Abstract
The cell cycle-dependent nucleocytoplasmic transport of proteins is predominantly regulated by CDK kinase activities; however, it is currently difficult to predict the proteins thus regulated, largely because of the low prediction efficiency of the motifs involved. Here, we report the successful prediction of CDK1-regulated nucleocytoplasmic shuttling proteins using a prediction system for nuclear localization signals (NLSs). By systematic amino acid replacement analyses in budding yeast, we created activity-based profiles for different classes of importin-α-dependent NLSs that represent the functional contributions of different amino acids at each position within an NLS class. We then developed a computer program for prediction of the classical importin-α/β pathway-specific NLSs (cNLS Mapper, available at http//nls-mapper.iab.keio.ac.jp/) that calculates NLS activities by using these profiles and an additivity-based motif scoring algorithm. This calculation method achieved significantly higher prediction accuracy in terms of both sensitivity and specificity than did current methods. The search for NLSs that overlap the consensus CDK1 phosphorylation site by using cNLS Mapper identified all previously reported and 5 previously uncharacterized yeast proteins (Yen1, Psy4, Pds1, Msa1, and Dna2) displaying CDK1- and cell cycle-regulated nuclear transport. CDK1 activated or repressed their nuclear import activity, depending on the position of CDK1-phosphorylation sites within NLSs. The application of this strategy to other functional linear motifs should be useful in systematic studies of protein–protein networks.
Keywords: CDK, computational method, nuclear localization signal, nuclear import, phosphorylation
Regulated nuclear import controls the nuclear function of many proteins that shuttle between the nucleus and cytoplasm or other cellular compartments in response to certain physiological conditions or extracellular stimuli (1). Nucleocytoplasmic shuttling comprises both nuclear import and nuclear export activities. Nuclear import is mediated by the interaction of nuclear localization signals (NLSs) with transport receptor importins, among which the importin-α family recognizes most major classes of NLSs with 1 or 2 basic stretches, called the classical NLSs (2–4). Six different classes of monopartite and bipartite NLSs bind to distinct binding grooves of importin-α (5–7). Nuclear export is mediated by nuclear export signals (NESs), which are recognized by Crm1/exportin or Msn5 in yeast (1, 8). The regulation of these 2 antagonistic activities directs a protein to either the nucleus or the cytoplasm. In general, these activities are regulated by the modification of the NLSs/NESs, by intermolecular or intramolecular interactions that lead to the structural masking of NLSs/NESs, or by the supply of an NLS/NES by another protein (9).
Phosphorylation within or around an NLS is the usual strategy for the regulated nuclear transport of a protein (4, 10–13). It has been shown that acidic amino acids adjacent to the basic core of an importin-α-dependent NLS inhibit the NLS activity (5, 14). Phosphorylation around an NLS mimics the inhibitory effect of acidic amino acids. Therefore, phosphorylation is inhibitory to NLS activity in most observed cases. Many phosphorylation-regulated NLSs have been identified, and a variety of protein kinases phosphorylate serine, threonine, or tyrosine residues within or around NLSs to regulate the NLS activities in response to some specific conditions, such as stress, nutrient availability, and the cell cycle (9). However, it is difficult to predict phosphorylation-regulated NLSs because neither the known consensus sequence of the NLS nor the protein phosphorylation site is stringent.
In this study, we have developed a highly accurate NLS prediction program (cNLS Mapper), which calculates NLS activity scores by using activity-based, but not sequence-based, profiles for different classes of importin-α-dependent NLSs. By combining this program and a known consensus phosphorylation site for CDK kinase, we predicted CDK1 (Cdc28)-regulated NLSs from budding yeast (Saccharomyces cerevisiae) databases. The CDK-dependent phosphorylation of NLSs confers cell cycle-dependent nucleocytoplasmic shuttling on proteins. In budding yeast, 6 proteins (Swi5, Swi6, Mcm3, Cdh1, Whi5, and Acm1) exhibit CDK1-regulated nucleocytoplasmic shuttling, and in 3 of these (Swi5, Swi6, and Mcm3), CDK1-regulated NLSs have been identified (15–20). These 6 proteins were all contained in our predicted protein fraction, and 5 proteins were newly identified as cell cycle-dependent nucleocytoplasmic shuttling proteins. This study presents an efficient and systematic identification of localization-regulated proteins by the prediction of composite regulatory motifs.
Results
Activity-Based Profiles of Different Classes of Importin-α-Dependent NLSs.
We have recently shown that, even within the same NLS class, the level of NLS activity varies depending on the NLS sequence (5, 14). To assess the functional contribution of various amino acids at every position in each of 3 monopartite NLS classes (classes 2–4) (5), all of the amino acid residues of the template NLSs, as representatives of these classes, were serially replaced with ≈20 other amino acid residues. The relative nuclear import activities of these altered sequences were assayed in yeast and ranked in 1 of the 10 levels based on the localization phenotype of the GFP reporter [see supporting information (SI) Fig. S1 for details], as described previously (14). The profiles of the 3 classes of NLSs are represented as scoring matrices based on the relative NLS activities (Fig. 1). These activity-based profiles highlight numerous specific-activity-affecting amino acid residues throughout more distal regions, particularly for the classical class 2 NLSs, than were appreciated previously. Moreover, comparison of the class 2 NLS profile with the property of the flanking sequence of class 1 NLS (5) (also see Fig. S2) indicates that class 1 NLSs are a subclass of class 2 NLSs and have lysine or arginine residues at position +3 (when the first lysine of the basic core is numbered as +1). Thus, the class 1 NLS profile is included in the class 2 NLS profile. The observation that every amino acid at every position within every NLS class contributes differently to its NLS activity highlights the difficulty in obtaining an accurate representation of the NLS consensus sequences without considering the contributions of all of the residues within an NLS.
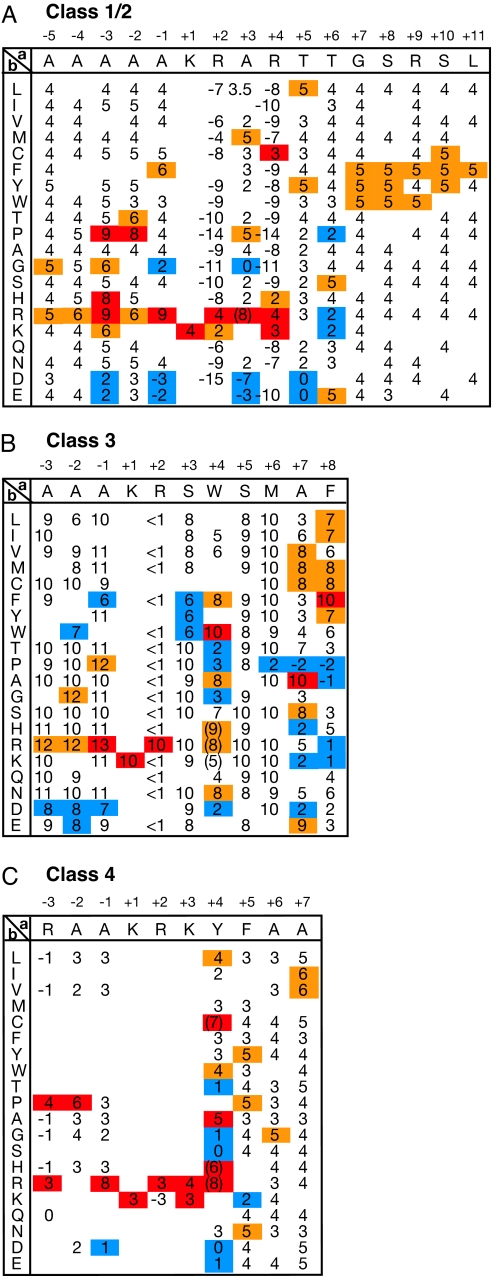
Fig. 1.
Activity-based profiles of classical and noncanonical classes of NLSs optimized for yeast. (A) A profile of a classical class 2 monopartite NLS. A single amino acid residue within the template sequence indicated at the top in the matrix was replaced with various other residues indicated in the left column, and the nuclear import activity was assayed in yeast. Activity scores were determined as in Fig. S1 -GUS-GFP reporters fused to NLSs with scores (1 and 2), (3–5), (6 and 7), and (8–10) exhibit nuclear, partially nuclear, nuclear plus cytoplasmic, and cytoplasmic phenotypes, respectively. This template NLS has an activity score of 4, a middle level of NLS activity. Scores with higher, slightly higher, and lower activities than an average value for each position are shown in red, orange, and blue, respectively. At several mutational positions, modified templates with a different level of basal activity were used to obtain more dispersed scores. Blanks represent undetermined scores. A value that may overlap the scores of class 4 NLSs is given in parentheses. (B) A profile of noncanonical class 3 monopartite NLS. Values that may overlap the scores of class 2 NLS are given in parentheses. (C) A profile of a noncanonical class 4 monopartite NLS.
Accurate Prediction of NLS Activity by Scoring with the NLS Profiles.
The amino acid substitution analyses of importin-α-dependent NLSs in our previous and present studies have demonstrated that the contribution of each residue to the overall NLS activity is, in most cases, independent and additive (14). Fig. S3A shows examples of the additive contribution of monopartite NLS residues, at least one of which is different between the NLSs at 3 positions (−3, +3, and +5). Thus, the total NLS score (relative activity level) can be calculated by adding the increased or decreased level of the score for each residue at each position given in the activity-based profiles. Because the scores in the profile represent the contribution of each residue to the entire NLS activity, the degree of the contribution corresponds to the increased or decreased level of the score at each position in the profile (for the class 2 NLS profile, a value obtained by subtracting the standard score 4.0 of the template NLS from a score of each residue) (Fig. S3B). Therefore, the sum of the increased or decreased level at each position represents a total contribution of amino acid exchanged from the template NLS, and a final score of an NLS can be obtained by adding the standard score to the summed value. We developed a computer program for NLS prediction, “cNLS Mapper,” to calculate NLS scores by using modified versions of these monopartite NLS profiles and a previously generated bipartite NLS profile (14). As shown in Fig. S4, several of the scores initially standardized by using a 10-point scale were not constantly increased by the addition of another weak NLS with a score of 1.0, indicating that the 10-point scale is not strictly linear. Thus, we modified the levels of the scores initially standardized to allow a more linear additive effect by adding or subtracting a half or 1 point level of score (see Materials and Methods for details).
We evaluated cNLS Mapper with synthetic monopartite NLSs (Fig. S2 and Figs. S5 and S6) and bipartite NLS sets previously screened from synthetic peptide libraries (5) as test sequences. In the monopartite NLS sets, each has mutations at least 3 different positions for each NLS class and an activity score measured in yeast. The program's prediction performance was compared with those of 2 other available methods, PSORT II (21) and PredictNLS (22), which are based on the traditional consensus sequence of the classical NLS and in silico mutagenesis analysis with several NLSs collected from the literature, respectively (Table 1). Calculations made with cNLS Mapper demonstrated that the scores for the test sequences correlated well (>90% in monopartite NLSs and >80% in bipartite NLSs) with their measured activity. The relatively low accuracy in the prediction of bipartite NLSs may be attributable to the higher capacities of bipartite NLSs to generate some overlapping motifs, including monopartite NLSs, and to form secondary structure, than those of monopartite NLSs, which could greatly influence their NLS activity. In contrast, the performance of the other 2 methods was considerably lower in terms of both sensitivity and specificity than that of cNLS Mapper (Table 1), although these earlier methods may be more effective for mammalian NLSs, and our method may be specific to yeast.
Table 1.
Comparison of the performance of cNLS Mapper with those of other prediction methods
| Predictor | NLS class | Prediction performance, % |
||
|---|---|---|---|---|
| Sens | Spec | Accur | ||
| cNLS Mapper | Class 1/2 | 99 | 94 | 98 |
| Class 3 | 100 | 100 | 100 | |
| Class 4 | 87 | 97 | 92 | |
| Bipartite | 87 | 82 | 85 | |
| PSORT II | Class 1/2 | 76 | 33 | 65 |
| Class 3 | 0 | 100 | 9 | |
| Class 4 | 39 | 79 | 62 | |
| Bipartite | 71 | 48 | 63 | |
| PredictNLS | Class 1/2 | 4 | 94 | 27 |
| Class 3 | 0 | 100 | 9 | |
| Class 4 | 0 | 100 | 56 | |
| Bipartite | 0 | 100 | 33 | |
The prediction accuracies of the cNLS Mapper, PSORT II (http://psort.nibb.ac.jp/form2.html), and PredictNLS (http://cubic.bioc.columbia.edu/predictNLS/) programs were determined by using test peptide sequences derived from synthetic NLS mutants (see Figs. S2, S5, and S6), in which at least 3 flanking residues were simultaneously mutated, and bipartite NLSs screened from biXs libraries (5). The test sequences were divided into 2 fractions, positive and negative sequences, based on the NLS activity determined in yeast: The former has a high or medium level of NLS activity, corresponding to scores of 6–10 (or GFP localization phenotypes N and Nc), and the latter has little NLS activity, corresponding to scores of 1 or 2 (or a phenotype C). Sequences with scores of 3–5 (or an NC phenotype) were omitted. We note that many of our negative sequences for class 2 and bipartite NLSs contain classical consensus sequences. The total number of positive and negative test sequences was 138 and 48 for class 1/2 NLSs, respectively, 51 and 5 for class 3 NLSs, 23 and 29 for class 4 NLSs, and 388 and 195 for bipartite NLSs. To increase the length of the test sequences, additional sequences comprising 6–9 residues, which were derived from the pTUE-GFP cloning site, were added to both termini of the test sequences. Both the prediction sensitivity (Sens) and specificity (Spec) achieved with cNLS Mapper were calculated as the percentages of true positives with calculated scores of >4 in the positive NLS fraction and true negatives with calculated scores of ≤4 in the negative NLS fraction. Prediction accuracy (Accur) was calculated as the percentage of the sum of true positives and true negatives.
We also searched the budding yeast genome database for the translated ORFs by using cNLS Mapper. Monopartite NLSs with a calculated score of ≥8 and bipartite NLSs with a calculated score of ≥7 were observed in 406 and 306 independent yeast proteins, respectively. Their localizations were nuclear (447), nonnuclear (133), and unknown (132), according to their annotations, suggesting nuclear function and/or subcellular localization data in the yeast GFP fusion localization database (http://yeastgfp.ucsf.edu/) (23). This analysis suggested that the apparent prediction specificity of cNLS Mapper was 77%. We randomly selected 30 monopartite NLSs predicted in nonnuclear proteins and assayed their NLS activities in yeast (Table S1). Twenty-nine NLSs (97%) were found to have a strong or medium level of NLS activity, although it is not clear whether they exert the NLS function in native proteins under some specific conditions. Only 1 NLS (Mnn10 NLS) exhibited weak activity, but mutational analysis suggested that its activity was most likely inhibited by the phosphorylation of the flanking serine residues (Table S1). We also randomly selected 10 putative bipartite NLSs in yeast nuclear proteins that matched the traditional consensus but had low scores when calculated with cNLS Mapper (Table S2). All these sequences had little NLS activity in yeast, in agreement with the cNLS Mapper prediction. These results demonstrate the highly accurate prediction performance of cNLS Mapper, at least when the NLSs are evaluated as isolated peptides. Implementation of cNLS Mapper is available online (http://nls-mapper.iab.keio.ac.jp/).
Prediction of CDK1-Regulated NLSs in Yeast Proteins.
The NLS profiles demonstrate that the activities of all classes of importin-α-dependent monopartite and bipartite NLSs are considerably reduced by the presence of acidic residues at positions −3 to −1 (when the first lysine of the basic core is numbered as +1). Serine or threonine at positions −3 or −2 can overlap with the consensus CDK kinase phosphorylation site (S/T)PX(K/R), and its phosphorylation mimics the action of acidic amino acids in repressing the NLS activity (Fig. 2A). To identify the NLSs regulated by CDK kinase, we searched for NLSs that overlap the consensus CDK site in the budding yeast genome database of translated ORFs by using cNLS Mapper. First, we focused on monopartite NLSs containing the CDK site because all CDK1-regulated NLSs (from Swi5, Swi6, and Mcm3) that have been previously identified in yeast are monopartite NLSs. A total of 11 proteins were predicted to have a CDK site-containing monopartite NLS with a score of ≥7 (Table S3). We also investigated the CDK site-containing NLSs from the top 200 substrates of CDK1 in yeast (24) and 35 CDK1 substrates identified by a proteomic study (25), and found 14 proteins carrying a CDK-site-containing NLS with a score of ≥4 (Table S3). These predicted fractions contained 5 (Swi5, Swi6, Mcm3, Whi5, and Acm1) of the 6 previously reported proteins whose nuclear localization is regulated by CDK1, indicating the high sensitivity of cNLS Mapper. Next, we searched for bipartite NLSs that overlap the consensus CDK site. Bipartite NLSs contain more potential CDK1 sites, which are predicted to be repressed or activated by phosphorylation, than do monopartite NLSs. In addition to the upstream flanking region, the terminal region of the linker sequence, at positions + 3, +4, +12, and +13, are sensitive to conversion to acidic residues (14). In contrast to these terminal regions repressed by acidic residues, the central linker region exerts an activating effect with the presence of acidic residues (5, 14). Therefore, we looked for bipartite NLSs containing the consensus CDK sequence in the upstream flanking and linker regions among the 235 reported CDK1 substrates (Fig. 2A), because this substrate set contains most of the reported CDK1-regulated proteins (Table S3). Ten proteins were predicted to have a CDK site-containing bipartite NLS with a score of ≥4, and this fraction contained another reported protein (Cdh1) (Table S3). Thus, our search for CDK1 site-containing NLSs produced a good predictive result, with the successful prediction of all known CDK1-regulated proteins and with a relatively low rate of false positives.
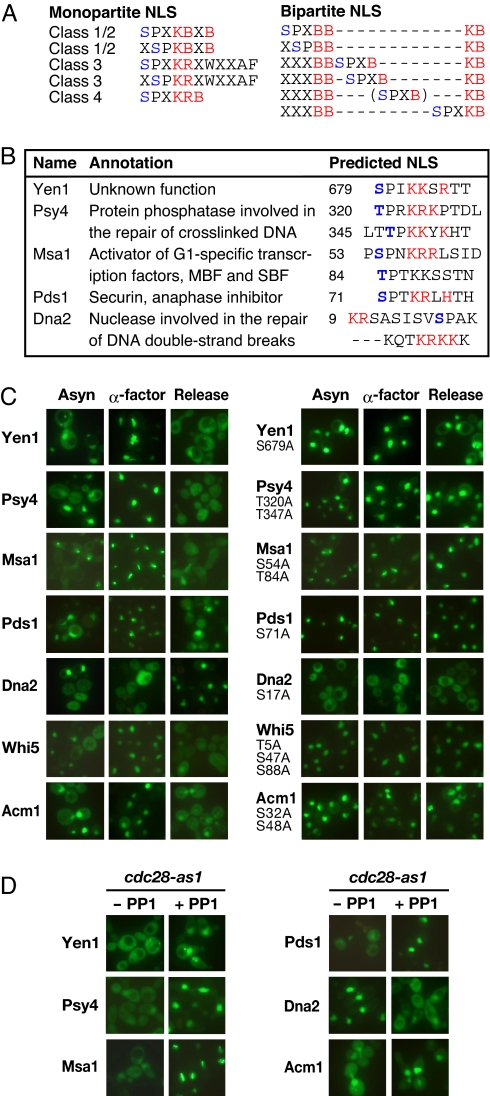
Fig. 2.
Previously unidentified cell cycle-regulated nucleocytoplasmic shuttling proteins. (A) Different patterns of importin-α-dependent NLSs overlapping the consensus CDK site. The core basic residues of the monopartite and bipartite NLSs and potential CDK phosphorylation sites are marked in red and blue, respectively. “B” represents Lys or Arg. (B) List of previously unidentified cell cycle-regulated nucleocytoplasmic shuttling proteins. (C) Cell cycle-regulated nuclear transport depending on predicted CDK1 sites. The subcellular localization of the indicated GFP-fused proteins was observed in exponentially growing (Asyn, asynchronous) and synchronized yeast cells (α-factor, arrested in G1 with α-factor; Release, cultured for 1 h after release of G1 arrest; Left). The previously reported Acm1 and Whi5 were analyzed to confirm our predicted NLS sites. The potential CDK phosphorylation sites within and around a predicted NLS were converted to alanine, and these mutants were similarly analyzed (Right). GFP-Pds1 was expressed from a galactose-inducible pYES-GFP vector to alleviate its overexpression-related toxic effects, and the induced yeast cells were arrested by 3-h incubation with nocodazole instead of the release of G1 arrest. The other GFP fusion proteins were expressed from a constitutive expression vector, pGAD-GFP or pAUA-GFP. (D) Cell cycle-regulated nuclear transport depends on CDK1 activity. cdc28-as1 cells expressing the GFP fusions of the indicated proteins were synchronized in S or G2 by arrest and release with the α-factor in the presence or absence of 1 μM 1NM-PP1, a specific inhibitor of Cdc28p-as1.
Newly Identified CDK1-Regulated Nuclear Transport Proteins.
The remaining 28 proteins (19 and 9 from the monopartite and bipartite NLS searches, respectively) in the predicted fractions were analyzed for their subcellular localization by fusing them with GFP. Five proteins (Yen1, Psy4, Pds1, Msa1, and Dna2) exhibited both nuclear and cytoplasmic localization in an asynchronous culture (Fig. 2 B and C), whereas many of the other proteins were exclusively and constantly localized in the nucleus, where several localized at the spindle poles or the nuclear periphery or in the cytoplasm (Table S3). Therefore, we further analyzed the 5 proteins as candidate cell cycle-dependent nucleocytoplasmic shuttling proteins. Four of the 5 proteins (Yen1, Psy4, Msa1, and Pds1) were predominantly nuclear in G1-arrested cells, but were cytoplasmically localized after release from G1 arrest, as well as the previously reported Whi5 and Acm1. In contrast, Dna2, which was predicted in the bipartite NLS search, was cytoplasmic in G1-arrested cells and nuclear in the S-to-M phases, indicating that its cytoplasmic localization is G1 specific (Fig. 2C).
To examine the role of a potential CDK1 site within a predicted NLS in its nuclear import activity, the phosphorylatable serine or threonine residue of the CDK1 site was substituted with a nonphosphorylatable alanine. All alanine substitution mutants, except a Dna2 mutant, showed either partial or complete nuclear localization throughout the cell cycle, and the simultaneous mutation of the nearby potential CDK1 sites in Psy4, Msa1, Whi5, and Acm1 enhanced their nuclear localization (Fig. 2C). In contrast, alanine substitution at a single site in Dna2 resulted in its complete cytoplasmic localization throughout the cell cycle (Fig. 2C). Furthermore, to confirm that the observed cell cycle-regulated nuclear localization depended on CDK1, we inhibited CDK1 activity by using a cdc28-as1 strain (Fig. 2D). Because this strain has a cdc28 allele, which expresses a Cdc28p that is sensitive to an ATP analogue (e.g., 1NM-PP1), treatment with this drug allows the selective inhibition of CDK1 (26). CDK1 inhibition with 1NM-PP1 caused the constitutive cytoplasmic localization of Dna2 and the constitutive nuclear localization of the other 6 proteins, including the previously reported Acm1. When considered together with the fact that most of these proteins are confirmed potential CDK1 targets, these results indicate that the cell cycle-dependent nuclear localization of these newly identified proteins is regulated by the direct phosphorylation of their NLSs by CDK1.
Activated Nuclear Localization of Dna2 by CDK1 Phosphorylation at the Central Linker Site of a Bipartite NLS.
Only the nuclear localization of Dna2 was activated by CDK1, whereas that of the other cell cycle-dependent nucleocytoplasmic shuttling proteins, including previously reported ones, was repressed by CDK1, indicating the opposite action of CDK1 on their NLSs. Because Dna2 was predicted to contain 2 putative overlapping bipartite NLSs, NLS1 and NLS2, which share a predicted CDK1 site in the linker and N-terminal flanking regions, respectively, we analyzed functional NLSs by alanine substitution in their core basic stretches (Fig. S7). The mutation of the first or third basic stretch led to a significant reduction in NLS activity, whereas the mutation of the second basic stretch had no detectable effect. This observation indicates that the nuclear import of Dna2 is promoted by the phosphorylation of NLS1 at its central linker site. Hence, the different actions of CDK1-mediated phosphorylation on the linker sequences of bipartite NLSs and the flanking sequences of monopartite NLSs are probably responsible for the different cell cycle specificity in the localization of Dna2 and the other CDK1-regulated proteins.
Nuclear Export Activities of CDK1-Regulated Nuclear Transport Proteins.
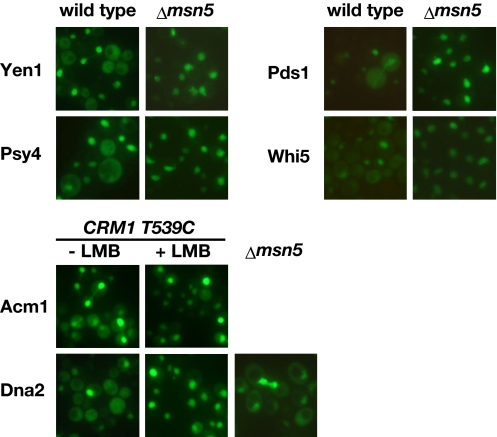
We examined whether the nucleocytoplasmic shuttling of the newly identified proteins involves their nuclear export activities by using mutants of known yeast nuclear exporters, Crm1 and Msn5 (Fig. 3). Yen1, Psy4, Pds1, and Whi5 were constitutively localized to the nucleus in an msn5Δ mutant, but not by treatment with leptomycin B (LMB), a Crm1 inhibitor, in a strain with a LMB-sensitive allele of Crm1 (T539C) (27), indicating that Msn5 mediates the nuclear export of these proteins. The nuclear import of Acm1 and Dna2 was enhanced after LMB treatment of the Crm1-T539C strain, indicating that Crm1 is a major export receptor for these 2 proteins. The nuclear export of Dna2 is also probably mediated by Msn5, because it was partially inhibited in the msn5Δ mutant. Conversely, no inhibition of the Msa1 nuclear export was observed in the mutants for these exportins, suggesting that the nuclear export of Msa1 is mediated by unknown transporters. These results indicate that the cell cycle-dependent nucleocytoplasmic shuttling proteins in yeast are regulated by the coordinated activity of CDK1-regulated nuclear import and the nuclear export mediated by different exportins.
Fig. 3.
CDK1-regulated nucleocytoplasmic shuttling involves nuclear export activities mediated by different export receptors. Subcellular localization of GFP-fused proteins with the indicated gene products was observed in exponentially growing cells of either the wild type, an msn5Δ mutant, or a Crm1-T539C strain treated with leptomycin B. The results for which there was no observed difference between the wild-type and mutant strains are not shown.
Discussion
This study shows that at least 11 proteins in budding yeast display CDK1-regulated nucleocytoplasmic shuttling. Five of them (Swi6, Whi5, Msa1, Mcm3, and Dna2) have been reported to control the G1–S transition, whereas 4 of them (Swi5, Cdh1, Acm1, and Pds1) regulate the M–G1 transition. Three of the G1–S regulators (Swi6, Whi5, Msa1) are components of the G1-specific transcription factors, MBF and SBF (28–30), and 3 of the M–G1 regulators (Cdh1, Acm1, and Pds1) regulate the anaphase-promoting complex (APC), an E3 ubiquitin ligase that controls anaphase initiation (18). The mRNA transcription and/or protein degradation of many of these regulators are also coupled to the cell cycle. The multiple levels of regulation of these proteins, involving nuclear import/export and their expression/degradation in the same signaling complexes and pathways, appear to ensure the timely onset of S phase or anaphase. Of the other identified proteins, Dna2 has been reported to function as a nuclease/helicase that is involved in Okazaki fragment processing in replication and in the repair of DNA double-strand breaks via homologous recombination (31, 32), and Psy4 is involved in the repair of cross-linked DNA (33), indicating a tight coupling of DNA repair to cell cycle control. Dna2 resects DNA double-strand-break ends in concert with other helicase and nucleases including Sae2 (34), which is regulated by CDK1 (35). These indicate that CDK1 targets not only Sae2 but also Dna2 to ensure their DNA processing activity specifically in S and G2 phases by controlling its localization.
Our success in comprehensively identifying CDK1-regulated nucleocytoplasmic shuttling proteins is the result of our strategy involving (i) the definition of potentially functional patterns for a CDK-site-overlapping NLSs and (ii) the development of a computational NLS prediction system. The activity-based profiles for several NLS classes allowed us to identify amino acid positions that are repressed or activated by acidic residues mimicking phosphorylated amino acids and to define several composite NLS patterns that are potentially regulated by CDK activities. Moreover, activity-based profiles were used to develop the computational prediction tool for importin-α-dependent NLSs, cNLS Mapper, which calculates NLS activities by using an additivity-based motif scoring algorithm. This algorithm is based on the observations that the activity-based NLS profile represents different levels of the contributions of various amino acid at each position within an NLS and that each residue within an NLS makes an independent and additive contribution to its entire activity. The additivity rule of NLS activity has been also supported by our recent study, demonstrating that NLS peptide inhibitors specific for the importin-α/β pathway can be designed by selecting the amino acids with the highest scores in an activity-based profile of the classical bipartite NLS (14).
The accurate prediction of NLSs by using this algorithm allowed us to systematically identify a composite regulatory motif containing an NLS and a CDK phosphorylation sites. In contrast, a search for CDK site-containing NLSs by using the traditional consensus sequences of the classical NLSs predicted a large number of apparently false-positive sites (145 hits in the yeast database of translated ORFs) and failed to predict 3 of the 5 previously unidentified nucleocytoplasmic shuttling proteins. A sequence similarity search using motif databases or defined consensus sequences has been the basis of a number of bioinformatics-based motif prediction methods, such as Prosite, ELM, and Scansite; however, these methods often involve low accuracy because the short length of the defined motif sequence generates functionally irrelevant false-positive motifs by chance and because the negative impact of underrepresented residues within a motif cannot be reflected in the defined consensus or profile. These disadvantages of the current prediction methods can be overcome by using the activity-based profiles of linear motifs. Nevertheless, the efficiency of our method for predicting CDK1–NLS motifs was still only ≈30%. The development of a more accurate prediction method for CDK phosphorylation sites would improve the true positive rate, because most true proteins were contained in the database of experimentally defined CDK1 substrates.
Although cNLS Mapper accurately predicts importin-α-dependent NLSs, it cannot predict other types of NLS that are directly recognized by β-type importins. Conversely, this raises the possibility that the sequences recognized by importin-β members have been generated in some NLS mutants tested in this study by chance, which might have affected the performance of cNLS Mapper. Another limitation is that cNLS Mapper evaluates NLS activities only as isolated peptide segments but not within the structural context of a native protein. This could lead to prediction of many false positives, which may be found in nuclear imported proteins mediated through importin-α-independent pathways (e.g., ribosomal proteins), nonnuclear compartment proteins (e.g., mitochondrial proteins), or internal protein regions that are usually inaccessible to importin-α. These limitations may be overcome by incorporating the prediction methods of other subcellular localization signals and of protein surface or disordered regions. Despite these problems, the analysis with cNLS Mapper demonstrates that proteins regulated via different signaling pathways can be identified by the prediction of composite motifs that overlap with their pathway-specific motifs. When activity-based profiles are created for other linear motifs, they could be used to determine the functional composite motif patterns and to develop an accurate motif prediction system to identify the motifs of the regulated proteins.
Materials and Methods
Plasmid Constructions.
Double-stranded oligonucleotides encoding an NLS were cloned into pTUE-GFP3 (5). For the localization analysis of yeast proteins fused to GFP, PCR-amplified yeast ORFs were cloned into pGAD-GFP, pAUA-GFP, or pYES-GFP vectors. Detailed descriptions are provided in SI Text.
Yeast Manipulation and Measurement of Nuclear Import Activities.
Yeast manipulations, including culture and transformation, were performed as described previously (14), and the strain SFY526 (Clontech) or YNN141 (MATa his3–532 trp1–289 ura3–1 ura3–2 ade2 leu2::HIS3) was used in most experiments. The cdc28-as1 and Crm1 (T539C) strains were produced as described previously (26, 36). Detailed descriptions are provided in SI Text.
Yeast Cell Cycle Arrest.
SFY526 or YNN141 cells were transformed with plasmids and grown to an optical density of 0.2 at 600 nm. To induce cell cycle arrest in G1 phase, the cells were washed once with sterile water and grown for 2 h in the YPD medium (pH 3.5) containing 5 μg/mL α factor (Sigma). Cell cycle arrest at G1 was released by washing the cells once with sterile water, and they were then grown in YPD medium (pH 6.0) containing 10 μg/mL pronase for 1 h to reach S or G2 phase. For the GAL1-promoter-driven constructs, cells grown in SD medium lacking tryptophan were transferred to YPD containing 0.1% galactose instead of 2% glucose and grown for 4–5 h. The cells were arrested in G1 after growth for 2 h in the same medium containing α-factor or were arrested in G2/M after growth for 3 h in medium containing 15 μg/mL nocodazole (Sigma). They were then released by washing and grown for an additional 1 h (1.5 h for Pds1). To repress the CDK1 activity derived from cdc28-as1, 1 μM 1NM-PP1 (Merck) was added 1 h after the addition of α-factor, and the cells were cultured for 2 h (2.5 h for Pds1).
Development of a Computational Program, cNLS Mapper, to Calculate NLS Scores.
To calculate the total score for an NLS, the increased or decreased scores corresponding to each position and residue in the NLS profiles were simply added because the contribution of each residue to the overall activity is independent and additive in most cases (Fig. S3). Before calculation, the scores categorized based on the GFP phenotypes were corrected to increase proportionally when added together. Fig. S4 shows the change in the activities of class 2 NLSs in the presence or absence of another weak NLS located on the same GFP molecule. Because the 2 NLSs located at the opposite termini of the GFP molecule function as independent monopartite NLSs, the N-terminal NLS (N-NLS) should exert an additive effect equally on any C-terminal NLS (C-NLS). An N-NLS (PAAKRTRAP) produced an average increase of 1.0 for the activities (scores) of C-NLSs. In the presence of the N-NLS, there was an increase of 2.0 on the C-NLSs with a score of 3 or 4 and no increase in the C-NLS with a score of 9, whereas the expected level (1.0) of increase was observed for the C-NLS with a score of 6 or 7 (Fig. S4). We thus corrected the levels of the scores initially standardized, so that the scores were constantly increased by the N-NLS (e.g., the observed scores 2, 3, 4, 5, 8, and 9 were corrected to 1.5, 2, 3, 4.5, 8.5, and 10, respectively.) To generate modified NLS profiles, the original scores were replaced with the corrected ones and the blanks that remained undetermined were filled with scores postulated from other NLS mutants or profiles for different NLS classes (Fig. 1 and Fig. S8) (5).
To calculate the total score (Ts) for an NLS, the standard score (St) was subtracted from the scores (Si) in the modified profiles corresponding to each position and residue of the NLS. The St is the background score for a template NLS used to generate the NLS profile (e.g., the St for the class 2 NLS profile is 4). The adjusted scores were added up and the St was then added, as shown by the following equation (Fig. S3).
Using the modified profiles for classes 2–4 and the bipartite NLSs, we developed a Perl program, cNLS Mapper, to calculate the total score of an NLS. This program scans the protein sequence with a window size of 16-aa residues and a shift size of 1 amino acid and calculates the total score for every sequence composed of 16 residues for monopartite NLSs. Some exceptional cases that failed to fit the calculation were observed, probably because there were multiple overlapping NLSs within the sequence of 16-aa residues. These poor correlations were corrected by a modification of scores or profiles. For example, 3 additional scores were added to the total score of class 1/class 2 NLSs containing the basic core KKRK. For the bipartite NLS, the program scans the protein with a window size of 26–28 residues within its internal region but with a window size ranging from 26 to 36 residues for both terminal regions containing 60-aa residues. Because the functional linker length of the bipartite NLS is between 10 and 20 aa in a structurally flexible context, the window size was expanded in the terminal regions, which are likely to be structurally disordered in many proteins. For bipartite NLSs containing the K(K/R)X(K/R) sequence in the C-terminal basic region, the score was calculated with another modified bipartite NLS profile, which was similar to the class 2 NLS profile in the region ranging from positions +12 to +20, with increased scores for K and R at positions +1 and +2.
Supplementary Material
Acknowledgments.
We thank Dr. Rintaro Saito for advising us on Perl programming. This work was supported by grants from Yamagata Prefectural Government and Tsuruoka City and from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project SY-1114).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900604106/DCSupplemental.
References
- 1.Ossareh-Nazari B, Gwizdek C, Dargemont C. Protein export from the nucleus. Traffic. 2001;2:684–689. doi: 10.1034/j.1600-0854.2001.21002.x. [DOI] [PubMed] [Google Scholar]
- 2.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 3.Lange A, et al. Classical nuclear localization signals: Definition, function, and interaction with importin α. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: A key control point in nuclear transport? BioEssays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 5.Kosugi S, et al. Six classes of nuclear localization signals specific to different binding grooves of importin α. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 6.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 7.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M. Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl Cell Differ. 2002;35:67–91. doi: 10.1007/978-3-540-44603-3_4. [DOI] [PubMed] [Google Scholar]
- 9.Poon IK, Jans DA. Regulation of nuclear transport: Central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 10.Xiao CY, Hubner S, Jans DA. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J Biol Chem. 1997;272:22191–22198. doi: 10.1074/jbc.272.35.22191. [DOI] [PubMed] [Google Scholar]
- 11.Fontes MR, et al. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-α. Biochem J. 2003;375:339–349. doi: 10.1042/BJ20030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harreman MT, et al. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J Biol Chem. 2004;279:20613–20621. doi: 10.1074/jbc.M401720200. [DOI] [PubMed] [Google Scholar]
- 13.Hubner S, Xiao CY, Jans DA. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- 14.Kosugi S, et al. Design of peptide inhibitors for the importin α/β nuclear import pathway by activity-based profiling. Chem Biol. 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Liku ME, Nguyen VQ, Rosales AW, Irie K, Li JJ. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol Biol Cell. 2005;16:5026–5039. doi: 10.1091/mbc.E05-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costanzo M, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Sidorova JM, Mikesell GE, Breeden LL. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enquist-Newman M, Sullivan M, Morgan DO. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol Cell. 2008;30:437–446. doi: 10.1016/j.molcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1) EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 21.Nakai K, Horton P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 22.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 24.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 25.Archambault V, et al. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 26.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 27.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 29.Ashe M, et al. The SBF- and MBF-associated protein Msa1 is required for proper timing of G1-specific transcription in Saccharomyces cerevisiae. J Biol Chem. 2008;283:6040–6049. doi: 10.1074/jbc.M708248200. [DOI] [PubMed] [Google Scholar]
- 30.Li JM, Tetzlaff MT, Elledge SJ. Identification of MSA1, a cell cycle-regulated, dosage suppressor of drc1/sld2 and dpb11 mutants. Cell Cycle. 2008;7:3388–3398. doi: 10.4161/cc.7.21.6932. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart JA, Miller AS, Campbell JL, Bambara RA. Dynamic removal of replication protein A by DNA2 facilitates primer cleavage during Okazaki fragment processing in Saccharomyces cerevisiae. J Biol Chem. 2008;283:31356–31365. doi: 10.1074/jbc.M805965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vazquez-Martin C, Rouse J, Cohen PT. Characterization of the role of a trimeric protein phosphatase complex in recovery from cisplatin-induced versus noncrosslinking DNA damage. FEBS J. 2008;275:4211–4221. doi: 10.1111/j.1742-4658.2008.06568.x. [DOI] [PubMed] [Google Scholar]
- 34.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic. 2008;9:2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.