Abstract
Rapid binding of peptides to MHC class II molecules is normally limited to a deep endosomal compartment where the coordinate action of low pH and HLA-DM displaces the invariant chain remnant CLIP or other peptides from the binding site. Exogenously added peptides are subject to proteolytic degradation for extended periods of time before they reach the relevant endosomal compartment, which limits the efficacy of peptide-based vaccines and therapeutics. Here we describe a family of small molecules that substantially accelerate the rate of peptide binding to HLA-DR molecules in the absence of HLA-DM. A structure-activity relationship study resulted in analogs with significantly higher potency and also defined key structural features required for activity. These compounds are active over a broad pH range and thus enable efficient peptide loading at the cell surface. The small molecules not only enhance peptide presentation by APC in vitro, but are also active in vivo where they substantially increase the fraction of APC on which displayed peptide is detectable. We propose that the small molecule quickly reaches draining lymph nodes together with the co-administered peptide and induces rapid loading of peptide before it is destroyed by proteases. Such compounds may be useful for enhancing the efficacy of peptide-based vaccines and other therapeutics that require binding to MHC class II molecules.
Keywords: MHC, Antigen Presentation, Peptides
Introduction
The MHC class II antigen presentation pathway serves an essential role in the induction of adaptive immune responses by CD4 T cells, and particular subsets of CD4 T cells control key aspects of the resulting response by provision of help to B cells (T follicular helper cells), coordination of responses to different classes of pathogens (Th1, Th2 and Th17 cells) and prevention of immune-mediated tissue damage (Foxp3+ regulatory T cells) (1, 2). The MHC class II pathway also plays a central role in the pathogenesis of many autoimmune diseases, and genome-wide studies have documented that the MHC class II region represents the most prominent susceptibility locus for a number of these diseases, including type 1 diabetes (3), multiple sclerosis (4) and rheumatoid arthritis (5). Much effort has been invested to develop therapeutics that act through the MHC class II pathway, but the cell biological mechanisms of peptide loading represent a major obstacle to efficient loading of MHC class II molecules with exogenously administered peptides or other types of therapeutics.
One of the central problems in peptide loading onto MHC class II molecules is that empty molecules quickly lose the ability to bind peptide (6–8). While a small fraction of molecules can slowly revert back to a peptide-receptive state, the majority of empty molecules lose functionality and aggregate (9). Prior to arrival of MHC class II molecules in the endosomal compartment, the peptide binding site is protected by the CLIP segment of invariant chain, and proteolytic degradation of invariant chain in endosomes then enables exchange of the CLIP peptide with peptides generated by proteolysis of other antigens (10, 11). The second major obstacle to efficient peptide loading is the long half-life of most MHC class II bound peptides (12), and rapid exchange thus requires a catalyst that accelerates dissociation of bound peptide. In the late endosomal peptide loading compartment, this function is executed by the HLA-DM (DM) protein, which accelerates dissociation of bound peptide and also facilitates association of new peptide by maintaining empty MHC class II molecules in a highly peptide-receptive state (13–19). Furthermore, the neutral pH at the cell surface considerably slows the dissociation of MHC class II bound peptides, and high affinity peptides are bound with half-lives of days to weeks under such conditions (12). Fast DM-catalyzed loading is thus restricted to a specialized low pH endosomal compartment (20), and exogenously administered peptides face prolonged exposure to proteases during transport to this site. Degradation of most of the administered peptide could be prevented by catalyzed loading at the cell surface or in an early endosomal compartment.
This challenge has spurred recent interest in identifying small molecules that can enhance the exchange of surface-presented peptides on APC with therapeutic peptides or proteins. Two structural mechanisms enable long-lived peptide display by MHC class II molecules: a series of hydrogen bonds between the MHC helices and the peptide backbone as well as occupancy of a set of MHC pockets by peptide side chains (21, 22). The hydrogen bonds between the MHC helices and the peptide backbone provide a sequence-independent binding mechanism that induces an extended conformation of the bound peptide. Important hydrogen bonds that contribute substantial binding energy are located close to the peptide N-terminus and involve backbone to backbone hydrogen bonds between the HLA-DR (DR) αchain (α51F and α53S) to the P-2 and P1 residues of the peptide as well as hydrogen bonds from DRβ side chains (β81H and β82N) to the backbone of the P-1 and P2 residues (22). How DM catalyzes peptide exchange is not fully understood, but it is known to interact with a lateral face of MHC class II close to the peptide N-terminus (23–25) and assumed to cause a conformational change that accelerates peptide dissociation (14, 26). Due to the large interaction area between DM and DR proteins, which spans the length of the extracellular domains of both proteins, small molecules with DM-like activity may help to define conformational changes in DR proteins that can result in peptide release.
Here we report a group of small molecules that substantially accelerates peptide loading of DR molecules. Similar to DM, these small molecules appear to act on DR proteins near the peptide N-terminus and accelerate both peptide dissociation and association. The small molecules are active over a broad pH range and can substantially enhance the display of peptides by APC both in vitro and in vivo. This class of small molecules may be useful for enhancing the therapeutic activity of DR binding peptides or for tethering proteins of interest to APC for long-lived display to T cells.
Materials and Methods
Fluorescence Polarization Assay
A previously described fluorescence polarization (FP) assay (16, 27) was used to study the kinetics of peptide binding to DR molecules. Soluble DR/CLIP complexes were expressed using Chinese hamster ovary (CHO) cell transfectants, and the linker connecting the CLIP peptide to the DRβ chain was cleaved with thrombin prior to peptide binding experiments (28). Three different DR/CLIP complexes were utilized: DR1 (DRA, DRB1*0101), DR15 (DRA, DRB1*1501) and DR4 (DRA, DRB1*0401). For assays involving DR4/CLIP, thrombin was added ~10 minutes prior to peptide exchange assays because this complex had a short half-life. For the FP assay with DR15, an Alexa488 labeled derivative of the human myelin basic protein (MBP) 85–99 peptide (MBP-488) was used in which the lysine at the P5 position of the peptide was modified to cysteine for attachment of a maleimide derivative of Alexa488 (ENPVVHFFC(Alexa-488)NIVTPR) (16). For the FP assay with DR1 and DR4, the influenza hemagglutinin (HA) 306–318 peptide was labeled with Alexa488 at the N terminus (HA-488, Alexa488-CGGGPKYVKQNTLKAT, cysteine and N-terminal linker underlined). Peptide binding assays were performed in 384-well plates in 40 μl volumes. Compounds in DMSO or DMSO controls were added in a 1 μl volume. Reactions were typically set up in a buffer containing 150 mM sodium chloride, 50 mM sodium citrate (pH 5.2), except for pH profiling experiments that used a citrate/phosphate buffer system (100 mM) in which pH steps were generated by mixing of equimolar stocks of citric acid and disodium hydrophosphate at different ratios. FP measurements were made using a Victor3 plate reader (PerkinElmer, Inc, Waltham MA), as previously described (16, 27) and fit to a single exponential binding function to extract rates (FP=FPmax*[1-exp(−rate*time)]). For peptide dissociation assays, a fluorescent DR15/MBP-488 complex was generated by incubation of DR15/CLIP (1 μM) with 500 nM MBP-488 for several hours at 37°C in sodium citrate buffer; unbound peptide was then removed with a gel filtration column (PD10; GE Healthcare, Piscataway NJ). Dissociation of MBP-488 from the DR15/MBP-488 complex (100 nM) was measured in the presence of J10 or J10-1 (100 μM) as well as unlabeled competitor peptide (50 μM).
Screening of small molecule libraries
The compound library consisted of approximately 83,000 small molecules, including compounds approved by the Food and Drug Administration (FDA), purified natural products as well as compounds purchased from Peakdale (High Peak, UK), Maybridge Plc. (Cornwall, UK), Cerep (Paris, France), Bionet Research Ltd. (Cornwall, UK), Prestwick (Ilkirch, France), Specs and Biospecs (CP Rijswijk, the Netherlands), ENAMINE (Kiev, Ukraine), Life Chemicals, Inc. (Burlington, Canada), MicroSource Diversity System’s NINDS custom collection (Gaylordsville, CT), Chembridge (San Diego, CA), ChemDiv (San Diego, CA), and several academic institutions. A series of computational filters, including filters for clogP and predicted solubility, were used to select compounds from these sources. In general, compounds adhered to Lipinski’s rules (i.e. molecular weight < 500, H-bond donors ≤ 5, H-bond acceptors ≤ 10 and logP < 5) and contained a low proportion of known toxicophores (i.e. Michael acceptors and alkylating agents) and unwanted functionalities (i.e. imines, thiols, and quaternary amines). The library was also optimized for molecular diversity.
The initial screening of modulators of peptide binding to DR15 was performed using 150 nM DR15, 30 nM MBP-488 and 20 nM DM in 150 mM sodium chloride, 50 mM sodium citrate (pH 5.2), while the subsequent evaluation of small molecule analogues was performed under the same conditions, but in the absence of DM. Soluble DM was expressed and purified as previously described (27) using the Baculovirus system (pAcDB3 plasmid with BaculoGold™ Baculovirus, BD Biosciences, San Jose, CA) and consisted of the α chain (signal peptide, residues α1-204 of the mature polypeptide followed by a FLAG tag) and β chain (signal peptide, residues β1-210 of the mature ploypeptide and a protein C tag) and had two of the three glycosylation sites mutated (αN165D and βN92D). DM was purified from Sf9 culture supernatants using an anti-Protein C column (Roche Applied Science, Indianapolis, IN).
All compounds had a unique screening code and were named in the text as follows: J10 (LDN-076340), J10-1 (LDN-134208), J10-2 (LDN-134151), J10-4 (LDN-112601), J10-5 (LDN-193172), J10-6 (LDN-074715), J10-7 (LDN-075399), J10-8 (LDN-134157), J10-9 (LDN-134177), J10-10 (LDN-112564), J10-11 (LDN-134148), J10-12 (LDN-134179), J10-13 (LDN-076344), J10-14 (LDN-075425), J10-15 (LDN-093678), J10-16 (LDN-093917), J10-17 (LDN-134259), J10-18 (LDN-134235), J10-19 (LDN-193150), J10-20 (LDN-187452), J10-21 (LDN-193152), J10-22 (LDN-193175), J10-23 (LDN-134234), J10-24 (LDN-193174), J10-25 (LDN-134203), J10-26 (LDN-193153).
Synthesis of J10 linked peptides
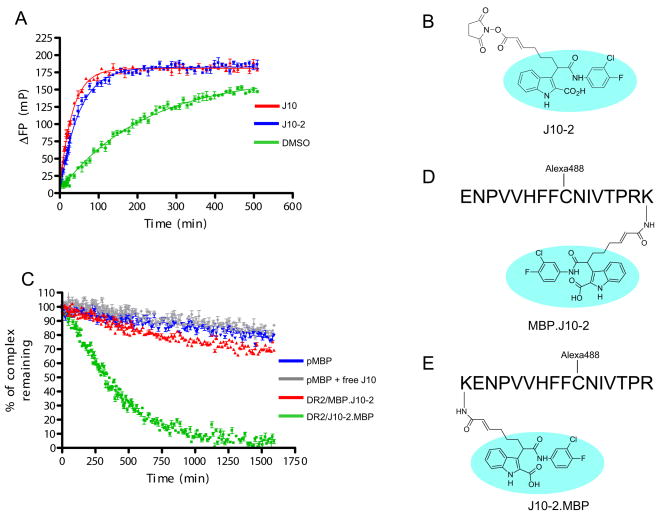
A succinimide ester derivative of J10 (J10-2) was attached to a lysine residue at the N-terminus of the MBP peptide (J10-2.MBP ; J10-2 – KENPVVHFFC(Alexa488)NIVTPR) or its C-terminus (MBP.J10-2; ENPVVHFFC(Alexa488)NIVTPRK – J10-2; JPT Peptide Technologies GmbH; Figure 4D & E). Alexa488 was attached using maleimide chemistry to a cysteine at the P5 position.
Figure 4.
N-terminal attachment of a J10 group to the peptide destabilizes the resulting DR/peptide complex. A. Linkage of a succinimide ester group to J10 (derivative J10-2) does not reduce J10 activity. 10 μM J10 (red) or J10-2 (blue) were incubated with 150 nM DR15/CLIP complex and 30 nM MBP-488 in 50 mM sodium citrate buffer, 150 mM NaCl, pH 5.2. ΔFP values were obtained by subtraction of FP values from wells containing only MBP-488. The binding curve of MBP-488 to DR15 in the absence of a small molecule (DMSO control) is shown in green. B. Structure of J10-2 illustrating the attachment site of the succinimide ester group. The J10 group is circled in light blue. C. The DR15/peptide complex with a J10 group linked to the peptide N-terminus has reduced stability. DR15 molecules were loaded with Alexa488 labeled peptides (D & E) carrying an N- or C-terminal J10 group (J10-2.MBP, MBP.J10-2). DR/peptide complexes (100 nM) with an N-terminal (green) or C-terminal (red) J10-2 group on the peptide were incubated in the presence of unlabeled pMBP competitor peptide (1 μM) and dissociation of the Alexa488 labeled peptides was tracked over time. Reactions with DR15/MBP-488 in the absence of J10 (blue) or in the presence of an equimolar concentration of free J10 (grey) are also shown. Free J10 showed no detectable activity at 100 nM. D & E. Structure of the J10 derivative covalently attached to lysine residues at the C-terminus (D) or N-terminus (E) of pMBP. An Alexa488 group was linked to these peptide at an internal cysteine residue.
Assessment of small molecule activity in cells
MBP peptide 85–99 (pMBP, ENPVVHFFKNIVTPR) presentation by MGAR cells (homozygous for the DR15 (DRB1*1501, DRB5*0101, DQB1*0602) haplotype; Health Protection Agency Culture Collections #88022014, Salisbury, UK) was measured using a recombinant antibody (MK16) that recognizes the DR15/pMBP complex (29). This antibody was modified by addition of a BirA biotinylation motif at the C-terminus of the heavy chain. The Fab fragment of this antibody was refolded from individual chains expressed in E. coli and the BirA site was biotinylated to generate a fluorescent MK16 tetramer with streptavidin-APC (Invitrogen, San Diego, CA). To directly measure peptide binding to DR15, 105 MGAR cells were incubated in a 96-well plate with pMBP at a concentration of 1–10 μM in the presence of J10, J10-1 or the inactive derivative J10-4 for the indicated times in DMEM, 10% FBS. Cells were then stained with either 1 μg of MK16 tetramerized with SA-APC or 100 ng of biotin labeled anti-DR mAb L243 and SA-APC in a volume of 100 μl for 1 hour on ice, washed twice with PBS, 0.5% FBS. The level of fluoresence was quantified by flow cytometry (FACSCalibur BD Biosciences). The background autofluorescence was determined by staining of MGAR cells that had not been exposed to pMBP.
To determine whether endocytosis is required for J10 function, MGAR cells were pre-fixed on ice using 1% formaldehyde in PBS for 5 minutes at 5×105 cells per ml and then washed three times with DMEM, 10% FBS. As a control, the same number of MGAR cells was incubated on ice in PBS without formaldehyde. 105 MGAR cells from each group were then incubated with 10 μM of pMBP in the presence of J10-1, J10-5 (both at 100 μM) or DMSO for 2 hours at 37°C in a volume of 100 μl. Peptide loading was assessed by MK16 staining as described above.
The effect of J10 and its derivatives on pMBP display was also assessed using a murine T cell hybridoma (7678, Dr. Lars Fugger, unpublished) that recognizes the DR15/pMBP complex. 100 μl of MGAR cells (5 × 105/ml) were pre-incubated with pMBP (4.57 to 123 nM) in the presence of small molecules (100 μM) for two hours at 37°C and washed twice to remove free peptide and small molecule. Peptide-pulsed MGAR cells (5 × 104) were then co-cultured overnight at 37°C with 7678 cells (5 × 104) and IL-2 release was assayed from 50 μl of supernatant using a Mouse IL-2 Flex Set Cytometric Bead Array (BD Biosciences, San Jose, CA). The same experimental design was also used to examine presentation of the influenza hemagglutinin 306–318 peptide (pHA, PKYVKQNTLKAT) by PRIESS cells (homozygous for the DR4 (DRB1*0401) haplotype; Health Protection Agency Culture Collections #86052111) to a human DR4 restricted T cell clone (clone HA:D7) specific for pHA. This T cell clone was generated by flow cytometric sorting of T cells stained with a DR4/pHA tetramer from a pHA responsive T cell line established from a healthy donor.
Assessment of in vivo activity
Transgenic mice that express DR15 as well as a DR15-restricted and pMBP-specific TCR (30) were used as a mouse model to assess the in vivo efficacy of J10-1. Biotinylated pMBP (bio-pMBP, biotin-SGSGENPVVHFFKNIVTPR) at a dose of 65 μg (36 nmol) was injected with or without J10-1 (5–20 nmol) in 10 mM phosphate buffer pH 7.4, 30 % DMSO in a total volume of 10 μl into the base of mouse ears, similar to a previously described procedure, using a 31 gauge insulin syringe (BD, Franklin Lakes NJ) (31, 32). DMSO was added to improve the solubility of injected compounds and did not cause alterations in bio-pMBP uptake, swelling at the injection site or differences in cell numbers recovered from draining lymph nodes (data not shown). 16 to 20 hours after injection, the draining superficial parotid lymph node (located dorsal to the junction between the superficial temporal and maxillary veins) (33) was removed, minced and cells were dispersed by collagenase D (Roche, Indianapolis IN) treatment for 30 minutes at 37°C with frequent agitation. Inguinal and non-draining contra-lateral parotid lymph nodes were used as negative controls. Lymph node cells (2 × 106 per condition) were stained for 1 hour on ice with 1 μg SA-APC to identify cells displaying bio-pMBP, 25 ng L243-FITC (which binds to the DRα chain), 1 μl anti-CD11c-PE/Cy7 (eBioscience) and 0.5 μl anti-CD19-Pacific Blue (BioLegend) to characterize APC populations as well as 2 μl anti-CD3-PerCP (BD Pharmingen) to exclude T cells. Cells were washed twice with PBS, 0.5% FBS and 10 μl ViaProbe (BD Pharmingen) was added to 300 μl of cells in PBS, 0.5% FBS just prior to analysis to exclude dead cells. Cells were analyzed by flow cytometry on a FACSAria (BD Biosciences). SA-APC staining was analyzed on B cells (CD19+) that expressed high levels of DR (L243hi, top 20% of L243+ cells), and the percentage of SA-APC positive cells in this population was calculated.
Enhancement of peptide presentation by J10-1 was confirmed by co-culture of lymph node cells from injected mice (2.5 × 104 to 4 × 105 cells per well) with the 7678 T cell hybridoma (5 × 104 cells/well) in DMEM supplemented with 10% FBS. Following overnight culture, supernatants were assayed for IL-2 secretion using a Mouse IL-2 Flex Set Cytometric Bead Array (BD Biosciences).
Toxicity studies
DR4 transgenic mice (34) (Taconic, Germantown NY) were injected subcutaneously with 10 mg/kg J10-1 (~500 nmol per mouse) or PBS alone (three mice per group) on a daily basis for 7 days. On day 8, mice were sacrificed and draining inguinal lymph nodes and spleens were removed for flow cytometric analysis of cell populations, using four sets of stains: 1. Cell populations of the adaptive immune system (CD4-Pacific Blue [Caltag Laboratories], CD8-APC [BD Pharmingen], CD19-PE [BD Pharmingen], CD49b-FITC [BD Pharmingen]); 2. T cell activation state (CD4-Pacific Blue, CD8-APC, CD44-bio:SA-APC [eBiosciences:Invitrogen], CD69-FITC [BD Pharmingen]). 3. Regulatory T cells (CD3-FITC [BD Pharmingen], CD4-Pacific blue, CD25-PE [BD Pharmingen], GITR-APC [eBioscience]). 4. Dendritic cell number and activation state (CD11c-PE/Cy7 [eBioscience], CD19-Pacific Blue [BioLegend], CD80-APC [eBioscience], CD86-bio:SA-PE [eBioscience, Invitrogen], L243-FITC). Mice were fixed in Bouin’s solution (Sigma, St. Louis, MO) and necropsy and histological analysis of 20 tissues was performed by the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core. The pathologist was not aware of the treatment status of the mice. All animal studies were approved by The Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Results
Small molecules that catalyze peptide loading
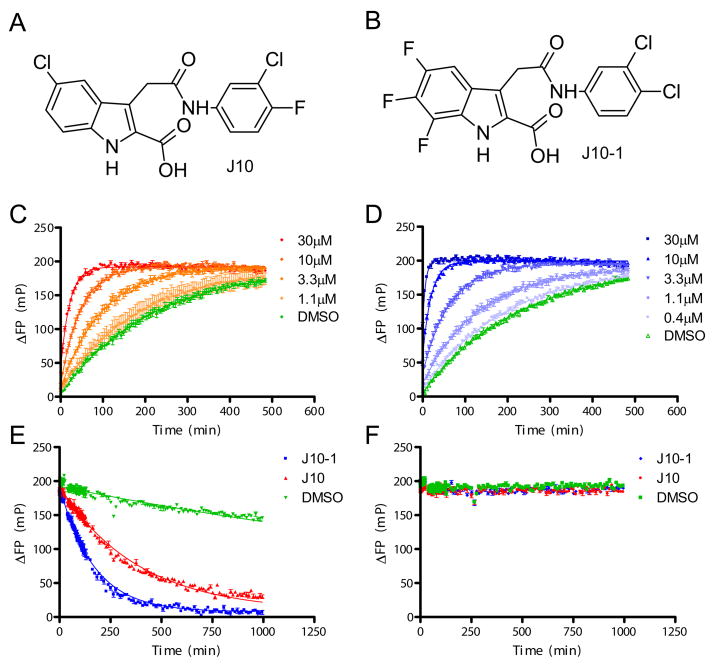
A high-throughput screening effort (83,000 compounds) was undertaken to identify small molecules that modulate peptide loading onto DR molecules. A class of small molecules (11 active analogues, data not shown) was identified that accelerated peptide exchange in the absence of DM (Figure 1) and the most active of these compounds, J10, was selected for further characterization. The ability of J10 to accelerate peptide loading was examined with a real-time peptide binding assay based on a fluorescence polarization (FP) readout. For that purpose, a peptide of myelin basic protein (pMBP, res. 85–99) that binds to DR15 (DRA, DRB1*1501) (35) was labeled with Alexa488 (MBP-488). In this FP assay, the small fluorescent peptide tumbles quickly in solution and thus gives low FP readings (~120mP), while the complex of fluorescent peptide and DR15 tumbles more slowly and produces higher FP readings (~350mP). In this manner binding curves could be measured in real time both in the presence and absence of small molecules and fit to a single exponential binding equation to extract rates. In this assay peptide loading could be accelerated 4.7 and 11.5 fold by J10 at concentrations of 10 and 30 μM, respectively.
Figure 1. Properties of the small molecule catalysts of peptide exchange.
A & B. Chemical structures of the original lead compound J10 and the most potent analog prepared, J10-1. C & D. Acceleration of peptide binding to DR15 by J10 (C) or J10-1 (D) measured using a fluorescence polarization (FP) assay. Increasing concentrations of J10 (1.1–30 μM) or J10-1 (0.4–30 μM) were incubated with 150 nM DR15/CLIP complex and 30 nM MBP-488 in 50 mM sodium citrate buffer, 150 mM NaCl, pH 5.2. ΔFP values (in millipolarization units, mP) were obtained by subtraction of FP values from wells containing only MBP-488. The binding curve of MBP-488 to DR15 in the absence of a small molecule is shown in green. E & F. J10 and J10-1 catalyze peptide dissociation from DR15. Dissociation of the MBP-488 peptide from the DR15/MBP-488 complex (100 nM) was measured over time by FP in the presence of 100 μM J10 (red line), 100 μM J10-1 (blue line) or no small molecule (DMSO control, green line). MBP-488 dissociation only occurred in the presence of 50 μM unlabeled pMBP competitor peptide (E), but not in its absence (F), indicative of rapid rebinding of MBP-488 to DR15.
A structure-activity relationship study was undertaken for which seventy-seven J10 analogs were prepared. A total of thirteen compounds had greater than 2-fold higher activity than J10 (Figure S2), and the most active compound, J10-1, had approximately 5-fold higher activity than J10 (Figure 1B & D). J10-1 could accelerate peptide exchange 15.9 fold at a concentration of 10 μM and 66.4 fold at 30 μM. These results demonstrate that derivatives of J10 greatly accelerate the loading of peptides onto DR15 molecules.
The extensive optimization efforts also allowed us to define the structural requirements for J10 activity (Figure S1). The placement of all hydrogen bond donors and acceptors was critical for J10 activity as removal or exchange (such as reversing the amide linker between the two ring structures) abrogated activity. The carboxylic acid moiety attached to the indole ring was also important for activity. When this group was converted to an ester (J10-4) activity was lost, while activity was retained upon replacement with a tetrazole, a well recognized carboxylic acid mimic (J10-9) (Figure S1A). Addition of electron withdrawing groups to either ring structure (except the 4-position of the indole or the 2-position of the phenyl) significantly improved activity, while electron donating groups (such as in J10-5) decreased activity (Figure S1B). The phenyl ring in J10 could be replaced with other heteroaromatic structures such as substituted isoxazoles, or substituted pyridines, but replacement of the phenyl ring with non-aromatic moieties such as a cyclohexyl or an isopropyl group abolished activity (Figure S1C).
Catalytic action on multiple DR allotypes
In cells, peptide binding is an exchange process involving dissociation of a DR-bound peptide and association of a new ligand. In the experiments shown above, this exchange reaction involves dissociation of the low affinity CLIP peptide from DR15 and binding of MBP-488. In order to directly investigate whether J10 and related compounds act on the first step, the dissociation of the DR-bound peptide, DR15 was loaded with MBP-488 and dissociation of the labeled peptide was followed in real time by FP. The complex between DR15 and MBP-488 used in this assay is much more stable than DR15/CLIP and thus provided a stringent test of J10 activity. In the absence of a small molecule catalyst, peptide dissociation was very slow (complex half life (t½) of 2097 minutes) even in the presence of a high concentration of competitor peptide. J10 and J10-1 greatly accelerated dissociation of MBP-488 (t½ of 269 and 116 minutes, respectively), indicating that they could induce dissociation of even a high affinity peptide from DR15 (Figure 1E). In contrast, a previously reported group of small molecules could only induce dissociation of low affinity peptides from DR molecules and this activity required much higher compound concentrations (millimolar range) (36).
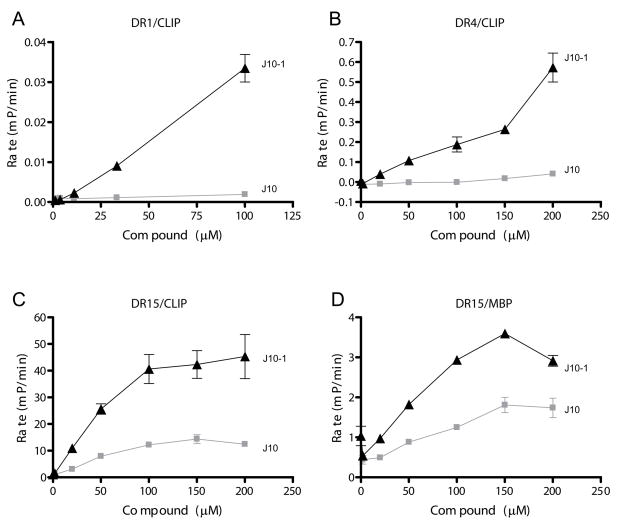
An important question was whether these compounds act only on DR15 or also on other DR allotypes. The analysis was therefore extended to DR1 (DRB1*0101) and DR4 (DRB1*0401), using the influenza hemagglutinin 306–318 peptide labeled with Alexa488 (HA-488) as a probe (Figure 2A & B). The overall rate of peptide binding in the absence of the small molecules differed substantially between these DR allotypes, and the slowest rate was observed for binding of HA-488 to DR1, which has the highest affinity for the CLIP peptide (37). Similarly, the rate of MBP-488 binding to DR15 was >10-fold faster when the low affinity complex of DR15 and CLIP rather than the high affinity complex of DR15 and pMBP was utilized (Figure 2C & D). The small molecule catalyst J10-1 had much higher activity than J10 with all three DR allotypes. Importantly, the activity of this small molecule was not restricted to a particular DR allotype (comparison of DR1, DR15 and DR4), a particular input DR/peptide complex (comparison of DR15/CLIP and DR15/pMBP) or a particular peptide moving into the site (comparison of MBP-488 or HA-488) (Figure 2). However, J10 and J10-1 had no discernible activity on a tested mouse MHC class II molecule (I-Ag7), even though human DM could accelerate peptide binding to this MHC protein (data not shown).
Figure 2.
The small molecules catalyze peptide binding to multiple DR molecules. The rate of peptide binding in the presence of increasing concentrations of J10 or J10-1 was determined for the interaction of HA-488 with DR1 and DR4 (DR1/CLIP and DR4/CLIP as input complexes, A & B, respectively) as well as the interaction of MBP-488 with DR15 (DR15/CLIP or DR15/pMBP as input complexes, C & D, respectively). Reactions were performed with DR/peptide complexes at 150 nM and fluorescent peptides at 30 nM in citrate buffer, pH 5.2 except for the assay with DR1 (200 nM DR1/CLIP and 100 nM HA-488). Initial rates of the reaction were calculated based on mP changes per minute in the FP assay (errors bars represent standard deviation of triplicate determinations). Rates of non-catalyzed peptide binding were very low for all DR/peptide complexes.
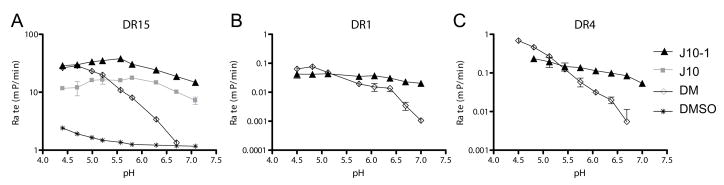
DM is most active at the acidic pH of the peptide loading compartment. In contrast, both J10 and J10-1 were active over a broad pH range (Figure 3), which may allow peptide exchange to occur in environments where peptides are more stable, such as the cell surface or early endocytic vesicles. For DR15, pH dependent peptide exchange was examined in the presence of DM, J10, J10-1 or in the absence of a catalyst (Figure 3A). The activity of DM declined substantially with increasing pH (acceleration of peptide binding of 15-fold at pH 4.7, 8-fold at pH 5.6; no activity at pH 7.1), while J10-1 remained active over this pH range (15.6-fold at pH 4.7, 28-fold at pH 5.6, 12.6-fold at pH 7.1). For the other two DR allotypes (DR1 and DR4), spontaneous exchange in the absence of DM was too slow to be reliably measured and the analysis therefore focused on a comparison of the pH profile of J10-1 and DM (Figure 3B & C).
Figure 3.
Small molecule catalysts are active over a broad pH range. The rate of fluorescent peptide binding (change in initial rate in mP/min) was determined in the presence of the small molecules J10 or J10-1 (both at 100 μM), DM (50 nM) or a solvent control (DMSO) using MBP-488 as a probe for DR15 (DR15/CLIP as input) and HA-488 as a probe for DR1 and DR4 (DR1/CLIP and DR4/CLIP as input complexes). The concentrations of DR/peptide complexes and fluorescent peptides were as reported in Figure 2. Reactions were performed in a 100 mM citric acid/phosphate buffer system adjusted to the indicated pH. Due to the slow binding kinetics of HA-488 to DR1 and DR4 only curves with DM and J10-1 are shown for these allotypes.
Destabilization of DR/peptide complex by a J10 group attached to the peptide N-terminus
In order to globally define the DR region to which J10 might bind, a J10 derivative was covalently attached to either the N- or C-terminus of pMBP (Figure 4). We reasoned that a DR/peptide complex may have reduced stability if the J10 binding site is located in the vicinity of one of the peptide termini because the linked J10 group could then engage its binding site and promote dissociation of the DR-bound peptide. This experimental approach required identification of a site on J10 to which a linker could be attached without a detrimental effect on activity. Chemically tractable sites such as the nitrogen of the indole, the carboxylic acid or the amide between the two rings were excluded as potential sites. However, a linker containing a succinimide ester group could be attached to the methylene carbon between the amide carbonyl and the indole without loss of activity (Figure 4B). This succinimide ester derivative of J10 (J10-2) retained full activity and catalyzed the binding of MBP-488 to DR15 (Figure 4A).
This chemistry was then used for covalent attachment of J10 to lysine residues placed at the N- or C-terminus of pMBP (Figure 4D and 4E). These peptides were also labeled with Alexa488 at an internal cysteine residue so that peptide dissociation could be directly compared to MBP-488 labeled at the same position. The linked J10 group was placed outside of the P1-P9 peptide core, at the P-5 position for N-terminal attachment and at P12 for C-terminal linkage, making it unlikely that it sterically interfered with peptide binding (38). MBP-488 and the fluorescent J10-linked peptides were preloaded into the DR15 binding site and the stability of the complexes was measured in the presence of an excess of unlabeled peptide to prevent rebinding of labeled peptide. When the J10 group was attached to the peptide C-terminus, only a small fraction of the fluorescent peptide dissociated during the observation period. However, N-terminal placement of the J10 group substantially reduced the stability of the complex (half life of only 300 minutes) (Figure 4C). The fluorescent DR/peptide complexes were used at a concentration (100 nM) at which little activity was observed with free J10, in order to exclude the possibility of J10 acting on neighboring molecules (Figure 4C grey line). These results indicate that the J10 group can access its binding site on the DR15 protein when it is linked to the peptide N-terminus.
J10 catalysts enhance peptide presentation to T cells in vitro
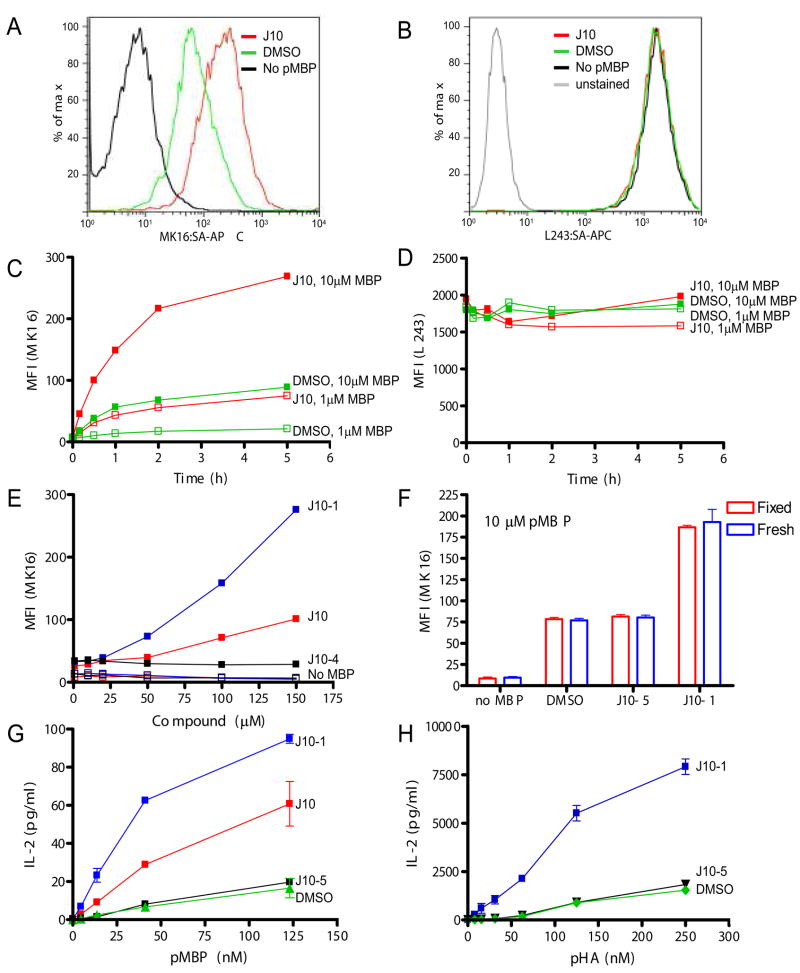
In vitro, DM acts as a typical catalyst that accelerates the rate of a reaction but does not alter the equilibrium (39), and the same behavior was observed for J10 in the biochemical assays described above. However, the situation may be different in cells because a substantial amount of the peptide may be degraded before equilibrium is reached or may not reach the compartment that contains DM. A mAb (MK16) (29) that binds to the DR15/pMBP complex but not to DR15 or free pMBP was used to directly quantify the effect of J10 catalysts on peptide presentation by APC. J10 substantially enhanced surface display of pMBP by MGAR cells, an EBV transformed B cell line homozygous for the DR15 haplotype (Figure 5A). The kinetics of pMBP display by MGAR cells were then tested over a five-hour time course at two peptide concentrations (1 or 10 μM) (Figure 5C). J10 not only accelerated pMBP binding to DR15 but also substantially increased the total amount of pMBP displayed on the cell surface. In fact, in the presence of J10 a 10-fold lower concentration of peptide was required for the same level of peptide display than in its absence. J10-1 had substantially higher activity than J10 in this cell based assay, consistent with the biochemical studies described above (Figure 5E). As a specificity control, a small molecule that was inactive in the biochemical assay (ester of J10, compound J10-4) was also tested, and this compound did not enhance peptide display by these APC (Figure 5E). Enhanced display of pMBP was also not due to an increase of total HLA-DR on the cell surface (Figure 5B and D).
Figure 5.
J10 and J10-1 are active in cells. A. J10 increases surface levels of DR15/pMBP in MGAR cells. MGAR cells were incubated with pMBP (1.7 μM) and J10 (100 μM, red line) or DMSO solvent control (green line) in DMEM media, 10% FCS for 30 minutes at 37°C. The cells were then labeled with a complex of biotinylated MK16 Fab and streptavidin-APC and analyzed by FACS; the MK16 Fab binds to the DR15/pMBP complex but not to either component alone (29). Cells not pulsed with peptide (black line) were used as a negative control to define background labeling of MK16. B. Staining of cells with the anti-DR mAb L243 showed that neither J10 nor pMBP affected the total amount of DR on the cell surface. C. J10 not only increases the kinetics of pMBP binding, but also the total amount of displayed peptide. MGAR cells were incubated with 1 μM or 10 μM pMBP and either 100 μM J10 (red) or DMSO solvent control (green). Cells were stained with the complex of biotinylated MK16 Fab and streptavidin-APC and the median fluorescence intensity (MFI) was measured by FACS over a course of five hours. D. The overall amount of DR on the cell surface (L243 staining) remained constant at all time points during the experiment shown in Figure 5C. E. J10-1 has increased activity in cells compared to J10. Cells were incubated with 1 μM pMBP (closed squares) for 30 minutes, either together with J10-1 (blue), J10 (red) or the inactive J10 analog J10-4 (black). Cells that were not pulsed with peptide (no pMBP) but still exposed to compounds were used as controls (open squares - J10 [red], J10-1 [blue] J10-4 [black]). F. J10-1 can enhance peptide loading at the cell surface. Fixed MGAR cells were incubated for 2 hours with pMBP (10 μM) in the presence of J10, J10-1, inactive analog J10-5 (100 μM) or DMSO solvent control and then stained with MK16. G. J10 increases presentation of pMBP to T cells. MGAR cells were pulsed with different concentrations of pMBP in the presence of 100 μM J10-1 (blue), 100 μM J10 (red), 100 μM J10-5 (black) or DMSO solvent control (green). Washed, peptide-pulsed B cells were then co-cultured with a DR15/pMBP specific T cell hybridoma and IL-2 production was quantified. H. J10-1 enhances peptide presentation to a human T cell clone specific for the DR4/pHA complex. PRIESS cells were incubated overnight with pHA at the indicated concentrations and 100 μM J10-1 (blue), 100 μM inactive analogue J10-5 or DMSO solvent control (green). Cells were washed, co-cultured with the human T cells and IL-2 production was quantified. The inactive J10-5 derivative was used for the experiments in D-F because J10-4 had been used up.
The broad pH spectrum of J10 activity may enable catalyzed peptide loading in compartments in which peptide exchange is normally very limited, such as the cell surface or early recycling endosomes. The ability of J10-1 to increase peptide presentation on the surface of fixed MGAR cells was tested because fixed cells are unable to internalize antigen through the endocytic pathway. Cells were fixed on ice in 1% formaldehyde before being incubated with 10 μM pMBP in the presence of J10-1, an inactive derivative J10-5 (both at 100 μM) or DMSO control for 2 hours at 37°C. J10-1 substantially increased pMBP binding to DR15 on these fixed APC indicating that it can catalyze peptide exchange at the cell surface (Figure 5F). No difference was seen in pMBP binding between fresh and fixed cells in any of the conditions tested. Nevertheless, these data do not exclude the possibility that J10 could also act in the endosomal pathway when used in conjunction with a more stable antigen.
As the next step, peptide presentation to T cells was investigated. MGAR cells were pulsed with pMBP in the presence or absence of small molecules (J10, J10-1 or inactive compound J10-5), and the cells were then co-cultured for 24 hours with a T cell hybridoma that recognizes the DR15/pMBP complex. The T cell hybridoma produced larger quantities of IL-2 when MGAR cells had been loaded with pMBP in the presence of J10 or J10-1, indicating that enhanced peptide loading by the small molecules results in a stronger T cell response (Figure 5G). The analysis was then extended to T cells specific for another DR/peptide combination using a human T cell clone specific for pHA bound to DR4 (DRB1*0401). Only J10-1 was tested in this experiment because J10 had limited activity on DR4 (Figure 2B). Again, co-culture of the T cells with B cells (PRIESS cells, homozygous for DR4) that had been pulsed with peptide in the presence of J10-1 enhanced IL-2 production by the T cells, while an inactive J10 analog (J10-5) had no effect (Figure 5H). Taken together, these results demonstrate that the J10 family of small molecules substantially increases peptide presentation by APC to CD4 T cells.
In vivo activity of the small molecules
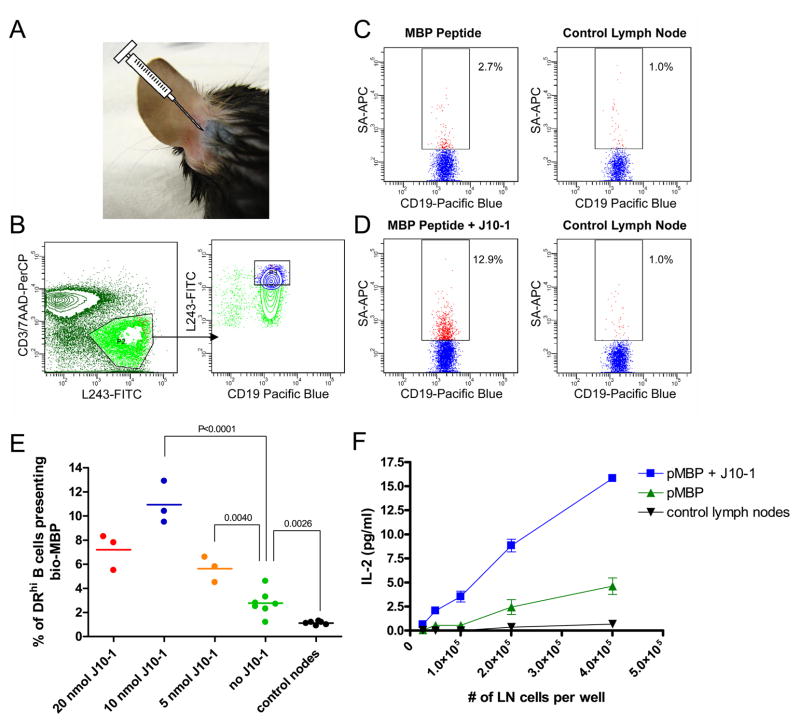
Vaccines are most commonly administered by subcutaneous injection and the resulting T cell response takes place in lymph nodes draining the injection site. Proteins and peptides flow through afferent lymphatic vessels from the subcutaneous tissue into the subcapsular sinus of lymph nodes. Electron microscopy studies have shown that the floor of the subcapsular sinus has small gaps through which proteins and peptides can rapidly reach follicular B cells (40). The Jenkins lab demonstrated that a fluorescent antigen can be readily detected in the draining lymph node after 0.5–3.5 minutes following intradermal injection into the mouse ear (32). This small space can accommodate approximately 10 μl of liquid (Figure 6A) and drains exclusively to the superficial parotid lymph nodes during the time course of the experiment (33). We therefore examined whether the small molecules could enhance DR15 binding of biotinylated pMBP in DR15/TCR transgenic mice (30) following injection at this site. This approach offered two advantages: 1) the mouse ear is delicate and any local inflammation or necrosis due to toxicity of the small molecule would be readily detected, 2) the peptide reaches a single, well-defined lymph node. The contra-lateral parotid lymph node was used as a control to examine whether presentation of pMBP was indeed confined to the draining lymph node. In initial experiments, we examined peptide binding by both CD19+ B cells and CD11c+ dendritic cells but found that the frequency of dendritic cells was too low (~1.5–2%) to permit detection of biotinylated peptide with streptavidin-APC. In subsequent experiments, we therefore focused the analysis on B cells, which can act as efficient APC (41, 42). The MK16 antibody could not be used for these experiments because of background binding to lymph node cells from mice not injected with pMBP.
Figure 6.
In vivo activity of J10-1. A. Illustration of the injection site. Trypan blue (10 μl) was injected at the base of the ear. B-D. J10-1 enhances binding of co-injected peptide in the draining superficial parotid lymph node, but not the contralateral lymph node. Biotinylated pMBP (36 nmol, 65 μg) was injected alone or in combination with J10-1 (10 nmol, 4.17 μg) at the base of the ear. The injectant (10 μl) was buffered with 100 mM sodium phosphate buffer, pH 7.5 and contained 30 % DMSO. Mice were sacrificed 16 to 20 hours post injection and lymph nodes were harvested. Cells were released from the lymph nodes by collagenase D treatment and stained with L243-FITC, CD3-PerCP, Viaprobe, streptavidin-APC, CD11c-PE/Cy7 and CD19-Pacific Blue. FACS analysis was performed on a BD Aria on 2×106 cells. B. Gating procedure. Cells were first gated on the lymphocyte population (not shown) and T cells (CD3+) and apoptotic cells (ViaProbe+) were excluded (B, left panel). B cells (CD19+) that were DRhi (top 20% of L243+ cells) were gated because peptide display was only detected on those cells (B, right panel). C & D. The biotinylated pMBP bound by these B cells was quantified with streptavidin-APC for injections with pMBP alone (C) and co-injection of pMBP with J10-1 (D). The parotid lymph node contra-lateral to the injection site was used as a negative control. E. Optimal J10-1 dose for enhancing peptide display in draining lymph nodes. Data were expressed as the percentage of CD19+/DRhi cells stained with streptavidin-APC for detection of surface presented biotinylated peptide. Each point represents data from a single draining lymph node. P values were calculated using an unpaired two tailed t test. F. Co-administration of J10-1 increases peptide presentation to T cells. Lymph node cells from injected mice were co-cultured with a T cell hybridoma that recognizes the DR15/pMBP complex and IL-2 production by T cells was quantified.
Injection of J10-1 did not cause any local inflammation or necrosis. We then injected the biotinylated pMBP (36 nmol, 65 μg) together with J10-1 (10 nmol, 4.17 μg) in a volume of 10 μl at this site and examined surface display of the biotinylated peptide by APC in the draining lymph node after 16–20 hours. This time frame was chosen to ensure that only biotinylated peptide bound to DR15 would be detected. When peptide was injected without the small molecule, a small population of B cells that displayed biotinylated pMBP could be detected which expressed a high level of HLA-DR (Figure 6B & C). A substantially larger population of streptavidin-APC labeled B cells was detected when peptide and small molecule were co-administered (11.6–12.9% of DRhigh B cells). In contrast, when the peptide was injected without the small molecule, the streptavidin-APC positive population was only modestly increased (2–4.6%) compared to control lymph nodes (0.7–1.8%). Neither peptide injection alone nor the combination of peptide and small molecule changed overall DR levels (L243 staining) on the surface of APC compared to non-draining lymph nodes. The small molecule was tested over a dose range of 5–20 nmol, and a dose of 10 nmol resulted in the highest level of pMBP staining on B cells (Figure 6E). Administration of J10-1 also increased presentation of the pMBP to T cells, as shown by increased IL-2 production of a pMBP specific and DR15 restricted T cell hybridoma co-cultured with draining lymph node cells isolated from mice co-injected with pMBP and J10-1 (Figure 6F). In the absence of hybridoma T cells, IL-2 could not be detected in cultures of draining lymph node cells even though the mice carried the transgene for a TCR that recognized pMBP presented by DR15; this result that may be explained by tolerance mechanisms. These results demonstrate that J10-1 is active in vivo and that it induces a substantial increase in peptide presentation by B cells in lymph nodes draining the injection site.
Absence of non-specific immune cell activation
While only small quantities of J10-1 were required to enhance the presentation of a co-injected peptide within the draining lymph node, it was nevertheless necessary to determine if J10-1 could cause toxicity or non-specific activation of any immune cell population during the time course of the experiments described above that might account for the observed increase in peptide presentation. To ensure mice could respond to J10-1 but also had a diverse T cell repertoire, DR4 transgenic mice (34) were injected subcutaneously on a daily basis for seven days with a high dose (10 mg/kg, ~500 nmol) of J10-1 in PBS or PBS as a control (two female and one male mouse per group). No changes in behavior or activity were noted during the one week observation period and the weight remained stable. Mice were sacrificed on day 8 and spleens and lymph nodes removed for FACS analysis. Necropsy and a comprehensive histological analysis of 20 organs were performed by a pathologist who was blinded of the treatment status of the mice. No differences were noted between the two groups; two males (one injected with J10-1 and one injected with PBS) showed very mild kidney abnormalities that were absent in the female mice; these abnormalities may be caused by the DR4 transgene.
We further examined whether small molecule administration altered the composition of immune cell subsets in lymph nodes draining the injection site or in the spleen by performing a comprehensive FACS analysis using the markers listed in Table S1. There were no differences in the composition of CD4 and CD8 T cell subsets, the frequency of memory or activated T cells or the frequency of regulatory T cells. Furthermore, no changes in the frequency of immature or mature dendritic cells, B cells or NK cells were observed. We conclude that there is no apparent acute toxicity even at a high dose of this small molecule as well as no evidence of non-specific immune system activation or depletion of a particular cell population. The increase in peptide display detected in the in vivo experiments is therefore due to specific action of J10-1 on DR molecules.
Discussion
This study describes a group of small molecules that significantly accelerate the rate of peptide loading onto DR molecules. An in depth medicinal chemistry effort led to a series of derivatives with substantially increased activity compared to the initial compound and demonstrated key features required for catalysis of peptide exchange. These compounds are active in vivo and substantially increase the display of co-injected peptide by local APC in lymph nodes draining the injection site.
For peptides to become good immunogens it is critical that fast loading is accomplished in environments with limited proteolysis. This concept is illustrated by the fact that proteins which are more resistant to proteolysis are better immunogens because they survive longer in the endocytic pathway (43). When injected in vivo, the time frame during which peptide can be loaded onto MHC molecules is short because of rapid degradation, and the J10 family of compounds described here act during this critical time frame by making MHC class II binding sites available. The broad pH activity range of these compounds enables exchange at the cell surface and in early endocytic compartments with a neutral or slightly acidic pH and limited protease activity.
Fluorescent protein has been shown to reach draining lymph nodes through afferent lymphatic channels within minutes following subcutaneous injection (31, 32). The co-injected peptide and small molecule may thus quickly reach the draining lymph node where the small molecule facilitates rapid loading of the peptide before it is degraded. The action of the small molecule is likely to be transient and local because the compound is diluted below the effective concentration range once it reaches the blood stream. However, the effect of the compound can be long-lasting in the local lymph node environment because it can substantially enhance display of peptides with a long half-life to T cells. The small dose that is required for this transient and local action in draining lymph nodes substantially reduces the potential for side effects that could be caused by toxicity or systemic changes of the peptide repertoire on APC. In fact, even repeated administration of 50-fold larger quantities of compound failed to reveal acute signs of toxicity or changes in the composition of immune cell populations.
Several other small molecules have been described that facilitate peptide loading onto DR molecules, but to our knowledge none of these have been shown to be active in vivo. The most active compound in the first set was parachlorophenol (pCP) (36) but very high concentrations of this compound were required for in vitro peptide loading (millimolar range), making it unlikely that it can be utilized in vivo. Also, the requirement for such high compound concentrations will make it difficult to identify the binding site of these molecules and their precise mechanism of action. The second set of compounds were adamantane derivatives (44) that were active at lower concentrations (100 μM for substantial activity), but only acted on a subset of DR molecules. They accelerated peptide exchange by DR molecules with a large P1 pocket (glycine at DRβ 86), but not DR molecules with a smaller P1 pocket (valine at DRβ 86). Modeling studies showed that the compound could fit into the P1 pocket of DR molecules with a glycine at DRβ 86, and the authors suggested that these compounds may facilitate peptide loading by preventing collapse of this hydrophobic pocket. The set of small molecules described here act on DR molecules with a valine (DRB1*1501) or glycine (DRB1*0101, DRB1*0401) at DRβ 86, and their mechanism of action may therefore be different from the adamantane derivatives with which they share no structural similarities. Short synthetic peptides have also been shown to facilitate peptide exchange at high concentrations and may prevent denaturation of empty DR molecules by transiently occupying the binding site (45).
The precise mechanism of action of DM remains unknown because it has thus far not been possible to crystallize the complex of DM and a MHC class II molecule. Thorough mutagenesis studies have identified a large interface between DM and DR that spans the length of the ectodomains of both proteins (23, 24). Small molecules may therefore be useful probes to study conformational states of MHC class II molecules that rapidly release the bound peptide. The functional studies revealed similarities between DM and the J10 family of compounds that are worth noting. Both facilitate peptide dissociation and association irrespective of peptide sequence and appear to bind in the vicinity of the peptide N-terminus (14–16, 24, 46). Mutagenesis studies have identified residues on the DRα chain close to the peptide N-terminus that are required for DM action (23, 25). The fact that J10-family compounds act on all tested DR molecules indicates that they could bind to the non-polymorphic DRα chain, but it remains possible that they bind to a surface feature conserved among the polymorphic DRβ chains. Structural studies will thus be required to determine whether J10 compounds bind to a similar site as DM and to elucidate conformational changes that facilitate peptide release from DR molecules.
The small molecule catalysts thus enable efficient loading of peptides in compartments that lack the natural catalyst DM and thereby address one of the critical problems of peptide-based therapeutics, their rapid degradation by proteases. Small molecules that accelerate peptide loading may be useful for enhancing the efficacy of peptide-based vaccines directed at cancer antigens and infectious agents. They could also potentially be used to enhance display of self-peptides for induction of T cell tolerance in autoimmune diseases and to improve presentation of Copaxone by HLA-DR proteins to T cells. Copaxone is a co-polymer of four amino acids that binds to HLA-DR molecules and induces regulatory CD4 T cells (47). It is currently used for the treatment of MS, but reduces relapses by only approximately 30% (48). Copaxone lacks a stable three-dimensional structure which renders it highly susceptible to proteolysis. Rapid loading by a DM-like catalyst at the cell surface could thus increase presentation of Copaxone-derived peptides. It may also be possible to covalently link J10 to such therapeutics using the chemistry reported here so that they will be equipped with a small molecule with DM-like activity that facilitates their binding to MHC class II molecules. The compounds reported here thus have a number of potential applications for MHC class II based therapeutics.
Supplementary Material
Acknowledgments
We would like to thank Jake Ni for his work optimizing the robotics during the high-throughput screening effort, Marcie Glicksman for her insightful discussion on this work as well as Rod Bronson and Anoop Kavirayani for their assistance in interpreting the histopathology results.
This work was supported by grants from the National Institutes of Health (RO1 NS044914, RO1 AI57493 and PO1 AI045757 to KWW). MJC, XX, GDC and RLS were supported by funds from the Harvard NeuroDiscovery Center.
References
- 1.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annual review of immunology. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SC, Jenkins SC, Palmer SM, et al. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 4.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. The New England journal of medicine. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 5.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natarajan SK, Assadi M, Sadegh-Nasseri S. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J Immunol. 1999;162:4030–4036. [PubMed] [Google Scholar]
- 7.Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 8.Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, Davis MM, McConnell HM. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 9.Joshi RV, Zarutskie JA, Stern LJ. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry. 2000;39:3751–3762. doi: 10.1021/bi9923656. [DOI] [PubMed] [Google Scholar]
- 10.Roche PA, Cresswell P. Proteolysis of the class II-associated invariant chain generates a peptide binding site in intracellular HLA-DR molecules. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:3150–3154. doi: 10.1073/pnas.88.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 12.Lanzavecchia A, Reid PA, Watts C. Irreversible association of peptides with class II MHC molecules in living cells. Nature. 1992;357:249–252. doi: 10.1038/357249a0. [DOI] [PubMed] [Google Scholar]
- 13.Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, Monaco JJ, Mellins E. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- 14.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 15.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 16.Grotenbreg GM, Nicholson MJ, Fowler KD, Wilbuer K, Octavio L, Yang M, Chakraborty AK, Ploegh HL, Wucherpfennig KW. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. The Journal of biological chemistry. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. The Journal of experimental medicine. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 19.vogt AB, Moldenhauer G, Hammerling GJ, Kropshofer H. HLA-DM stabilizes empty HLA-DR molecules in a chaperone-like fashion. Immunology letters. 1997;57:209–211. doi: 10.1016/s0165-2478(97)00061-8. [DOI] [PubMed] [Google Scholar]
- 20.Sanderson F, Kleijmeer MJ, Kelly A, Verwoerd D, Tulp A, Neefjes JJ, Geuze HJ, Trowsdale J. Accumulation of HLA-DM, a regulator of antigen presentation, in MHC class II compartments. Science (New York, N Y. 1994;266:1566–1569. doi: 10.1126/science.7985027. [DOI] [PubMed] [Google Scholar]
- 21.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 22.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 23.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 24.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 25.Stratikos E, Mosyak L, Zaller DM, Wiley DC. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-DM catalysis. The Journal of experimental medicine. 2002;196:173–183. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullrich HJ, Doring K, Gruneberg U, Jahnig F, Trowsdale J, van Ham SM. Interaction between HLA-DM and HLA-DR involves regions that undergo conformational changes at lysosomal pH. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13163–13168. doi: 10.1073/pnas.94.24.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson MJ, Moradi B, Seth NP, Xing X, Cuny GD, Stein RL, Wucherpfennig KW. Small molecules that enhance the catalytic efficiency of HLA-DM. J Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, Bell S, Harcourt G, Walker BD, Klenerman P, Wucherpfennig KW. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. The Journal of clinical investigation. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogsgaard M, Wucherpfennig KW, Cannella B, Hansen BE, Svejgaard A, Pyrdol J, Ditzel H, Raine C, Engberg J, Fugger L. Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA)-DR2-MBP 85–99 complex. The Journal of experimental medicine. 2000;191:1395–1412. doi: 10.1084/jem.191.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellmerich S, Mycko M, Takacs K, Waldner H, Wahid FN, Boyton RJ, King RH, Smith PA, Amor S, Herlihy AH, Hewitt RE, Jutton M, Price DA, Hafler DA, Kuchroo VK, Altmann DM. High incidence of spontaneous disease in an HLA-DR15 and TCR transgenic multiple sclerosis model. J Immunol. 2005;174:1938–1946. doi: 10.4049/jimmunol.174.4.1938. [DOI] [PubMed] [Google Scholar]
- 31.Itano AA, McSorley SJ, Reinhardt RL, Ehst BD, Ingulli E, Rudensky AY, Jenkins MK. Distinct dendritic cell populations sequentially present antigen to CD4 T cells and stimulate different aspects of cell-mediated immunity. Immunity. 2003;19:47–57. doi: 10.1016/s1074-7613(03)00175-4. [DOI] [PubMed] [Google Scholar]
- 32.Pape KA, Catron DM, Itano AA, Jenkins MK. The humoral immune response is initiated in lymph nodes by B cells that acquire soluble antigen directly in the follicles. Immunity. 2007;26:491–502. doi: 10.1016/j.immuni.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Van den Broeck W, Derore A, Simoens P. Anatomy and nomenclature of murine lymph nodes: Descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. Journal of immunological methods. 2006;312:12–19. doi: 10.1016/j.jim.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 34.Ito K, Bian HJ, Molina M, Han J, Magram J, Saar E, Belunis C, Bolin DR, Arceo R, Campbell R, Falcioni F, Vidovic D, Hammer J, Nagy ZA. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. The Journal of experimental medicine. 1996;183:2635–2644. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL, Hafler DA. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. The Journal of experimental medicine. 1994;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin-Esteban V, Falk K, Rotzschke O. "Chemical analogues" of HLA-DM can induce a peptide-receptive state in HLA-DR molecules. The Journal of biological chemistry. 2004;279:50684–50690. doi: 10.1074/jbc.M407598200. [DOI] [PubMed] [Google Scholar]
- 37.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. The Journal of experimental medicine. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith KJ, Pyrdol J, Gauthier L, Wiley DC, Wucherpfennig KW. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. The Journal of experimental medicine. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogt AB, Kropshofer H, Moldenhauer G, Hammerling GJ. Kinetic analysis of peptide loading onto HLA-DR molecules mediated by HLA-DM. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9724–9729. doi: 10.1073/pnas.93.18.9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Ewijk W, Brekelmans PJ, Jacobs R, Wisse E. Lymphoid microenvironments in the thymus and lymph node. Scanning microscopy. 1988;2:2129–2140. [PubMed] [Google Scholar]
- 41.Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314:537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 42.Lanzavecchia A. Antigen presentation by B lymphocytes: a critical step in T-B collaboration. Current topics in microbiology and immunology. 1986;130:65–78. doi: 10.1007/978-3-642-71440-5_6. [DOI] [PubMed] [Google Scholar]
- 43.Delamarre L, Couture R, Mellman I, Trombetta ES. Enhancing immunogenicity by limiting susceptibility to lysosomal proteolysis. The Journal of experimental medicine. 2006;203:2049–2055. doi: 10.1084/jem.20052442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hopner S, Dickhaut K, Hofstatter M, Kramer H, Ruckerl D, Soderhall JA, Gupta S, Marin-Esteban V, Kuhne R, Freund C, Jung G, Falk K, Rotzschke O. Small organic compounds enhance antigen loading of class II major histocompatibility complex proteins by targeting the polymorphic P1 pocket. The Journal of biological chemistry. 2006;281:38535–38542. doi: 10.1074/jbc.M606437200. [DOI] [PubMed] [Google Scholar]
- 45.Gupta S, Hopner S, Rupp B, Gunther S, Dickhaut K, Agarwal N, Cardoso MC, Kuhne R, Wiesmuller KH, Jung G, Falk K, Rotzschke O. Anchor side chains of short peptide fragments trigger ligand-exchange of class II MHC molecules. PLoS ONE. 2008;3:e1814. doi: 10.1371/journal.pone.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 47.Illes Z, Stern JN, Reddy J, Waldner H, Mycko MP, Brosnan CF, Ellmerich S, Altmann DM, Santambrogio L, Strominger JL, Kuchroo VK. Modified amino acid copolymers suppress myelin basic protein 85–99-induced encephalomyelitis in humanized mice through different effects on T cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11749–11754. doi: 10.1073/pnas.0403833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson KP, Brooks BR, Cohen JA, Ford CC, Goldstein J, Lisak RP, Myers LW, Panitch HS, Rose JW, Schiffer RB. Copolymer 1 reduces relapse rate and improves disability in relapsing-remitting multiple sclerosis: results of a phase III multicenter, double-blind placebo-controlled trial. The Copolymer 1 Multiple Sclerosis Study Group. Neurology. 1995;45:1268–1276. doi: 10.1212/wnl.45.7.1268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.