacterized cyanobacterial proteins (possibly, prophage-derived) | No significant similarity | hypothetical DNA-directed RNA polymerase, Bacillus phage 0305phi8-36 (1e-14, 14%' detected in 2nd PSI-BLAST iteration) | Putative DNA-dependent RNA polymerases |
Human Dicer protein sequence (Q9UPY3.2) was employed as the query for searching the NCBI non-redundant protein sequence database using BLASTP
Human Argonaute protein sequence (NP_036331) was used as the query
Human PIWI-like protein sequence (Q96J94) was used as the query
The C. elegans RdRP sequence (CAA91312) was used as the query
The second key protein, Ago, also displays apparent archaeal roots, since archaeal Ago homologs seem to be more closely related to the eukaryotic Ago than the bacterial homologs are [26]. The biological functions of the archaeal and bacterial Ago homologs remain unclear, although there is some evidence to suggest that they are involved in chromatin remodeling [27, 28]. Ultimately, Ago proteins probably originated from basal DNA replication machinery, as suggested by the structure of the RNAse (Slicer) domain which possesses a derived RNAse H fold. The presence of multiple Argonaute-Piwi paralogs in metazoa indicates that animals possess multiple RNA-induced silencing complexes (RISCs) that carry out related but distinct biological functions [5]. Ago proteins are ubiquitously expressed and bind to siRNAs or miRNA. In contrast, expression of Piwi proteins is mostly restricted to the germ line, and these proteins associate with Piwi-interacting RNAs (piRNAs) to facilitate silencing of mobile genetic elements and to provide adaptive defense in the transposon arms race [29].
The third key component of the RNAi machinery, the RdRP, appears to share a common origin with the second largest, catalytic subunit of DNA-dependent RNA polymerases [30, 31]. The direct ancestors of the eukaryotic RdRPs are likely to be uncharacterized bacteriophage proteins that, possibly, function as DNA-dependent RNA polymerases ([30, 31]; Table 2).
With the exception of some highly derived versions of Ago, all three key proteins of RNAi show high levels of sequence conservation among eukaryotes. By contrast, the relationships of these proteins with their prokaryotic homologs are quite different. The two enzymatic domains of Dicer show highly significant sequence similarity to their prokaryotic orthologs that are ubiquitous and, by implication, essential in archaea (helicase) or bacteria (RNAse III). By contrast, the prokaryotic homologs of Ago and, especially, RdRP show scattered distributions in archaea and bacteria, and limited, not easily detectable sequence similarity to the eukaryotic RNAi components (Table 2). Conceivably, during the emergence of eukaryotic RNAi, the latter two proteins have undergone major mechanistic changes, perhaps linked to the transition from DNA-dependent to RNA-dependent activities, which led to substantial acceleration of evolution.
In conclusion, the evolution of the eukaryotic RNAi machinery seems to have followed the same general principle that governs evolution of other novel eukaryotic functional systems, such as the nuclear pore or the spliceosome [13, 32, 33]. In each of these cases, the eukaryotic system was pieced together from diverse building blocks of prokaryotic origin, with a substantial change in the specific function of each component. The RNAi protein apparatus is particularly notable in that it seems to have been assembled from three distinct prokaryotic sources, archaeal (the helicase domain of Dicer and Ago), bacterial (the RNAse domain of Dicer), and viral (RdRP). Like in other eukaryotic systems, the earliest stages of RNAi evolution involved domain fusion and shuffling that led to the emergence of Dicer via the fusion of the helicase and RNAse III domain, and acquisition of the RNA-binding PAZ domain from Piwi. Subsequently, each of these signature eukaryotic systems evolved via a series of duplications, some of which antedate the divergence of the major eukaryotic lineages and some of which are lineage-specific [8, 13, 32, 33] (Table 2).
In addition, the fact that the eukaryotic RNAi machinery seems to have been assembled from prokaryotic proteins involved in DNA repair and RNA processing pathways unrelated to RNAi points towards an independent origin of RNAi in eukaryotes. Indeed, while prokaryotes possess RNAi-type systems which are functionally similar to eukaryotes, these systems are constructed from very different, non-homologous proteins (Box 2). It thus appears that, although both prokaryotes and eukaryotes possess multiple RNAi-type pathways, the systems evolved independently.
Box 2. RNAi silencing systems of prokaryotes.
RNAi-like mechanisms do exist in prokaryotes and seem to show functional analogies both to the miRNA and the siRNA pathways of eukaryotes, even though the proteins involved in these processes are non-homologous. A growing repertoire of small, non-coding regulatory RNAs (~60 in E. coli) have been discovered in bacteria and archaea [76], although the repertoire of such segments is relatively small compared to complex eukaryotes. Similarly to the eukaryotic miRNAs, these regulatory RNAs show partial complementarity to the target mRNAs and either trigger their degradation or inhibit translation. The action of bacterial small RNAs is mediated by the RNA chaperone Hfq which is homologous to the core domain of an eukaryotic spliceosome protein rather than to any protein involved in RNAi [71].
In addition to the miRNA-like, Hfq-dependent pathways, a siRNA-type antiphage system has been recently discovered in numerous bacteria and archaea, first by computational prediction [77] and then experimentally [78, 79]. The detailed mechanism of this system, named CAS (CRISPR-Associated System), remains to be elucidated, but it seems clear that CAS functions via the siRNA mechanism although with an important difference from the eukaryotic RNAi. The functioning of this system involves integration of fragments of foreign genes into CRISPR repeats in prokaryotic genomes, providing heritable immunity to the respective agents. These regions are apparently transcribed yielding siRNA-like molecules that fold into the miRNA-like stable hairpins [77, 78]. The piRNA system in animals, with small RNAs originating from clustered master loci [80] also bears resemblance to the prokaryotic CRISPR system. The CRISPR loci are associated with a large number of genes that encode proteins with predicted activities involved in the siRNA-like pathway including a distinct polymerase and helicase as well as several predicted nucleases and RNA-binding proteins [77]. The activities of some of these proteins are thought to be analogous to those of Dicer, Slicer, and RdRP, but there are no direct homologies (except for the generic relationship between the respective helicases).
Evolution of miRNAs
Although thousands of miRNA genes have been discovered, their origin and evolution remain largely obscure. Conserved miRNA families have been identified in many metazoans, indicating that their functions could persist throughout the evolution of animals. Thirty miRNA gene families are present throughout bilaterians [34, 35]. However, only three of these miRNA families have been identified in the sequenced genome of and early-branching animal, the sea anemone Nematostella vectensis [34]. The dramatic expansion of the miRNA repertoire in bilaterians suggests that increased miRNA-mediated gene regulation accompanied and, probably, substantially contributed to the emergence of complex, organ-containing animal body plans [36]. New miRNAs seem to contribute to functional innovation at later stages of evolution as well [37, 38]. An example is a recently described X-linked miRNA cluster with multiple copies in primates but not in rodents or dog [39].
Similar evidence for early evolutionary origin exists for plant miRNAs. A member of the miR166-miR165 family is highly conserved in species ranging from angiosperms to liverworts and hornworts. Thus, the miR166 miRNA likely dates back to the last common ancestor of the land plants [40].
The two oldest conserved miRNA families, miR166-miR165 in plants and let-7 in animals, are both involved in crucial symmetry-establishing and differentiation events during plant and animal development. Indeed, such evolutionary conservation is expected for loci involved in essential biological functions. Generally, it should be expected that the more fundamental a process regulated by a miRNA is, the greater is the likelihood that a common ancestor existed and can be reconstructed. However, in contrast to the conservation of several miRNA families within plants and animals, no compelling cases of cross-kingdom conservation of miRNA are known (with a single potential exception [41]).
The major differences in the biogenesis and mechanisms of action between animal and plant miRNAs (Box 1, Table I) has led to the hypothesis that the two systems originated and evolved independently [42]. However, the mechanism of miRNA generation and action is dictated by their secondary structures in animals and plants, so selection for specific structures could have played an important role in miRNA evolution (Box 1 Figure I, Box 2). The advent of a RNA silencing system mediated by miRNAs required co-evolution of three complementary or homologous sequences, i.e., the two miRNA hairpin strands and the target sequence in plants and animals.
Box 1, Table I.
A comparison of the miRNA systems of plants and animals.
| miRNA features | Plants | Animals |
|---|---|---|
| Secondary structure | Stem-loop | Stem-loop |
| Thermodynamic stability | Stable | Stable |
| dsRNA duplex | Strand asymmetry | Strand asymmetry |
| Number of miRNA genes | 100 - 200 | 100-500 |
| Location within genome | Intergenic regions (mostly), introns | Intergenic regions, introns |
| Clusters of miRNAs | Not common | Common |
| Biogenesis | Dicer-like | Drosha (or splicing), Dicer |
| Repression mechanism | mRNA cleavage | Translation repression |
| Degree of complementarity to target | High | Partial, relatively low |
| Location of miRNA targets | Open reading frames (mostly) | 3' untranslated regions (mostly) |
| Number of miRNA target sites | One (generally) | Multiple (generally) |
| Function of target genes | Enzymes, regulators of development | Enzymes, regulators of development, structural proteins) |
| miRNA methylation | 3'-end methylated | Not methylated |
Box 1, Fig I.
MiRNA biogenesis pathways in a) animals and b) plants. The canonical and mirtronic biogenesis pathways are shown for animal miRNAs (upper left panel). Mature miRNAs are shown in red, the antisense strands are shown in blue.
a) In animals, sequential cleavages of miRNA primary transcripts by two RNase-III enzymes, Drosha and Dicer, lead to the formation of small, imperfect dsRNA duplexes (miRNA:miRNA*) that contain both the mature miRNA strand and its complementary strand (miRNA*). Drosha is predominantly localized in the nucleus and cleaves the flanks of pri-miRNA to liberate an ~70-100-nucleotide stem-loop, the precursor miRNA (pre-miRNA) [81]. An alternative pathway for miRNA biogenesis occurs when intronic miRNA precursors (mirtrons) enter the miRNA-processing pathway without a Drosha-mediated step [68, 69]. The pre-miRNA is exported from the nucleus to the cytosol by Exportin5/RanGTP that specifically recognizes the characteristic structural features of pre-miRNAs [63]. Dicer is a cytosolic RNase III that functions in a complex with a dsRBD protein partner. Dicer makes two cuts that define the other end of the mature miRNA, producing a ~19-21nt RNA duplex [82, 83]. The miRNA: miRNA* duplex is further processed in the cytosol by the RNA-induced silencing complex (RISC). Only one strand, the miRNA, from the duplex enters the RISC, the other miRNA*strand is degraded. The complementarity between animal miRNAs and their mRNA targets is usually restricted to the 5' region and binding of the RISC blocks translation of the target mRNA into protein [63].
b) The maturation of miRNAs in plants differs from miRNA maturation in animals. Plants lack an ortholog of Drosha, and it is thought that the function of Drosha is carried out by one or more of the specialized Dicer paralogs found in plants. In particular, DICER-LIKE1 (DCL1) is localized in the plant nucleus and is required for both processing steps that are performed in animals by Drosha and Dicer [84], suggesting that DCL1 has the Drosha-like activity [85].
As in animals, the miRNA: miRNA* duplex is further processed in the cytosol by RISC, although the subunit composition of the complex differs between plants and animals. When the one strand RNA guide in the RISC pairs to a target mRNA, the RISC functions as an endonuclease and cleaves the mRNA between the target nucleotides paired to bases 10 and 11 of the miRNA [63].
There is no definitive answer to the critical unresolved question: “Did miRNA regulation in plants and metazoans evolve before or after the divergence of these two fundamentally different types of multicellular life?” Below, we discuss several hypotheses and potential mechanisms that have been proposed recently for the evolution of miRNA genes. These are likely to help our understanding of the origin and evolution of eukaryotic miRNA systems.
Evolution of miRNAs by duplication and mutation in animals
The evolution of at least some miRNA genes could be explained by a duplication-mutation scenario whereby miRNAs with new binding specificities evolve by duplication of an existing miRNA followed by mutations in the target recognition region. If tandem duplication was the dominant mode of miRNA-family evolution, clustering of paralogous miRNA genes in the genome would be expected [41]. Indeed, approximately 37% of the human miRNA genes [28] and ~ 50% of Drosophila miRNA genes are clustered, i.e., form arrays of miRNA genes without intervening genes [6]. Some of the clusters contain multiple miRNA genes that can be transcribed as a single primary transcript [43]. Furthermore, the clusters are likely to be maintained during evolution by selection for co-regulation of miRNA expression [6, 28]. Thus, clustering of miRNA genes in animal genomes is certainly greater than previously thought and expected by chance, and could reflect the evolutionary mechanisms responsible for spreading of miRNA genes in genomes under the duplication-mutation model.
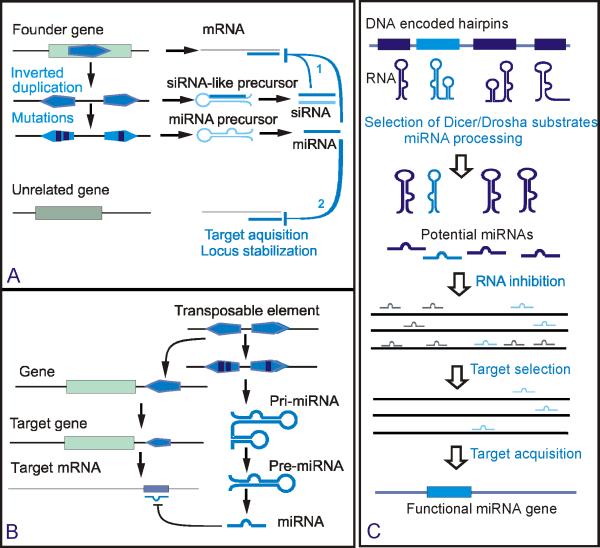
Inverted duplication model of the origin of plant miRNA genes
MiRNA gene families in plants are much larger than in animals. However, plant miRNAs are usually scattered over the genome and only a few members are occasionally clustered within a range of several kilobases. A possible explanation of miRNA genes scattering is that plant genomes have undergone extensive shuffling since the amplification of ancient miRNA families [44].
For several miRNA genes in Arabidopsis, not only the mature miRNAs but also adjacent regions of pre-miRNAs are complementary to their mRNA target. This could be expected if the miRNA genes have originated from inverted duplication of their target loci [45, 46]. If this is the case, the genes might be expected to be evolutionarily young. Indeed, sequence analyses of miRNA precursors revealed evidence of recent origins of 16 miRNA loci by inverted duplication of protein-coding sequences [47]. Although the inverted duplication model (Fig. 2a) is an attractive explanation for the evolution of perfectly pairing miRNAs in plants, it seems to apply only to nonconserved plant miRNA genes, because no such similarity has been found between conserved miRNA genes and their targets [44]. Furthermore, this model appears irrelevant for animal miRNAs where complementary sequences in miRNA-binding sites are much shorter than in plants.
Figure 2.
Genesis and evolution of miRNA loci. a) Inverted duplication model of the origin of plant miRNAs [45] and potential pathways of stabilization of new miRNA loci in plants [4]. A new miRNA is selected with the capacity to regulate the progenitor or a family member gene (1), or unrelated gene (2). b). Origin of miRNAs from genomic repeats or transposable elements in animals [48-52]. c) Random selection model of miRNA origin [53]. Potential miRNA genes are selected from hairpins encoded in the genome. Random targeting of transcripts by potential miRNAs could be deleterious with only a few targets being selectively neutral or advantageous. Acquisition and expression of a potential novel miRNA gene can occur only when, by chance, it is not strongly deleterious [41].
Origin of miRNAs from transposable elements and repeats
Many animal miRNAs appear to be derived from repeats and transposable elements (TEs). For example, approximately 20% of human miRNA genes share sequences with transposable elements [48-50]. In particular, some of the human miRNAs exhibit typical short seed complementarity with a specific site within Alu repeats which are highly conserved within 3′ untranslated regions of human mRNAs [48]. Similarly, it has been proposed that a subset of mammalian miRNA genes originated from MIR/LINE-2 TEs [49]. A recently discovered family of human miRNA genes, hsa-mir-548, was derived from miniature, transposable inverted repeats with a high potential to form extremely stable miRNA-like hairpin structures [51]. Another case in point is the primate- specific hsa-mir-548 family that is represented by numerous paralogs in the human genome. Their palindromic organization suggests origin from actively transcribed TEs that produce transcripts with characteristic miRNA-like hairpins [51]. Thus, insertion of TEs into new genomic sites could be one of the driving forces of miRNA evolution in animals [52, 53] (Fig. 2b). Recently, many plant TE insertions that encode both siRNAs and miRNAs have been discovered, suggesting that this route of evolution could be important in plants as well [54].
Random selection model
According to this model, spuriously produced potential miRNAs form a pool from which they are randomly drawn to silence potential mRNA targets ([53], Fig 2 c). It has been argued that for many miRNAs no mRNA targets have been identified. Therefore, the number of functional miRNAs could be much smaller than the number of potential miRNAs that could be randomly produced by Drosha and Dicer via cleavage of suitable miRNA-like hairpin precursors. This random selection model does not explain how “successful” random miRNAs that are efficient silencers are fixed in the lineage. However, a recent genome-wide bioinformatic screen showed that the human genome encodes millions of potential hairpins, mostly located in the transcribed part of the genome [55]. Thus, a modified random selection model could be envisaged whereby target-specific miRNAs emerge from the pool of hairpin transcripts via random mutation, after which the miRNA-target pair is locked by purifying selection. Indeed, a scenario of this kind is implicit in the transcriptional control model of miRNA evolution, which is further described below.
Transcriptional control model for the emergence of new miRNAs
When a new animal miRNA emerges, it is likely to non-specifically target many mRNAs by chance, because the minimal binding site is short. The transcriptional control model [41] postulates that, to avoid an adverse effect on the fitness of the organism, the initial transcription level of a novel miRNA should be low. With time, provided that the regulation elicited by the new miRNA is physiologically important, purifying selection would purge deleterious off-target sites from the genome, while maintaining physiologically beneficial sites and/or fixing newly emerging ones. After that, expression of the miRNA can increase without deleterious consequences. This scenario is in good agreement with the observation that short 3′ untranslated regions of a large set of genes involved in basic cellular processes are specifically depleted of miRNA-binding sites, which allows them to avoid miRNA regulation [56]. This model incorporates aspects of the random selection model by postulating that numerous, randomly transcribed hairpins serve as a pool of novel miRNA genes that have the potential to eventually acquire regulatory function. This is compatible with the available human miRNA expression data that reveal a strong, positive correlation between the estimated evolutionary age of a miRNA and its expression level [41, 57]. This model seems less relevant in plants than in animals due to the higher specificity of the interaction between plant miRNAs and their targets.
Conclusions and perspective
Comparative genomic analysis shows that the protein machinery of RNAi is conserved in all major eukaryotic lineages, independent loss in many unicellular forms notwithstanding. Thus, it appears most likely that LECA possessed relatively complex RNAi machinery. At a minimum, this primordial RNAi machinery consisted of an Argonaute-like protein, a Piwi-like protein, a Dicer, and an RNA-dependent RNA polymerase. Comparison of these proteins with their prokaryotic homologs suggests that, during eukaryogenesis, the protein machinery of RNAi was pieced together from archaeal, bacterial and phage proteins that performed diverse functions in DNA repair and RNA processing, unrelated to RNA interference.
Although aspects of biogenesis and mechanisms of miRNA-mediated gene silencing are substantially different in plants and animals, the plant and animal miRNAs share a common processing pathway and certain structural features. Thus, although it remains a distinct possibility that LECA possessed only siRNA-mediated pathways involved in defense against parasites, the alternative, namely, that a proto-miRNA system also existed at that early stage of evolution, cannot be ruled out. Considering the constrained structure of miRNAs, it seems likely that the primordial proto-miRNAs evolved from inverted repeats, perhaps derived from transposable elements, and were processed via a siRNA-processing mechanism.
Crucial questions on origins and evolution of the miRNA system remain open, in particular, what were the structure and mechanism of biogenesis of ancestral miRNAs, if any such existed, and what is the predominant path(s) of new miRNA generation during the further course of eukaryotic evolution. A comprehensive search for miRNAs in all walks of eukaryotic life and comparative-genomic analysis of miRNA genes are indispensable to address these questions. For such searches to be productive, improved experimental and computational methods for miRNA identification and comparison are required.
Acknowledgements
The number of publications on miRNA is vast, and space limitations preclude the citation of many relevant ones. We apologize to all colleagues whose work is not cited, solely, for this reason. We thank Nikolay Spiridonov for helpful discussions and critical reading of the manuscript and Yuri Merezhuk for help with the preparation of the figures. The authors' research was supported by the Intramural Research Program of the NIH, NLM.
Glossary
- microRNAs (miRNAs)
an abundant class of small (21-22 nucleotides) non-protein-coding RNAs that regulate translation in eukaryotes
- siRNAs
small interfering RNAs that are involved in defense response to alien nucleic acids (e.g., primary anti-viral defense) and are fully complementary to their targets
- piRNAs
PIWI-interacting RNAs, 24-30 nucleotide long RNAs found in germ cells of animals. piRNAs are unique among small silencing RNAs in that they require neither an RdRP nor Dicer for their production
- RNAi
RNA interference, a mechanism that inhibits gene expression at the stage of translation or transcription of specific genes. RNAi consists of two major branches, namely, small interfering (si)RNA-mediated gene silencing and miRNA-based pathways that are involved, respectively, in defense against viruses and transposable elements, and in regulation of eukaryotic gene expression
- Drosha
nuclear endonuclease that cuts stem-loop precursor miRNAs from primary miRNA transcripts in animals
- Dicer
ribonuclease that produces ~22 nucleotide long mature miRNAs containing 5′ phosphate and 3′ hydroxy termini
- Slicer
ribonuclease of the Ago family that cleaves the target mRNA in siRNA pathways
- Mirtrons
pre-miRNAs that are located in introns of different genes and processed first by the premRNA splicing machinery in the nucleus, rather than Drosha, and then by Dicer in the cytoplasm
- Seed complementarity
complementarity of the target to the nucleotides 2-7 of the miRNA
- Bilaterians
members of the animal kingdom exhibiting bilateral symmetry (two similar sides with definite upper and lower surfaces and anterior and posterior ends)
References
- 1.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai EC. microRNAs: runts of the genome assert themselves. Curr Biol. 2003;13:R925–936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Jones-Rhoades MW, et al. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 8.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matranga C, Zamore PD. Small silencing RNAs. Curr Biol. 2007;17:R789–793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 11.Shi H, et al. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. Rna. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Makarova KS, et al. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005;33:4626–4638. doi: 10.1093/nar/gki775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 15.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 16.Li HW, Ding SW. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 18.Waterhouse PM, et al. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 19.Sijen T, et al. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 20.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, et al. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komori K, et al. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J Biol Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- 24.Nishino T, et al. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 25.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Aravind L, et al. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma JB, et al. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altuvia Y, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aravin AA, et al. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 30.Iyer LM, et al. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salgado PS, et al. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:e434. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mans BJ, et al. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 33.Collins L, Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol. 2005;22:1053–1066. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- 34.Prochnik SE, et al. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- 35.Hertel J, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sempere LF, et al. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, et al. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol. 2008;25:929–938. doi: 10.1093/molbev/msn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, et al. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, et al. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 41.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 42.Millar AA, Waterhouse PM. Plant and animal microRNAs: similarities and differences. Funct Integr Genomics. 2005;5:129–135. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- 43.Guddeti S, et al. Molecular evolution of the rice miR395 gene family. Cell Res. 2005;15:631–638. doi: 10.1038/sj.cr.7290333. [DOI] [PubMed] [Google Scholar]
- 44.Li A, Mao L. Evolution of plant microRNA gene families. Cell Res. 2007;17:212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- 45.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 46.Rajagopalan R, et al. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fahlgren N, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Aravin AA, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 51.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piriyapongsa J, et al. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svoboda P, Di Cara A. Hairpin RNA: a secondary structure of primary importance. Cell Mol Life Sci. 2006;63:901–908. doi: 10.1007/s00018-005-5558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. Rna. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 56.Stark A, et al. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Berezikov E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 58.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 59.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez A, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 62.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 63.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 64.Shabalina SA, et al. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2006;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silva JM, et al. Free energy lights the path toward more effective RNAi. Nat Genet. 2003;35:303–305. doi: 10.1038/ng1203-303. [DOI] [PubMed] [Google Scholar]
- 66.Matveeva O, et al. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 68.Ruby JG, et al. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okamura K, et al. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berezikov E, et al. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 72.Carmel L, et al. Patterns of intron gain and conservation in eukaryotic genes. BMC Evol Biol. 2007;7:192. doi: 10.1186/1471-2148-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 74.Molnar A, et al. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 75.Brennecke J, et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Majdalani N, et al. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 77.Makarova KS, et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 79.Sorek R, et al. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 80.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 82.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 84.Tang G, et al. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]