Abstract
The respective pro- and antiapoptotic functions of the transcription factors p53 and nuclear factor κB (NF-κB), and their potential impact on tumorigenesis and response to tumor therapy are well recognized. The capacity of the RelA(p65) subunit of NF-κB to specify a pro-apoptotic outcome in response to some stimuli is less well recognized, but needs to be understood if rational manipulation of the NF-κB pathway is to be deployed in cancer therapy. In this report, we provide evidence that the growth-responsive nuclear protein, proenkephalin (Penk), is required, in part, for apoptosis induction, in response to activation or overexpression of p53 and RelA(p65). We describe UV-C-inducible physical associations between endogenous Penk and p53 and RelA(p65) in mammalian cell lines. Depletion of Penk by RNA interference (RNAi) substantially preserves viable cell number following exposure to UV-C irradiation or hydrogen peroxide and confers transient protection in cells exposed to the genotoxin etoposide. In virally transformed and human tumor cell lines, overexpression of nuclear Penk with overabundant or activated p53, or RelA(p65) even in the absence of p53, enhances apoptosis to the point of synergy. We have further shown that Penk depletion by RNAi substantially derepresses transcription of a range of antiapoptotic gene targets previously implicated in repression-mediated apoptosis induction by NF-κB and p53. Physical association of endogenous Penk with the transcriptional co-repressor histone deacetylase suggests that it may be a component of a transcriptional repression complex that contributes to a pro-apoptotic outcome, following activation of the NF-κB and p53 pathways, and could therefore help to facilitate an antitumor response to a broad range of agents.
Keywords: apoptosis, NF-κB, proenkephalin
The importance of the pro-apoptotic function of p53 in its role as a tumor suppressor is well established. p53 transactivates a number of pro-apoptotic genes including Noxa, Puma and Bax.1 p53-mediated transcriptional repression2 and transcription-independent3 mechanisms have also been proposed in the induction of apoptosis by p53. No single gene product has been shown to be absolutely required for p53-mediated apoptosis; thus, it seems likely that a number of factors and mechanisms collaborate in the cellular decision to undergo apoptosis following activation of the p53 pathway.
In the context of the apoptotic response to antitumor agents, p53 can play an important role, but the response to many cancer therapies is further modulated by other molecules and pathways such as the nuclear factor κB (NF-κB) pathway. Ionizing radiation and certain chemotherapeutics activate NF-κB in an antiapoptotic manner, which contributes to resistance to such agents.4 Thus, pharmacological efforts to inhibit the NF-κB pathway and thereby enhance the efficacy of anticancer drugs are underway. On the other hand, NF-κB can in some circumstances mediate a pro-apoptotic effect5,6 and can even suppress tumor formation.7 An understanding of how NF-κB mediates these opposing functions will therefore be crucial in helping to inform pharmacological interventions to manipulate the NF-κB pathway, particularly in the context of cancer therapy.
We have previously described localization of the opioid precursor, proenkephalin (Penk), within the nucleus of a range of neural and non-neural cell types. The capacity of Penk to undergo subnuclear reorganization in response to growth arrest and differentiation cues suggests its involvement in decision-making events in growth control.8 Opioid pathways have been implicated in the regulation of cell death and survival,9,10 and morphine has been shown to have antitumor activity in vivo, mediated in part through phosphorylation and activation of p53.10
In this study, we describe inducible physical associations between endogenous nuclear Penk and two transcription factors: p53 and the RelA(p65) subunit of NF-κB. Penk knockdown by RNA interference (RNAi) attenuates apoptosis induced by UV irradiation and hydrogen peroxide, and diminishes apoptosis to a lesser extent, following treatment with the genotoxin etoposide. Transiently overexpressed nuclear Penk cooperates independently with transfected wild-type p53 and RelA(p65) to cause significant enhancement of apoptosis. Penk overexpression also represses activity of transiently expressed p53 and NF-κB-dependent promoter–reporter constructs. Penk knockdown by RNAi in turn increases basal transcription from a range of endogenous p53 and NF-κB-repressed antiapoptotic gene targets. This implicates endogenous Penk in tonic repression of prosurvival genes. Physical association of endogenous Penk with the co-repressor histone deacetylase (HDAC), taken together with its association with p53 and NF-κB, suggests its participation in transcriptional repression complexes involved in apoptosis induction. If nuclear Penk facilitates an apoptotic outcome in response to activation of both the p53 and NF-κB pathways it could conceivably assist the antitumor response to a range of therapies in p53 wild-type and p53-null tumors which may warrant its investigation as a target for therapy.
Results
Endogenous Penk reorganizes and associates with p53, following UV irradiation
Given that Penk and p53 share the capacity to be reorganized into nuclear bodies following change in growth state or cellular stress,8,11 we investigated whether Penk and p53 interact in vitro and as endogenous proteins in living cells. Baculovirus-expressed wild-type p53 bound to in vitro translated Penk (Figure 1a), and human embryonic kidney (HEK) 293-expressed nuclear-targeted Penk bound to in vitro translated p53 (Figure 1b). Thus, Penk and p53 physically associate in vitro.
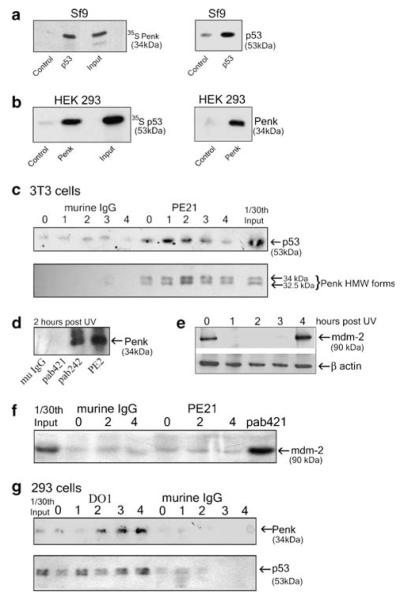
Figure 1.
Penk physically associates with p53, in vitro and in vivo, in response to UV-C irradiation. (a) Penk- and baculovirus-expressed wild-type p53 undergo physical association in vitro. Insect (Sf9) cells were infected with a human wild-type p53 baculovirus construct and lysates prepared 48 h after infection. Lysates were incubated with the p53 antibody DO1 and protein G agarose beads and in vitro translated 35S-labeled Penk. The products were subjected to 10% PAGE, and then autoradiographed (left panel). Uninfected cell lysates were used as control. Immunoblot for p53 (right panel) was used to confirm the expression of wild-type p53 in baculovirus-infected cells. (b) Mammalian cell-expressed nuclear-targeted Penk associates with in vitro translated p53. Plasmid vector encoding Penk minus its signal peptide (PenkΔsig) was transfected into HEK 293 cells. Cell lysates prepared after 48 h were incubated with a Penk antibody (PE18), or with murine isotype IgG control antibody, and in vitro translated 35S-labeled p53 (left hand panel). As an additional control, non-transfected cells were incubated with anti-Penk antibody (not shown). Immunoblot for Penk (right panel) confirmed low levels of Penk in control (transformed, non-transfected) HEK 293 cells compared with transfected cells. (c and d) Endogenous Penk inducibly associates with p53 in murine fibroblasts, following UV-C irradiation. 3T3 cells were exposed to 50 J/m2 of UV-C irradiation and whole-cell lysates prepared at the indicated times. (c) Lysates were subjected to co-immunoprecipitation assays using protein G agarose, and either the Penk antibody PE21 or isotype control antibody murine IgG. Immunoprecipitation products were immunoblotted for p53 using the anti-murine p53 polyclonal antibody CM5 (top panel); efficiency of precipitation was checked by immunoblotting against Penk monoclonal antibodies PE 14 and 19 (lower panel). (d) Two hours post UV treatment, lysates were subjected to co-immunoprecipitation assays using protein G agarose and isotype control antibody murine IgG, anti-p53 monoclonal antibodies to murine p53 – pab 421 and pab 242 – or the Penk antibody PE2. Immunoprecipitation products were immunoblotted for Penk, using the anti-Penk monoclonal antibodies PE 14 and 19. (e and f) Mdm-2 is transiently decreased following UV irradiation (e) but does not associate with Penk (f). After 0, 2 and 4 h of UV treatment, lysates were immunoblotted for mdm-2 (e) and also subjected to co-immunoprecipitation assays (f), using protein G agarose and a Penk antibody PE21, isotype control antibody murine IgG or anti p53 antibody pab 421 (as positive control). Immunoprecipitation products were immunoblotted for mdm-2 using the anti-mdm-2 antibody SMP14. (g) Endogenous Penk inducibly associates with p53 in HEK 293 cells following UV-C irradiation. HEK 293 cell lysates were prepared at the indicated times following 50 J/m2 of UV-C irradiation and co-immunoprecipitations were carried out as above. Immunoprecipitation was performed using the p53 monoclonal antibody DO1 or isotype control antibody murine IgG. The products were immunoblotted for Penk using the anti-Penk monoclonal antibody PE14 (top panel). Immunoprecipitation products were also immunoblotted for human p53, using the anti-p53 polyclonal antibody CM1, to check efficiency of precipitation (lower panel)
A UV-stimulated association between endogenous Penk and p53 was also revealed by Penk immunoprecipitation in murine fibroblasts (3T3 cells) (Figure 1c, upper panel). Immunoprecipitation with the anti-p53 antibody, pab242, also revealed association between p53 and Penk (Figure 1d); however, a different anti-p53 antibody, pab421, failed to reveal the association (Figure 1d). pab421 binds to the C-terminus of p53, suggesting that this region may be involved in binding to Penk following UV irradiation.
To address potential downstream gene targets that may be involved in a UV-induced Penk–p53 interaction, we immunoblotted extracts from UV-irradiated 3T3 cells for mdm-2, a known p53-regulated gene target. Somewhat surprisingly, mdm-2 levels were transiently decreased at 1–3 h following UV irradiation but had recovered by 4 h (Figure 1e). This is consistent with a recent report that mdm-2 protein levels decline following UV, due to self-ubiquitination and degradation.12 Interaction between Penk and mdm-2 was not involved in the UV-induced degradation of mdm-2, as Penk antibodies, unlike p53 antibodies, failed to precipitate mdm-2 before and after a UV stimulus (Figure 1f).
A UV-inducible association between Penk and p53 was also revealed in HEK 293 cells by p53 antibody immunoprecipitation (Figure 1g). Repeat analyses revealed that although the UV-inducible nature of the physical association was a consistent finding, the precise time at which it was revealed appeared to be influenced by culture conditions and growth state of the cells.
Penk knockdown using RNAi preserves cell viability following UV irradiation
The UV-inducible association between Penk and p53 led us to address whether depletion of Penk might compromise the apoptotic response to UV, which is at least partly dependent on the correct functioning of the p53 protein. Penk siRNA achieved a reduction in Penk mRNA of greater than 70% compared with control mRNA (glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) (Figure 2a); knockdown was also confirmed by immunoblotting (Figure 2b), demonstrating that a variety of high-molecular-weight Penk forms, previously described,13 were efficiently depleted.
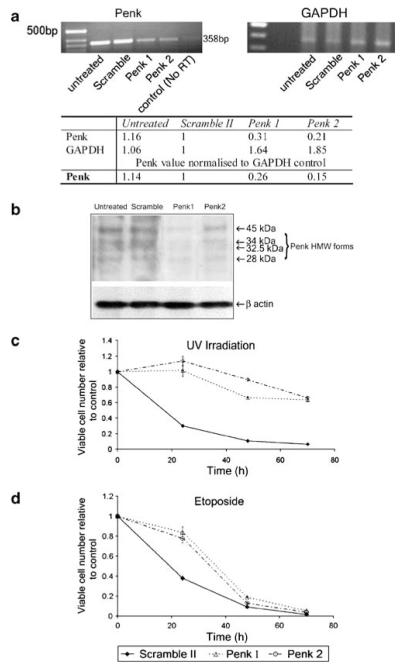
Figure 2.
HEK 293 cells depleted of Penk by siRNA are protected from apoptotic cell death. (a–d) 293 cells were transfected with two siRNA constructs directed against Penk and a scrambled oligonucleotide as control (Penk 1, Penk 2 and Scramble II). Cells were harvested after 3 days and either lysed for RNA extraction to confirm Penk depletion or re-plated for assays of viable cell number (MTS assay), following treatment with 50 J/m2 UV-C irradiation or 25 μM etoposide. (a) RT-PCR of RNA from 293 cells transfected with Scramble II, Penk 1 and Penk 2 siRNA confirmed depletion of Penk with siRNA. RNA isolated from 293 cells transfected with Scramble II, Penk 1 and Penk 2 siRNA was subjected to first strand synthesis using 1 μg of each RNA, and then reverse transcribed using oligo-(dT15) and AMV reverse transcriptase. PCR was carried out using 25 ng of RNA (Penk, top left panel) and 1 ng (GAPDH, top right panel): densitometry of PCR bands was conducted to quantify PCR products (lower panel). (b) Whole-cell extracts of untreated 293 cells and 293 cells transfected with Scramble II, Penk 1 and Penk 2 siRNA were immunoblotted for Penk, using Penk antibodies (PE14 and PE19). Both Penk 1 and Penk 2 depleted a range of high-molecular-weight forms of Penk,13 although Penk 1 appeared to be more efficient. Actin immunoblot of same lysates confirmed equivalent protein loading. (c) Penk siRNA substantially preserves cell viability following UV irradiation. Change in cell viability in siRNA-transfected cells was measured in the MTS assay over a 72-h time course, following exposure to UV-C irradiation. Each data set was obtained from a representative experiment performed at least three times. Data points represent mean values (±S.E.M.) from wells in triplicate relative to control values (from non-irradiated, mock-transfected cells). (d) Penk siRNA delays cell death following exposure to the genotoxin etoposide. Change in cell viability over a time course following addition of etoposide was measured in the MTS assay, as above
Following UV irradiation, there was a substantial reduction in viable cell number in cell populations transfected with scrambled oligonucleotide, whereas in cell populations that had been transfected with siRNA to Penk (Penk 1 and Penk 2) there was a substantial preservation of viable cell number (Figure 2c). In contrast, cells exposed to the genotoxin etoposide were afforded only transient protection by Penk siRNA (Figure 2d). This suggests that apoptotic stimuli may be differentially dependent on the presence of Penk, but that UV-C irradiation is one stimulus that appears to require Penk for optimal apoptosis induction.
Overexpressed nuclear Penk cooperates with activators of p53 and with overexpressed p53 to enhance cell killing
Given that a reduction in nuclear Penk can alleviate cell killing in response to some apoptotic stimuli, the question then arose whether overexpression of nuclear Penk would enhance apoptosis induction. We have previously demonstrated that a transfected deletion mutant of Penk, from which the signal peptide sequence has been removed, circumvents the secretory pathway and is targeted to the nucleus;8 furthermore, nuclear import of the transfected protein occurs in a regulated manner. Transiently overexpressed nuclear-targeted Penk enhanced apoptosis induced by etoposide (Figure 3a and b) and cotransfected p53 (Figure 3c and d). Assay of viable cell number by the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay revealed a striking reduction in viable cells, in the combined presence of Penk and p53 (Figure 3e). The apparently greater effect on cell number, as revealed by phase microscopy and MTS assay compared with fluorescence-activated cell sorter (FACS) analysis, would be consistent with Penk, providing a cooperative stimulus to p53’s known capacity to promote phagocytic engulfment of apoptotic cells in HEK 293 cells, as we have previously reported.14 Overexpressed Penk also enhanced apoptosis when cotransfected with p53 in human tumor cell lines including osteosarcoma (U2-OS) cells (Figure 3f), H1299 (p53-null lung carcinoma) cells and MCF-7 (mammary carcinoma) cells (not shown).
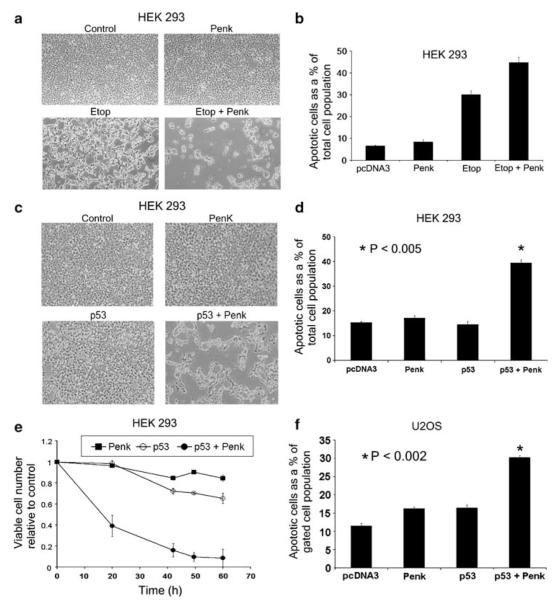
Figure 3.
Overexpressed nuclear-targeted Penk cooperates with p53 in apoptosis induction. (a and b) Transiently transfected nuclear Penk enhances etoposide-induced apoptosis and cell clearance. HEK 293 cells were transfected with vector encoding nuclear-targeted Penk (PenkΔsig) or parent vector (pcDNA3), to which etoposide (50 μM) or drug vehicle was added after 18 h. Transfectants were left for a further 48 h and examined by phase microscopy (a), or assayed for apoptotic cell numbers by FACS analysis of propidium iodide-stained, permeabilized cells (to reveal sub-G0 cells with sub-normal DNA content) (b). Apoptotic cell numbers are expressed as a percentage of the total cell population (sample size; n>15 000). Values are expressed as means of samples in triplicate (±) S.E.M. (c–e) Overexpressed nuclear Penk synergizes with overexpressed wild-type p53 in apoptosis induction and cell clearance. HEK 293 cells were transfected with Penk and p53, singly and in combination. Forty-eight hours after transfection, the cells were examined by phase microscopy (c), or harvested for FACS analysis of propidium iodide-stained cells (d), as above. Values expressed as mean±S.E.M. P-value calculated by Students t-test comparing the Penk/p53 combination with p53 alone (n = 6). (e) Cell viability (MTS) assays were also conducted. Viable cell number relative to control (parent vector transfected) cells was assayed at time intervals. Values expressed as mean±S.E.M. from wells in triplicate. (f) Nuclear Penk and p53 cooperate in apoptosis induction when overexpressed in human tumor cells. Human osteosarcoma U2-OS cells were transfected with vectors encoding Penk and wild-type p53, singly and in combination. Apoptotic cell numbers, as a percentage of the transfected rather than total cell population, were estimated at 48 h post-transfection by bivariate FACS analysis: propidium iodide-stained cells with a sub-normal DNA content were assayed within a gated cell population that had been cotransfected with GFP (sample size, n>15 000). P-value was calculated by Students t-test, comparing p53 transfectants with the p53/Penk combination (n = 4)
UV-C-inducible association of Penk with the RelA (p65) subunit of NF-κB
The greater protective effect of Penk knockdown following UV-C irradiation, compared with etoposide treatment, raised the possibility that Penk knockdown was also modulating the outcome of NF-κB pathway activation. Transcriptional cross-talk between the p53 and NF-κB pathways, due in part to competition between p53 and RelA (p65) for binding to the transcriptional coactivator p300, has previously been described.15
Similar to p53, Penk bound to RelA(p65) in vitro (Figure 4a). Furthermore, a UV-inducible association between endogenous Penk and RelA was also revealed by Penk immunoprecipitation in 3T3 cells – not shown – and in HEK 293 cells (Figure 4c). RelA antibody also precipitated Penk from HEK 293 cells (not shown). Penk antibodies were unable to precipitate the IκB protein, another component of the NF-κB pathway, despite the capacity of anti-RelA antibodies to do so (not shown). Thus, endogenous Penk is stimulated to associate with RelA(p65) following UV irradiation.
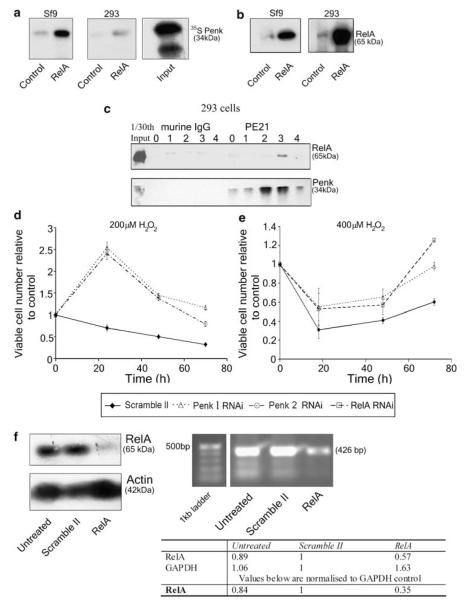
Figure 4.
Penk inducibly associates with RelA(p65) following UV-C irradiation and is required for hydrogen peroxide-mediated cytotoxicity. (a and b) in vitro association between Penk and RelA(p65). (a) Lysates of insect (Sf9) cells infected with a human RelA baculovirus construct or HEK 293 cells transfected with vector encoding RelA were prepared after 48 h. Lysates were incubated with a RelA/p65 antibody sc109 and protein G agarose beads, and in vitro translated 35S-labeled Penk. The products were subjected to 10% PAGE, and then autoradiographed. Uninfected or untransfected cell lysates were used as control. (b) Immunoblot for RelA(p65) using the polyclonal antibody sc372 was carried out to confirm overabundance of RelA(p65)in the baculovirus-infected and transfected 293 cells. (c) Endogenous Penk inducibly associates with RelA(p65) in HEK 293 cells following UV-C irradiation. HEK 293 cells were exposed to 50 J/m2 of UV-C irradiation and whole-cell lysates prepared at the indicated intervals (lanes 0–4: 0–4 h post UV treatment). Lysates were subjected to co-immunoprecipitation assays using protein G agarose, and either the Penk antibody PE21 or isotype control antibody murine IgG. Immunoprecipitation products were immunoblotted for RelA using the anti-RelA polyclonal antibody sc372 (top panel) Immunoblotting using the anti-Penk monoclonal antibodies PE 14 and 19 confirmed efficiency of immunoprecipitation (lower panel). (d and e) Penk or RelA(p65) knockdown by siRNA produced a comparable attenuation of hydrogen peroxide-mediated cytotoxicity. (d) 293 cells were transfected separately with Penk 1, Penk 2 or control (Scramble) oligonucleotides. Seventy-two hours post-transfection, cells were re-seeded for MTS cell viability assay, in the presence and absence of 200 μM H2O2. Viable cell number (mean±S.E.M.) is expressed as the ratio of MTS values in hydrogen peroxide-treated, siRNA-transfected (Scramble or Penk) compared to control, mock-transfected cells that had not been exposed to hydrogen peroxide. (e) 293 cells at high cell density were transfected with Penk 1, RelA or Scramble siRNAs, and then subjected to MTS assay using the same procedure as above, but in the presence and absence of 400 μM H2O2. (f) RelA knockdown by siRNA produces efficient depletion of RelA protein (upper panel left). Untransfected 293 cells and cells transfected with RelA or Scramble siRNAs were lysed and immunoblotted for RelA(p65) using sc372. β-actin immunoblot of the same lysates confirmed equivalent protein loading. (f, upper right panel) RT-PCR of RNA from 293 cells transfected with Scramble II or RelA siRNA confirmed depletion of RelA with siRNA. RNA isolated from 293 cells transfected with Scramble II or RelA siRNA was subjected to first strand synthesis using 1 μg of each RNA, and then reverse transcribed using oligo-(dT15) and AMV reverse transcriptase. PCR was carried out using 5 ng of RNA. PCR products were quantified by densitometry (f, lower panel)
Penk knockdown by RNAi preserves cell viability following oxidant-mediated activation of the NF-κB pathway in pro-apoptotic mode
Perkins and co-workers have previously described the activation of NF-κB in an apparently pro-apoptotic mode by UV-C irradiation.6 The physical association between Penk and RelA(p65), following UV-C irradiation (Figure 4), coupled with the marked preservation of cell viability following UV irradiation in the face of Penk depletion (Figure 2c), suggested that Penk may be enhancing the ability of RelA(p65) to instruct an apoptotic fate following a UV stimulus. In common with UV-C, hydrogen peroxide has also been reported to activate NF-κB in a pro-apoptotic manner.16
Penk knockdown substantially preserved cell viability following exposure to hydrogen peroxide (Figure 4d). At higher cell density (Figure 4e), a higher concentration of hydrogen peroxide was required to produce cell death; even under these conditions, Penk knockdown still provided moderate protection from cell death. RelA(p65) depletion using an siRNA construct previously described by Perkins and co-workers6 protected cells to a degree similar to that achieved by Penk knockdown (Figure 4e). This suggests that Penk may be required in at least some circumstances for RelA-mediated induction of cell death.
Nuclear Penk provides a p53-independent pro-apoptotic ‘switch’ to activated NF-κB or overexpressed RelA(p65)
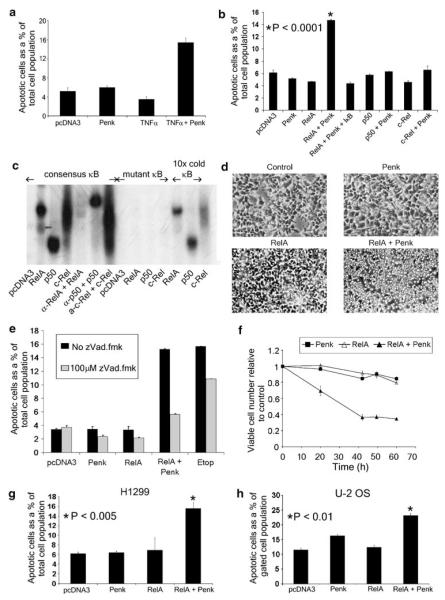
The requirement for Penk in death induced by some atypical activators of NF-κB raised the question whether Penk may be involved in the cellular decision to survive or die following NF-κB pathway activation, in general. Tumor necrosis factor α (TNFα) differs from UV-C and hydrogen peroxide, in engaging the NF-κB pathway in a more typical antiapoptotic mode, an outcome that depends at least partly on RelA(p65).17 To address whether Penk may have the capacity to alter the outcome of NF-κB activation in favor of apoptosis, we overexpressed nuclear Penk in the presence of a low, sub-lethal concentration of TNFα. In the presence of Penk or TNFα alone, cells remained viable; however, in their combined presence, there was a more than three-fold induction of apoptosis above baseline (Figure 5a). Thus, an increase in nuclear Penk levels appears to uncover apoptosis in the presence of a more typical activator of NF-κB. Overexpressed nuclear Penk also revealed apoptosis when coexpressed with the RelA(p65) subunit of NF-κB but not with the p50 or c-Rel subunits; furthermore, the apoptotic induction was prevented in triple transfectants that expressed a super-repressor form of IκB (Figure 5b). Expression of NF-κB subunits in functional forms capable of binding to DNA (consensus κB site containing oligonucleotides) was confirmed by electrophoretic mobility shift assay (EMSA), and shown to be specific18,19 (ND Perkins, unpublished) (Figure 5c). The atypical behavior of c-Rel in the EMSA (which revealed a less discrete band and absence of supershift) may reflect its poor ability to bind κB consensus sites as a homodimer. Thus, overexpressed nuclear Penk cooperates specifically with the RelA subunit of NF-κB to promote apoptosis in HEK 293 cells.
Figure 5.
Overexpressed Penk provides a specific pro-apoptotic switch to the NF-κB subunit RelA(p65). (a) Transiently transfected nuclear Penk synergizes with a sub-lethal concentration of TNFα in apoptosis induction. Parent vector- or Penk-transfected HEK 293 cells were exposed 18 h post-transfection to 10 ng/ml TNFα, and then left for a further 30 h before harvesting for FACS analysis. Apoptotic cell numbers (propidium iodide-stained cells with sub-normal DNA content) are expressed as a percentage of the total cell population (mean±S.E.M.; sample size, n>15 000). (b–d) Penk specifically provides a pro-apoptotic switch to the RelA(p65) subunit but not to other NF-κB subunits. (b) Apoptotic scores (obtained by FACS analysis as above) from HEK 293 cells transfected with RelA(p65), c-Rel and p50, singly and in combination with Penk; cells were also transfected in the presence and absence of IκB super-repressor (double serine mutant) to act as a dominant inhibitor of NF-κB. P-value was obtained by comparing Penk/RelA-transfected cells with vector-transfected controls in Student’s t-test (n = 6). (c) To confirm expression of the NF-κB subunits, 293 cells were transfected with 10 μg of the control plasmid pcDNA3 or RSV-driven plasmids containing one of three NF-κB subunits: RelA, p50 or c-Rel. After 48 h, nuclear extracts were prepared and subjected to EMSA to detect subunit binding to 32P-labeled consensus NF-κB DNA binding site; specificity of binding was confirmed by incubation with mutant NF-κB DNA binding site and incubation with non-mutant consensus κB site, in the presence of supershift antibodies or a 10-fold excess of competing cold consensus oligo. All the subunits were expressed and bound to the NF-κB consensus DNA binding site. Anti-p50 antibody (Ab-1, CN Bioscience) revealed a supershift of the bound p50, whereas anti-RelA (sc109, Santa Cruz) abolished binding of RelA(p65). (d) Phase micrographs of HEK 293 cells 48 h after transfection with Penk and RelA(p65), singly or in combination. (e) Co-overexpression of Penk and RelA(p65) induces caspase-dependent apoptosis. HEK 293 cells transfected with Penk or RelA, singly and in combination, and in the presence and absence of the caspase inhibitor zVAD.fmk, were compared with cells that had been treated with 50 μM etoposide, also in the presence and absence of zVAD.fmk. Apoptotic scores were obtained by FACS analysis, as above. (f) Co-overexpressed Penk and RelA(p65) synergize to reduce viable cell number. HEK 293 cells transfected with Penk or RelA(p65), singly and in combination, were subjected to MTS assay at the indicated times. Viable cell number was compared with control cells that had been transfected with parent vector. (g) The cooperativity between Penk and RelA(p65) does not require p53. p53-null H1299 human lung carcinoma cells were transfected with Penk or RelA, singly and in combination. Forty-eight hours post-transfection, apoptotic cell numbers were estimated as a percentage of total cell numbers by FACS analysis of propidium iodide-stained cells, as above. The P-value was obtained from Student’s t-test by comparing RelA/Penk transfectants with cells transfected with vector alone (n = 5). (h) Penk and RelA cooperate to induce death in osteosarcoma cells. U2-OS cells transfected with Penk or RelA(p65), singly and in combination, and also with GFP, were subjected to bivariate FACS analysis of propidium iodide- and GFP-stained cells – to reflect apoptotic cell scores within the transfected cell population. P-value obtained from Student’s t-test, comparing the RelA(p65)/Penk combination with vector-transfected cells (n = 4)
Despite the rather different appearance of the Penk/RelA-transfected cells (Figure 5d), compared with those transfected with Penk and p53 (Figure 3c), death was attenuated by a cell-permeable broad-spectrum caspase inhibitor, N-benzyloxy-carbonyl-valyl-alanyl-aspartyl-fluoromethylketone (zVAD.fmk), and therefore at least partly caspase dependent (Figure 5e). This difference would be consistent with our previous report that when apoptosis is engaged via a p53- or DNA damage-dependent pathway, there is coordinate induction of phagocytic engulfment but not when apoptosis is triggered in other ways.14 Assays of cell number confirmed a substantial decline in viability in cell populations transfected with Penk and RelA, compared to control cell populations (Figure 5f).
It has been suggested that NF-κB may be required for p53-mediated apoptosis20 and cross-talk between the NF-κB and p53 pathways is well recognized.15,20 However, cooperative death induction by RelA and Penk was revealed in H1299 lung carcinoma cells, that lack p53 protein, and therefore was independent of p53 (Figure 5g); similarly cooperativity between p53 and Penk was not blocked by IκB, indicating a lack of dependence on NF-κB activation (data not shown). Apoptosis induction in a RelA/Penk-transfected cell population was confirmed by coexpression with green fluorescent protein (GFP) in U2-OS osteosarcoma (U2-OS) cells (Figure 5h).
Taken together, these data indicate that nuclear Penk can be expressed to high levels without appreciably compromising cell viability; however, when nuclear Penk levels are elevated in the presence of an NF-κB activating stimulus, or overexpressed RelA(p65), this uncovers a significant apoptotic drive.
Penk can act as a transcriptional repressor
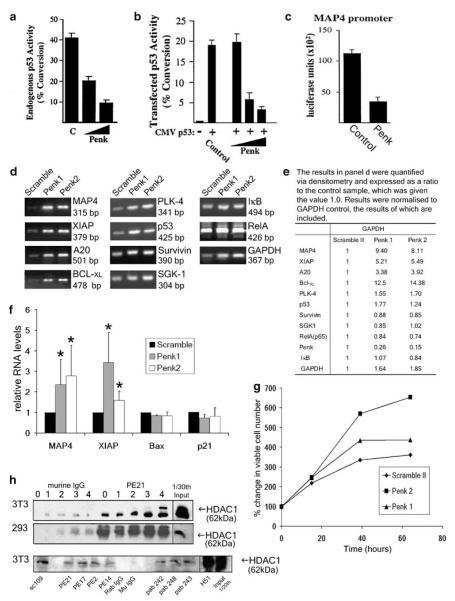
Penk possesses structural features that suggest it may act as a transcription factor.21 To address this, we conducted a series of transcriptional assays. A preliminary analysis revealed that a fusion protein comprised of Penk sequence fused to a DNA targeting motif can repress transcription from a heterologous promoter (data not shown). Unfused Penk was also able to repress a p53-regulated synthetic promoter (PG13) in human foreskin fibroblast (HFF) cells grown at high density to activate endogenous p53 (Figure 6a), and in the presence of cotransfected p53 in U2-OS cells (Figure 6b). An authentic gene target purported to be involved in p53-mediated transcriptional repression (MAP4) was also repressed (Figure 6c). Microtubule-associated protein 4 (MAP4) has been shown to be downregulated in p53-dependent death and overexpression of MAP4 protects cells from apoptotic stimuli.2 In contrast MAP4 is not downregulated in p53-independent death.2
Figure 6.
Penk represses p53- and NF-κB-regulated antiapoptotic gene targets, possibly through association with a transcriptional co-repressor. (a–c) Penk alone can repress transcription from authentic (MAP4) as well as synthetic promoters. (a) Expression from a PG13CAT reporter, when activated by endogenous p53 in HFF cells grown at high cell density, is repressed by Penk in a dose-dependent manner. HFF cells were transfected with 3 μg of a multimerized p53 response element reporter plasmid (PG13CAT) and 5 or 7.5 μg of pcDNA3:Penk. Lysates were subjected to assay for conversion of chloramphenicol to its acetylated form, as a transcriptional reporter assay. (b) Penk represses p53-stimulated Bax-CAT reporter gene expression in a dose-dependent manner. U2-OS cells were transfected with 3 μg of the Bax-CAT reporter plasmid, 2 ng weight of cytomegalovirus (CMV):p53 expression plasmid and 2, 5 and 7.5 μg of the indicated pcDNA3:Penk expression plasmid. (c) Penk represses transcription from the MAP4 promoter. U2-OS cells were transfected with 3 μg of the MAP4 luciferase reporter plasmid and 7.5 μg of the indicated pcDNA3:Penk expression plasmid. Control cells were transfected with 7.5 μg of pcDNA3. (d) Penk siRNA transfection derepresses antiapoptotic genes in 293 cells. RNA isolated from 293 cells transfected with Scramble II, Penk 1 and Penk 2 siRNA was subjected to first strand synthesis using 1 μg of each RNA and reverse transcribed using oligo-(dT15) and AMV reverse transcriptase. PCR was carried out using 10 ng of RNA (MAP4, XIAP), 5 ng of RNA (BclXL, A20, p53, PLK4 and RelA) and 1 ng (Survivin, SGK1 and GAPDH). PCR was also carried out using three serial dilutions of the RNA, to ensure that the relationship between RNA input and product was linear (data not shown). (e) PCR products were quantified via densitometry and normalized to GAPDH control, confirming that MAP4, XIAP, A20 and BclXL are derepressed in Penk siRNA-transfected cells; similar results were observed when normalized to a β-actin control (data not shown). (f) Real-time PCR: RNA from siRNA treated 293 cells was reverse transcribed and subjected to real-time PCR, using primers and probes against MAP4, XIAP, bax, p21 and β-actin as a control. mRNA levels were normalized to β-actin and expressed as a percentage of control (Scramble siRNA transfection). Values are means±S.D. of four experiments. Student’s t-test compared RNA levels in Penk siRNA-transfected cells with those in scrambled oligonucleotide-transfected cells. P-values at less than 0.05 are indicated by *. (g) Penk siRNA transfection confers an increase in cell viability. Cell viability (MTS) assays were conducted on 293 cells transfected with Scramble II, Penk 1 and Penk 2 siRNA. Viable cell number was assayed at time intervals. Values expressed as mean from wells in triplicate. (h) Penk constitutively associates with histone deacetylase 1 (HDAC1). 3T3 and 293 cells were plated out and left overnight to achieve approximately 50% confluence, the cells were then exposed to 50 J/m2 of UV-C irradiation and then whole-cell lysates were prepared at 0, 1, 2, 3 and 4 h following irradiation. The lysates were precleared with protein G agarose beads and murine IgG, and aliquots then used in co-immunoprecipitations (co-IPs) using Penk antibodies (PE21) and protein G agarose. The products were immunoblotted with the anti-HDAC1 antibody Ab-1 (CN Biosciences). This revealed a constitutive interaction between Penk and HDAC1 in both 3T3 (top panel) and 293 (middle panel) cells. Two hour post UV treatment, treated 3T3 lysates were subjected to co-IP assays using protein G agarose, and either the Penk antibodies PE21, PE17, PE2 and PE14, isotype control antibody murine IgG, an anti-RelA polyclonal antibody sc109 (Santa Cruz) or anti-p53 monoclonal antibodies pab242, pab248 and pab243 (lower panel). Immunoprecipitation products were immunoblotted for HDAC1 with anti-HDAC1 antibody Ab-1 (CN Biosciences)
Campbell et al.6 elucidated a potential mechanism for the pro-apoptotic function of NF- κB that is due, at least in part, to RelA-mediated active repression of antiapoptotic genes. We therefore reasoned that Penk may be involved in p53 and/or NF-κB-mediated repression of antiapoptotic genes. Semi-quantitative RT-PCR analysis of HEK 293 cells, in which Penk was knocked down by siRNA (as above, Figure 3a and b), revealed derepression of the p53-regulated target MAP-42 and a number of NF-κB-regulated targets: A20,22 X-linked inhibitor of apoptosis (XIAP)6 and BclXL6 (Figure 6d and e). Polo-like kinase 4 (PLK4)23 and p53 itself were moderately derepressed in Penk siRNA cells, whereas survivin,24 serum and glucocorticoid-regulated kinase 1 (SGK1),25 RelA(p65) and inhibitor of κB (IκB)26 were not derepressed (Figure 6d and e). The alleviation of repression of MAP4, XIAP, A20 and BclXL mRNA levels was not due to depletion of p53 or RelA, as p53 and RelA mRNA levels were not repressed in Penk siRNA-transfected cells (Figure 6d). XIAP and BclXL have previously been shown to be repressed in a RelA-dependent manner,6 which would point to a role for Penk in NF-κB-mediated transcriptional repression as part of a pro-apoptotic pathway. Although MAP4,2 survivin, PLK4 and SGK1 have all been reported to be repressed in a p53-dependent manner, only MAP4 and, to a lesser extent PLK4, were derepressed by depleting Penk.
To confirm the apparent derepression of antiapototic genes by Penk knockdown, RNA from 293 cells transfected with Penk siRNA was subjected to real-time polymerase chain reaction (PCR), using Taqman primers and probes to a p53-repressed target gene (MAP4) and an NF-κB target gene (XIAP); this also demonstrated relief of transcriptional repression at these target genes, in the presence of Penk siRNA. On the other hand, genes that are transactivated by p53 – bax and p21 – were not derepressed by Penk knockdown (Figure 6f). The tonic repression of pro-survival genes by endogenous Penk may be functionally significant even in untreated cells, as Penk-depleted cells are present in greater numbers compared to control cells 48–60 h after treatment (Figure 6g).
Given that p53 and RelA have both been reported to interact physically with a member of the HDAC group of transcriptional co-repressors (HDAC1),27,28 and that Penk may reside in high-molecular-weight nuclear complexes with p53 and RelA, we investigated whether Penk could also bind HDACs. A constitutive interaction between endogenous Penk and HDAC1 in irradiated and non-irradiated 3T3 and 293 cells was revealed (Figure 6h). A glutathione-S-transferase (GST)-Penk fusion protein expressed in bacteria also bound HDAC, precipitated from HeLa cell extracts, with high affinity (not shown). In contrast, mSin3a, a co-repressor that also binds HDAC1,28 did not bind even with low affinity to GST-Penk. Provisionally, therefore, Penk may exist in a stable association with HDAC and be targeted to promoters following inducible associations with p53 and NF-κB to assist repression at antiapoptotic gene targets.
Discussion
In this paper, we describe a role for the nuclear protein, Penk,8 in regulating apoptosis induction by p53 and NF-κB. Endogenous Penk inducibly associates with p53 and RelA(p65) following UV-C irradiation and cooperates with both to enhance apoptosis induction, when co-overexpressed. Penk is also required in part for the apoptotic response to a number of cellular stressors including UV irradiation, etoposide and hydrogen peroxide. Together, these data suggest that nuclear Penk stimulates stress-activated apoptosis through inducible physical associations with wild-type p53 and the RelA(p65) subunit of NF-κB.
Whereas a role for p53 as an inducer of apoptosis is well established, the potential for the NF-κB subunit RelA(p65) to promote rather than repress apoptosis is more contentious.29 What is emerging is that an apoptotic function of RelA is only revealed in certain cellular contexts or in response to particular initiating stimuli. In the context of cancer therapy, a number of chemotherapeutics, as well as ionizing radiation, activate NF-κB in an anti- rather than pro-apoptotic mode,30 now recognized as likely to be an important mechanism for de novo or acquired resistance to therapy. Thus, it is of great practical relevance to understand the nature of any cellular and molecular events that decide the biological outcome of NF-κB activation, and whether these might be manipulated for therapeutic gain.
Given that RelA has been shown to mediate active repression of antiapoptotic genes,6 we determined the effect of Penk knockdown by RNAi on transcription from a number of κB-regulated gene targets. Interestingly, the level of transcription from three known antiapoptotic κB-regulated gene targets – A20, XIAP and BclxL, – was substantially increased when endogenous Penk was inhibited by RNAi. This suggests that Penk may tonically inhibit an antiapoptotic arm of NF-κB function and thereby help to prime cells for apoptosis.
p53-mediated gene repression has also been implicated in apoptosis induction. For example, in hypoxia there is p53-dependent repression of a number of genes such as β-tubulin.31 Interestingly, the only p53-regulated gene found to be transactivated under hypoxic conditions is c-fos, which is a component of the activating protein 1 (AP-1) transcription factor complex known to upregulate Penk expression.32 p53-dependent transcriptional repression has also been reported in non-hypoxic apoptosis. For example, MAP-4 has been reported to be repressed during p53-dependent apoptosis and MAP-4 overexpression can rescue p53-dependent apoptosis.2 p53 has also been shown to repress the pro-survival gene survivin.24 It was therefore of interest to investigate whether Penk may contribute to p53-dependent repression at genes implicated in apoptosis. Penk knockdown alleviated transcriptional repression of MAP-4, whereas survivin was not significantly affected. This suggests that there may be a degree of selectivity in Penk’s action at p53-repressed gene targets.
In addition to physical association of Penk with p53 and RelA(p65) we have also demonstrated constitutive, high-affinity association of Penk with the transcriptional co-repressor HDAC1, which suggests it may be stably incorporated into transcriptional repression complexes. On the other hand, Penk transiently associates with p53 and NF-κB following stress. Transient dissociation and reassociation of RelA(p65) with the inhibitor protein I-κB is well recognized to be instrumental in the activation and termination of the NF-κB response. Furthermore, proteasome-mediated degradation of promoter-bound RelA(p65) is essential for termination of the response.33 Transient physical associations made by p53 are also recognized, for example, in its association with DNA repair complexes that is revealed in different subnuclear compartments.34 We have previously reported transient revelation of Penk antigenic domains in different subnuclear compartments, during decision-making events in cells undergoing growth arrest and differentiation,8 and we have also shown similar transient subnuclear reorganization of Penk in response to UV irradiation and DNA damage. It is therefore tempting to speculate that transient associations made by Penk with RelA(p65) and p53 in response to apoptotic stimuli cause a Penk–HDAC complex to be targeted to selected p53 and κB-regulated gene targets. Once delivery of the repressor complex to the relevant targets has been achieved, the association is no longer necessary.
This study has uncovered a new participant in apoptosis regulation, the opioid precursor Penk, that appears to stimulate the apoptotic function of both p53 and the RelA(p65) subunit of NF-κB, and thereby assists stress-activated apoptosis. Cancer therapies can act in part through stimulation of p53-dependent apoptosis, but this is offset by activation of NF-κB in antiapoptotic mode, now recognized as an important mechanism of resistance to therapy. Even in tumors that lack functional p53 protein, therapy resistance due to activation of NF-κB is emerging. Thus, therapeutic strategies designed to elevate levels of nuclear Penk might have the potential to enhance response to a range of cancer therapies in p53 wild-type and p53-null tumors.
Materials and Methods
Plasmids
Nuclear-targeted Penk expression plasmid was generated by PCR deletion of the NH2-terminal signal peptide (PenkΔsig), as previously described.8 pCMV:p53 and pT7:p53 were supplied by Carol Midgely and David Lane (University of Dundee). The Rous sarcoma virus (RSV) expression plasmids containing RelA(p65), p50, p52, c-Rel and RSV:IκBMSS have previously been described.15,18
Antibodies
Human p53 was detected using DO1 monoclonal antibody (Invitrogen, Carlsbad, CA, USA) or CM-1 polyclonal antibody (Novocastra Laboratories Ltd., Newcastle Upon Type, UK). Murine p53 was detected using CM-5 polyclonal antibody (Novocastra). Rabbit anti-RelA polyclonal sc109 (Santa Cruz Biotechnology Inc., CA, USA) was used to immunoprecipitate RelA, whereas anti-RelA polyclonal sc372 (Santa Cruz) was used to detect RelA in Western blots. Penk antibodies used were as described previously.8 Actin was detected using rabbit anti-actin polyclonal (Sigma A 2066). HDAC1 was detected by Ab-1 (CN Bioscience, San Diego, CA, USA) and H51 (Santa Cruz).
Transfections
HEK 293 at a cell density of 2.5 × 105 cells per milliliter were transfected by calcium phosphate, as previously described.35 H1299 and U2-OS cells at a cell density of 1 × 105 cells per milliter were transfected using Fugene-6 (Roche), according to the manufacturer’s recommendations.
RNAi
This was carried out essentially as described previously.35 Penk siRNA oligo sequences (MWG Biotech) were as follows (sense strand only): Penk 1 GCAGAGCUUCCUCAAGAUG and Penk 2 CCUGCAAGGAGCUCCUGCA. The Scramble II siRNA oligo (Dharmacon) that had no sequence homology to any known sequence was used as a control. RelA siRNA oligo sequence was as previously described.35
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was carried out as previously described,6 except that densitometry was carried out using a Syngene Genegenius (Synoptics Ltd), using the GeneSnap and GeneTools programs.
Primers for RT-PCR
| Forward | Reverse | |
|---|---|---|
| Penk | GCTGTCCCAAACCAGAGCTTC | TGAAGCCCCCATATCTCTTG |
| Map4 | CACAGGCCTTCCTTCTTCTG | AGCCAAAGATGTTCCACCAC |
| XIAP | TGGCAATATGGAGACTCAGC | TGCACTTGGTCACCAATACC |
| RelA | GGCGAGAGGAGCACAGATAC | ATCTTGAGCTCGGCAGTGTT |
| p53 | CCTCACCATCATCACACTGG | TCTGAGTCAGGCCCTTCTGT |
| PLK4 | GTTGGCATTGTTGTCTGTGG | GGTCAGCCACTCCCAAATAA |
| Bcl-XL | GAGCTGTTTATGGCCTCAGC | CCAGCAGCTCCTCACACATA |
| Survivin | TTTCTGCCACATCTGAGTCG | TGTCGAGGAAGCTTTCAGGT |
| SGK1 | CGAGGCTTTCCTAGCCTTTT | GGCACTCTAAACGCTCGTTTC |
| IκBα | CTGAGCTCCGAGACTTTCGAGG | CGTCCTCTGTGAACTCCGTG |
| A20 | CACGAGCCCGAGCTGATGAGG | CTTCCCCTTGCTCGTCACTG |
| GAPDH | GGTCGTATTGGGCGCCTGGT CACC |
ACACCCATGACGAACATGG GGGC |
Real-time PCR
Primers and probes were designed using Primer Express software (Applied Biosystems, Foster City, CA, USA) and obtained from Sigma-ProLigo. RNA was isolated from 293 cells transfected with Scramble II, Penk 1 and Penk siRNA, using SV Total isolation kit (Promega, Madison, WI, USA) and real-time PCR was performed essentially as described.36
Primers for real-time PCR are as follows: MAP4: forward: GGCCTTCCATCTTACCTTCAAA, reverse: CCGCTTCTCAGGAGCCTTT; XIAP: forward: CCTGCAGACATCAATAAGGAAGAA, reverse: CGGTATCTCCTTCACCAGTATAAAGAA.
Probes used for real-time PCR are as follows: MAP4: 5′ 6-FAM-ACGTGAAGCCAAAGCCCATTGCA-TAMRA 3′; XIAP: 5′ 6-FAM-TTCAGCATCAACACTGGCACGAGCA-TAMRA 3′.
DNA sequencing
PCR products were sequenced at the DNA Analysis Facility, Ninewells Hospital, Dundee.
Sf9 cells
Sf9 (Spodoptera frugiperda) cells (American Tissue Culture Collection, ATCC#CRL1711) were used to propagate recombinant baculovirus stocks. These cells were grown in Ex Cell 400™ medium with L-glutamine (JRH Bioscience) and supplemented with 1 U/ml penicillin and 1 μg/ml streptomycin.
Recombinant protein expression
HEK 293 cells were transfected with either pCMV:p53, pcDNA3:Penk or pRSV:RelA(p65), and 48 h after transfection, either whole cell or nuclear extracts were prepared. Nuclear extracts (RelA(p65)) were prepared essentially as previously described.37 Whole-cell extracts were prepared as previously described.38 Baculovirus-expressed p53 and RelA(p65) were prepared as previously described.39
in vitro association assays
in vitro translation of full-length and mutant Penk and in vitro association assays were essentially carried out as described elsewhere,35 except that for baculovirus-expressed p53, and RelA- and HEK 293-expressed p53 and Penk, whole-cell extracts were used instead of nuclear extracts.
Immunoprecipitation and Western blotting
These were carried out essentially as previously described.15
Cell viability/proliferation assay (transfection experiments)
The ‘MTS’ Cell Titre 96RAQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) was performed essentially as previously described,40 except that cell viability at each time point was expressed as a ratio of control values.
EMSA
These assays were performed essentially as described.37 c-Rel antibody was a kind gift from Sonia Rocha (Dundee).
Dual-fluorescence flow cytometry for detection of apoptotic cells
FACS analysis was carried out as described previously.14
Acknowledgements
We would like to thank Carol Midgeley and the past and present members of David Lane’s laboratory for generous provision of p53 pathway reagents. This work was supported by Wellcome Trust grant 033790 and Breast Cancer Research Scotland grant 2000/167.
Glossary
Abbreviations
- HDAC
histone deacetylase
- RNAi
RNA interference
- CMV
cytomegalovirus
- RSV
Rous sarcoma virus
- HEK
human embryonic kidney
- U2-OS
U2-osteosarcoma
- Penk
proenkephalin
- MAP4
microtubule-associated protein 4
- XIAP
X-linked inhibitor of apoptosis
- PLK4
Polo-like kinase 4
- SGK1
serum and glucocorticoid-regulated kinase 1
- NF-κB
nuclear factor κB
- IκB
inhibitor of κB
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- EMSA
electrophoretic mobility shift assay
- FACS
fluorescence-activated cell sorter
- MTS
3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- TNF
tumor necrosis factor
- GFP
green fluorescent protein
- HFF
human foreskin fibroblast
- PCR
polymerase chain reaction
- GST
glutathione-S-transferase
- AP-1
activating protein 1
- S.E.M.
standard error of the mean
- zVAD.fmk
N-benzyloxycarbonyl-valyl-alanyl-aspartyl-fluoromethylketone
References
- 1.Yu J, Zhang L. The transcriptional targets of p53 in apoptosis control. Biochem Biophys Res Commun. 2005;331:851–858. doi: 10.1016/j.bbrc.2005.03.189. [DOI] [PubMed] [Google Scholar]
- 2.Murphy M, Hinman A, Levine AJ. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 3.Yan Y, Shay JW, Wright WE, Mumby MC. Inhibition of protein phosphatase activity induces p53-dependent apoptosis in the absence of p53 transactivation. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Kotamraju S, Konorev E, Kalivendi S, Joseph J, Kalyanaraman B. Activation of nuclear factor-kappaB during doxorubicin-induced apoptosis in endothelial cells and myocytes is pro-apoptotic: the role of hydrogen peroxide. Biochem J. 2002;367:729–740. doi: 10.1042/BJ20020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell KJ, Rocha S, Perkins ND. Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol Cell. 2004;13:853–865. doi: 10.1016/s1097-2765(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 7.Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 8.Bottger A, Spruce BA. Proenkephalin is a nuclear protein responsive to growth arrest and differentiation signals. J Cell Biol. 1995;130:1251–1262. doi: 10.1083/jcb.130.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tegeder I, Geisslinger G. Opioids as modulators of cell death and survival – unraveling mechanisms and revealing new indications. Pharmacol Rev. 2004;56:351–369. doi: 10.1124/pr.56.3.2. [DOI] [PubMed] [Google Scholar]
- 10.Tegeder I, Grosch S, Schmidtko A, Haussler A, Schmidt H, Niederberger E, et al. G protein-independent G1 cell cycle block and apoptosis with morphine in adenocarcinoma cells: involvement of p53 phosphorylation. Cancer Res. 2003;63:1846–1852. [PubMed] [Google Scholar]
- 11.Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- 12.Lee MH, Lee SW, Lee EJ, Choi SJ, Chung SS, Lee JI, et al. SUMO-specific protease SUSP4 positively regulates p53 by promoting Mdm2 self-ubiquitination. Nat Cell Biol. 2006;8:1424–1431. doi: 10.1038/ncb1512. [DOI] [PubMed] [Google Scholar]
- 13.Vindrola O, Chervin A, Vitale M, Mella AN, Aloyz R, Basso A. Elevated proenkephalin-derived peptide levels in ACTH-producing adenomas: nucleus and cytoplasm localization. Endocrine. 1998;8:231–240. doi: 10.1385/ENDO:8:3:231. [DOI] [PubMed] [Google Scholar]
- 14.Wiegand UK, Corbach S, Prescott AR, Savill J, Spruce BA. The trigger to cell death determines the efficiency with which dying cells are cleared by neighbours. Cell Death Differ. 2001;8:734–746. doi: 10.1038/sj.cdd.4400867. [DOI] [PubMed] [Google Scholar]
- 15.Webster GA, Perkins ND. Transcriptional cross talk between NF-kappaB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaltschmidt B, Kaltschmidt C, Hofmann TG, Hehner SP, Droge W, Schmitz ML. The pro- or anti-apoptotic function of NF-kappaB is determined by the nature of the apoptotic stimulus. Eur J Biochem. 2000;267:3828–3835. doi: 10.1046/j.1432-1327.2000.01421.x. [DOI] [PubMed] [Google Scholar]
- 17.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 18.Perkins ND, Schmid RM, Duckett CS, Leung K, Rice NR, Nabel GJ. Distinct combinations of NF-kappa B subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci USA. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins ND, Agranoff AB, Pascal E, Nabel GJ. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 21.Bakalkin G, Ponomariev D, Sarkisyan RA, Terenius L. Sequence similarity between opioid peptide precursors and DNA-binding proteins. FEBS Lett. 1991;282:175–177. doi: 10.1016/0014-5793(91)80471-e. [DOI] [PubMed] [Google Scholar]
- 22.He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22:6034–6045. doi: 10.1128/MCB.22.17.6034-6045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Tan M, Li L, Pamarthy D, Lawrence TS, Sun Y. SAK, a new polo-like kinase, is transcriptionally repressed by p53 and induces apoptosis upon RNAi silencing. Neoplasia. 2005;7:312–323. doi: 10.1593/neo.04325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 25.Maiyar AC, Phu PT, Huang AJ, Firestone GL. Repression of glucocorticoid receptor transactivation and DNA binding of a glucocorticoid response element within the serum/glucocorticoid-inducible protein kinase (sgk) gene promoter by the p53 tumor suppressor protein. Mol Endocrinol. 1997;11:312–329. doi: 10.1210/mend.11.3.9893. [DOI] [PubMed] [Google Scholar]
- 26.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucharczak J, Simmons MJ, Fan Y, Gelinas C. To be, or not to be: NF-kappaB is the answer – role of Rel/NF-kappaB in the regulation of apoptosis. Oncogene. 2003;22:8961–8982. doi: 10.1038/sj.onc.1207230. [DOI] [PubMed] [Google Scholar]
- 30.Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 31.Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonnenberg JL, Rauscher FJ, III, Morgan JI, Curran T. Regulation of proenkephalin by Fos and Jun. Science. 1989;246:1622–1625. doi: 10.1126/science.2512642. [DOI] [PubMed] [Google Scholar]
- 33.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restle A, Janz C, Wiesmuller L. Differences in the association of p53 phosphorylated on serine 15 and key enzymes of homologous recombination. Oncogene. 2005;24:4380–4387. doi: 10.1038/sj.onc.1208639. [DOI] [PubMed] [Google Scholar]
- 35.Anderson LA, Perkins ND. Regulation of RelA (p65) function by the large subunit of replication factor C. Mol Cell Biol. 2003;23:721–732. doi: 10.1128/MCB.23.2.721-732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, et al. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 37.Anderson LA, Perkins ND. The large subunit of replication factor C interacts with the histone deacetylase HDAC1. J Biol Chem. 2002;277:29550–29554. doi: 10.1074/jbc.M200513200. [DOI] [PubMed] [Google Scholar]
- 38.Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23:4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kernohan NM, Hupp TR, Lane DP. Modification of an N-terminal regulatory domain of T antigen restores p53-T antigen complex formation in the absence of an essential metal ion cofactor. J Biol Chem. 1996;271:4954–4960. doi: 10.1074/jbc.271.9.4954. [DOI] [PubMed] [Google Scholar]
- 40.Spruce BA, Campbell LA, McTavish N, Cooper MA, Appleyard MV, O’Neill M, et al. Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 2004;64:4875–4886. doi: 10.1158/0008-5472.CAN-03-3180. [DOI] [PubMed] [Google Scholar]