Abstract
Aquaporin 6 (AQP6) is an anion channel that is expressed primarily in acid secreting α-intercalated cells of the kidney collecting duct. In addition, AQP6 anion channel permeability is gated by low pH. Inspection of the N-terminus of AQP6 revealed a putative calmodulin binding site. AQP6-expressing CHO-K1 cell lysates were mixed with calmodulin beads and AQP6 was pulled down in the presence of calcium. Mutagenesis of the N-terminal calmodulin binding site in full length mouse AQP6 resulted in a loss of calmodulin binding activity. Mouse and human AQP6 calmodulin binding site peptides bound dansyl-calmodulin with a dissociation constant of approximately 1 μM. The binding of AQP6 to calmodulin may be an important key to determining the physiological role of AQP6 in the kidney.
Keywords: AQP6, aquaporin, calmodulin, calcium
Introduction
Aquaporin 6 (AQP6) is expressed primarily in intracellular vesicles in α-intercalated cells in the collecting duct of the kidney [1]. Recently AQP6 has also been observed in parotid gland acinar cells [2] and in the inner ear [3, 4]. AQP6 has low water permeability and primarily transports anions [5]. In addition, the anion permeability of AQP6 is increased at least 5-fold by exposure to low pH [5]. The expression of AQP6 in acid-secreting intercalated cells, the localization of AQP6 in vesicles that contain the V-type H+-ATPase, and the gating of AQP6 by low pH, together suggest that AQP6 functions to promote urinary acid secretion.
Aquaporin 0 (AQP0) is another aquaporin with a low water permeability that has a permeability that is regulated. Calmodulin binds the C-terminus of AQP0 in a calcium dependent manner [6] and inhibits AQP0 permeability [7]. In addition a peptide from the C-terminus of AQP0 was shown to bind calmodulin in a calcium dependent manner [8, 9] with a dissociation constant (KD) of 0.48 μM [10]. Studies of calmodulin binding to AQP0 were followed by studies in Xenopus oocytes showing the inhibition of AQP0 water channel activity by calmodulin [11]. However, the physiological role of calmodulin binding to AQP0 in the lens is still not clear.
Here we report a calmodulin binding site in the N-terminus of mouse, rat and human AQP6. The calmodulin binding site in AQP6 binds calmodulin in a calcium-dependent manner with a dissociation constant of approximately 1 μM. Mutagenesis of the calmodulin binding site in the N-terminus abolishes binding of AQP6 to calmodulin. Our studies reveal that AQP6 may be regulated through the binding of calmodulin which may be crucial for identifying its physiological role in the kidney.
Materials and Methods
Plasmid construction
Standard methods were used [12]. Putative calmodulin binding sites were found using the Calmodulin Target Database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html). PCR (Expand High Fidelity System, Roche) was used to perform site directed mutagenesis of the putative calmodulin binding sites in human and mouse AQP6. Sense and anti-sense PCR primers were made complementary to the target region with the desired point mutations. Each mutagenesis primer was paired with the appropriate upstream or downstream primer so that either the 5′ or 3′ half of hAQP6 or mAQP6 from before or after the mutation could be amplified. The two halves of the gene were combined in a PCR reaction so that they would overlap at the mutation site. The inserts were first cloned into pCR2.1 using the TOPO TA cloning kit (Invitrogen) for selection and sequencing, then digested with BamHI for hAQP6 or EcoRI and NotI for mAQP6, and ligated into pcDNA 3.1/Zeo (+) (Invitrogen).
Transfection, calmodulin binding assay and immunoblotting
CHO-K1 cells were obtained from the America Type Culture Collection (Manassas, VA) and maintained in Kaighn’s modification of Ham’s F12 medium (F12K) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37 °C and 5% CO2. CHO-K1 cells were transfected with wild-type human, mouse or rat AQP6, or one of five different mutants of mouse AQP6, using Lipofectamine 2000 (Invitrogen). One day following transfection, cells were solubilized in PBS containing 0.5% Triton and a Complete Protease Inhibitor Cocktail tablet (Roche). Cell lysate was mixed with calmodulin-agarose beads (Sigma) in the presence of either 1 mM Ca2+ or 2.5 mM EGTA at 4 °C for four hours to assay for calmodulin binding. The supernatant was removed and beads were washed and centrifuged twice in PBS with 0.1% Triton and either Ca2+ or EGTA to remove unbound protein. Lastly, beads were incubated with Laemmli Sample Buffer (1% SDS) for 30 minutes at 37 °C. Membrane protein was run on 12% acrylamide SDS-PAGE, transferred to Immun-Blot PVDF membrane (BioRad), and probed overnight at 4 °C with rabbit anti-AQP6 antibody (Alomone, Israel) diluted 1:500 in 4% BSA, 0.05% Tween-20, 1 mM NaN3 in PBS. Human AQP6 was detected with anti-human AQP6 antibody (Alpha Diagnostic). Blots were washed and probed with goat anti-rabbit HRP-conjugated secondary antibody (Amersham) for 1 hour at room temperature and visualized using ECL Plus (Amersham).
Fluorometric measurements using dansyl-calmodulin
Dansylation of calmodulin was carried out as described [13]. Briefly, bovine brain calmodulin (1 mg, Sigma) was dissolved in 1 ml 0.1M NaHCO3 pH 9.0, and 60 μl 10 mg/ml dansyl chloride (Invitrogen) in DMF was added. The reaction mixture was incubated with shaking at room temperature for one hour. Calmodulin was re-purified using size-exclusion by centrifugation in Nanosep 10K columns (Pall), and washed using 0.1M NaHCO3 pH 9.0 buffer, before being re-dissolved in 100 mM TRIS 50 mM NaCl pH 7.5 buffer to a final volume of 1 ml. Final concentration was determined by BCA protein assay (Pierce) to be 60 μM. Degree of incorporation was determined to be ~9 mol dansyl/mol calmodulin molecules using an extinction coefficient of €= 3400 M−1cm−1 at 340 nm.
Fluorescence emission spectra were obtained at 23 °C using a Hitachi F-4500 fluorescence spectrophotometer set at an excitation wavelength of 340 nm and detection set in emission scan mode from 400 to 600 nm. The excitation slit width was set at 5.0 nm and the emission slit width at 10.0 nm. Dansyl-calmodulin (200 nM) was titrated using 21-aa synthetic peptides of AQP6 (Biopeptide) at concentrations from 0 to 20 μM in a reaction buffer consisting of 100 mM TRIS, 50 mM NaCl pH 7.5, 1 mM EGTA and 1.1 mM CaCl2 to produce a calcium-buffered environment of 100 μM free Ca2+. Calcium-buffer calculations were conducted using WebMaxCLite v. 1.15 (http://www.stanford.edu/~cpatton/webmaxc/webmaxclite115.htm).
Fluorometric determination of Kd
Titrations of dansyl-calmodulin were conducted in the same reaction buffer as described above, with a dansyl-calmodulin concentration of 0.20 μM and peptide concentrations between 0 and 20 μM. The apparent dissociation constant Kd was determined by fitting the normalized fluorescence at 495 nm to the following equation, described before [14], using KaleidaGraph 2.1 (Synergy Software):
where F is the relative fluorescence, F0 is the fluorescence in the absence of peptide, and F∞ is the fluorescence at saturation; [peptidet] is the total peptide concentration, [dCaMt] is the total calmodulin concentration, and Kd the apparent dissociation constant.
Results and Discussion
Interaction of wild-type AQP6 with calmodulin
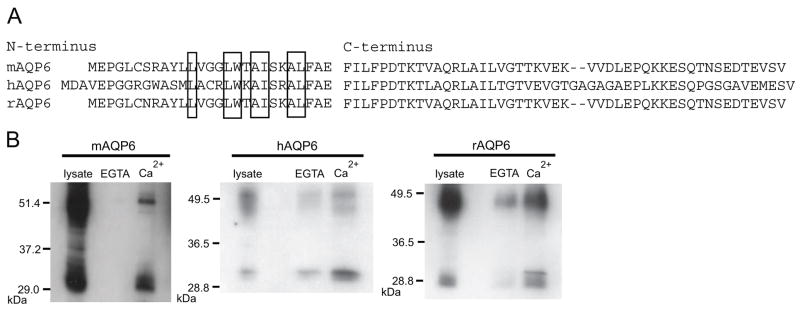
The N and C-termini of mouse and rat AQP6 are almost identical while human AQP6 differs more substantially. However, putative calmodulin binding sites exist in the N-termini of mouse, rat, and human AQP6 (Figure 1A). Lysates from CHO-K1 cells expressing human, mouse or rat AQP6 were tested for their interaction with calmodulin by pull-down with calmodulin beads in the presence of Ca2+ or EGTA. All three polypeptides demonstrated binding to calmodulin in the presence of calcium which was inhibited in the presence of EGTA (Figure 1B).
Figure 1.
A, Amino acid sequences from the N and C-termini of mouse, human and rat AQP6. The residues of the putative calmodulin binding region are boxed. B, Immunoblots of samples from CHO-KI cells expressing mAQP6, hAQP6, or rAQP6. Lysates were mixed with calmodulin-agarose beads and either 2.5 mM EGTA or 1 mM Ca2+. The samples that bound the beads were loaded (EGTA and Ca2+).
Manipulation of putative calmodulin binding site in mouse aquaporin 6
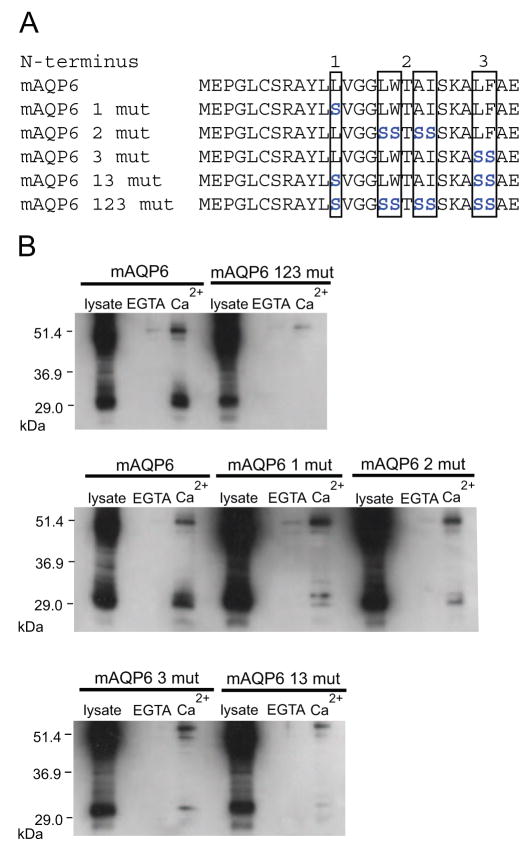
The putative calmodulin binding site in AQP6 conforms to a Type A 1-8-14 motif with hydrophobic residues at positions 1, 5, 8, and 14 [15]. Due to the additional hydrophobic residues adjacent to the 1, 5, 8, and 14 positions, we selectively replaced several residues with serine, as shown in Figure 2A. Expression of the mutants in CHO-K1 cells and binding assays with the mutated forms showed that while binding was disrupted to varying degrees by each of the mutations, only the combination of all three sets of mutations resulted in nearly complete elimination of calmodulin binding (Figure 2B).
Figure 2.
A, Amino acid sequences of mouse AQP6 N-terminus calmodulin binding site mutants. Residues that are mutated are blue. B, Immunoblots of samples from CHO-KI cells expressing full length mAQP6 or mAQP6 mutants. Lysates were mixed with calmodulin-agarose beads and either 2.5 mM EGTA or 1 mM Ca2+. The samples that bound the beads were loaded (EGTA and Ca2+).
Fluorometric determination of binding affinity of mouse AQP6
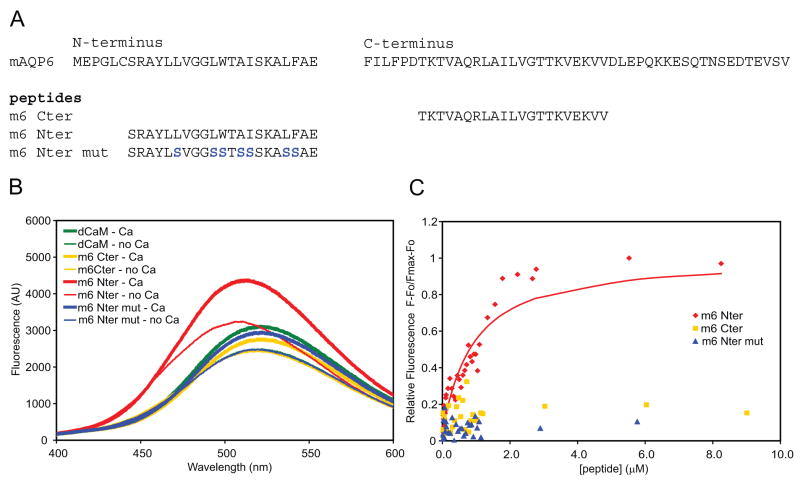
Synthetic peptides were obtained for the regions comprising the putative site from the N-terminus of mouse AQP6, a mutated version of the N-terminal site, and a putative calmodulin binding site from the C-terminus of mouse AQP6 (Figure 3A). We used fluorometry to test the calmodulin binding affinity of the peptides. Sample spectra in Figure 3B show that the mAQP6 N-terminal peptide (Nter) binding to dansyl-calmodulin not only enhances fluorescence, but also results in a blue-shift in its emission spectrum, from a λmax of ~521 nm for dansyl-calmodulin alone to ~512nm in the presence of the peptide. An increase in dansyl-calmodulin fluorescence and a blue-shift in emission spectrum is characteristic of peptide binding [16]. The fluorescence of dansyl-calmodulin in the presence of peptide was increased in 100 μM free Ca2+ compared to 0 μM free Ca2+. By titrating dansylated calmodulin with increasing concentrations of the peptides, the mAQP6 N-terminal peptide (Nter) yielded an apparent Kd of 0.79 μM ± 0.08μM, while the mAQP6 C-terminal and mAQP6 N-terminal mutant peptides showed no binding to calmodulin (Figure 3C).
Figure 3.
A, Amino acid sequences of wild-type mAQP6 N- and C-termini and of the three synthetic 21-aa peptides tested. B, Sample fluorometric emission spectra of 200 nM dansyl-Calmodulin in 0 or 100 μM Ca2+, in the presence of 3 μM of the peptides. Reactions were conducted at 23 °C in 100 mM Tris-HCl buffer (pH 7.5) containing 50 mM NaCl, using an EGTA/CaCl2 buffer to regulate the Ca2+ concentration. C, Titration of 200 nM dansyl-calmodulin with 0 to 9 μM of the three mAQP6 peptides in the presence of Ca2+. The fluorescence was measured at the emission wavelength of 495 nm.
Fluorometric determination of calmodulin binding affinity of human AQP6
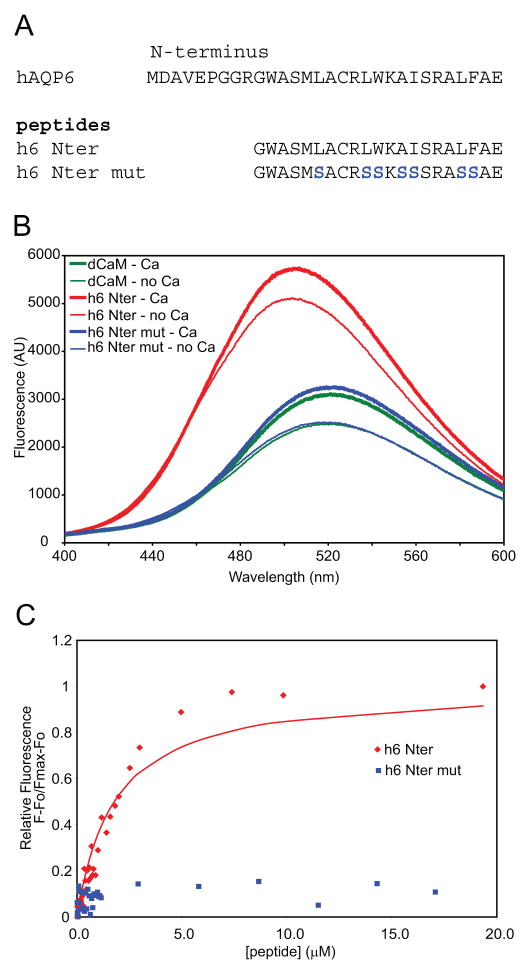
Similar to the studies of mouse AQP6, peptides were obtained for the putative calmodulin binding site of human AQP6 (h6 Nter) and for its mutant version (h6 Nter mut) and used to titrate dansylated calmodulin for fluorometry (Figure 4A). As with the mAQP6 N-terminal peptide, the hAQP6 N-terminal peptide both enhanced fluorescence significantly and resulted in a shift in emission to a λmax of ~506 nm (Figure 4B). The hAQP6 N-terminal peptide yielded an apparent Kd of 1.78 μM ± 0.13 μM, while the mutant peptide showed no binding affinity for dansyl-calmodulin (Figure 4C). The Kd for mAQP6 and hAQP6 peptides binding to dansylated calmodulin is similar to the Kd for portions of channels such as TRPV1 [17], a cyclic nucleotide-gated channel [18], the serotonin 5-HT1A receptor [19], and a P/Q-type calcium channel [20] binding to calmodulin.
Figure 4.
A, Amino acid sequences of wild type hAQP6 N-terminus and two synthetic peptides tested. B, Sample fluorometric emission spectra of 200 nM dansyl-Calmodulin in 0 or 100 μM Ca2+, in the presence of 3 μM of the peptides. Reactions were conducted at 23 °C in 100 mM Tris-HCl buffer (pH 7.5) containing 50 mM NaCl, using an EGTA/CaCl2 buffer to regulate the Ca2+ concentration. C, Titration of 200 nM dansyl-calmodulin with 0 to 20 μM of the two hAQP6 peptides in the presence of Ca2+. The fluorescence was measured at the emission wavelength of 495 nm.
We have shown that AQP6 binds calmodulin in a calcium-dependent manner in vitro. The ability of AQP6 to bind calmodulin has been maintained during evolution despite some divergence in the sequence of the N-terminus of AQP6 across species. The binding of AQP6 to the N-terminus is interesting because the N-terminus is thought to be important for AQP6 localization to intracellular vesicles [21]. It is possible that calmodulin binding to the N-terminus of AQP6 could affect its localization in the cell.
Future studies should focus on the role of calmodulin binding AQP6 in vivo. Since an increase in intracellular calcium is often involved during vesicular trafficking, calmodulin may bind AQP6 during transport of AQP6 and H+-ATPase containing vesicles to the plasma membrane. It is possible that the binding of calmodulin to AQP6 inhibits ion channel activity at a time when AQP6 is presumably about to enter the plasma membrane. Studies of isolated and perfused collecting ducts comparing wild type and AQP6 null mice may be useful in these studies. Understanding the role of calmodulin in regulating AQP6 activity is crucial for determining its physiological role in intercalated cells in the kidney.
Acknowledgments
This work was supported in part by National Institutes of Health grant DK065098.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yasui M, Kwon TH, Knepper MA, Nielsen S, Agre P. Aquaporin-6: An intracellular vesicle water channel protein in renal epithelia. Proc Natl Acad Sci USA. 1999;96:5808–5813. doi: 10.1073/pnas.96.10.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuki-Fukushima M, Hashimoto S, Shimono M, Satoh K, Fujita-Yoshigaki J, Sugiya H. Presence and localization of aquaporin-6 in rat parotid acinar cells. Cell Tissue Res. 2008;332:73–80. doi: 10.1007/s00441-007-0558-4. [DOI] [PubMed] [Google Scholar]
- 3.Lopez IA, Ishiyama G, Lee M, Baloh RW, Ishiyama A. Immunohistochemical localization of aquaporins in the human inner ear. Cell Tissue Res. 2007;328:453–460. doi: 10.1007/s00441-007-0380-z. [DOI] [PubMed] [Google Scholar]
- 4.Taguchi D, Takeda T, Kakigi A, Okada T, Nishioka R, Kitano H. Expression and immunolocalization of aquaporin-6 (Aqp6) in the rat inner ear. Acta Otolaryngol. 2008;128:832–840. doi: 10.1080/00016480701765691. [DOI] [PubMed] [Google Scholar]
- 5.Yasui M, Hazama A, Kwon TH, Nielsen S, Guggino WB, Agre P. Rapid gating and anion permeability of an intracellular aquaporin. Nature. 1999;402:184–187. doi: 10.1038/46045. [DOI] [PubMed] [Google Scholar]
- 6.Peracchia C, Girsch SJ. Is the C-terminal arm of lens gap junction channel protein the channel gate? Biochem Biophys Res Commun. 1985;133:688–695. doi: 10.1016/0006-291x(85)90959-3. [DOI] [PubMed] [Google Scholar]
- 7.Girsch SJ, Peracchia C. Lens cell-to-cell channel protein: I. Self-assembly into liposomes and permeability regulation by calmodulin. J Membr Biol. 1985;83:217–225. doi: 10.1007/BF01868696. [DOI] [PubMed] [Google Scholar]
- 8.Girsch SJ, Peracchia C. Calmodulin interacts with a C-terminus peptide from the lens membrane protein MIP26. Curr Eye Res. 1991;10:839–849. doi: 10.3109/02713689109013880. [DOI] [PubMed] [Google Scholar]
- 9.Reichow SL, Gonen T. Noncanonical binding of calmodulin to aquaporin-0: implications for channel regulation. Structure. 2008;16:1389–1398. doi: 10.1016/j.str.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose KM, Wang Z, Magrath GN, Hazard ES, Hildebrandt JD, Schey KL. Aquaporin 0-calmodulin interaction and the effect of aquaporin 0 phosphorylation. Biochemistry. 2008;47:339–347. doi: 10.1021/bi701980t. [DOI] [PubMed] [Google Scholar]
- 11.Németh-Cahalan KL, Hall JE. pH and calcium regulate the water permeability of aquaporin 0. J Biol Chem. 2000;275:6777–6782. doi: 10.1074/jbc.275.10.6777. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- 13.Bertrand B, Wakabayashi S, Ikeda T, Pouysségur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J Biol Chem. 1994;269:13703–13709. [PubMed] [Google Scholar]
- 14.Leclerc E, Corti C, Schmid H, Vetter S, James P, Carafoli E. Serine/threonine phosphorylation of calmodulin modulates its interaction with the binding domains of target enzymes. Biochem J. 1999;344:403–411. [PMC free article] [PubMed] [Google Scholar]
- 15.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 16.Malencik DA, Anderson SR. Binding of simple peptides, hormones, and neurotransmitters by calmodulin. Biochemistry. 1982;21:3480–3486. doi: 10.1021/bi00257a035. [DOI] [PubMed] [Google Scholar]
- 17.Grycova L, Lansky Z, Friedlova E, Obsilova V, Janouskova H, Obsil T, Teisinger J. Ionic interactions are essential for TRPV1 C-terminus binding to calmodulin. Biochem Biophys Res Commun. 2008;375:680–683. doi: 10.1016/j.bbrc.2008.08.094. [DOI] [PubMed] [Google Scholar]
- 18.Grunwald ME, Yu WP, Yu HH, Yau KW. Identification of a domain on the beta-subunit of the rod cGMP-gated cation channel that mediates inhibition by calcium-calmodulin. J Biol Chem. 1998;273:9148–9157. doi: 10.1074/jbc.273.15.9148. [DOI] [PubMed] [Google Scholar]
- 19.Turner JH, Gelasco AK, Raymond JR. Calmodulin interacts with the third intracellular loop of the serotonin 5-hydroxytryptamine1A receptor at two distinct sites: putative role in receptor phosphorylation by protein kinase C. J Biol Chem. 2004;279:17027–17037. doi: 10.1074/jbc.M313919200. [DOI] [PubMed] [Google Scholar]
- 20.Lee A, Wong ST, Gallagher D, Li B, Storm DR, Scheuer T, Catterall WA. Ca2+/calmodulin binds to and modulates P/Q-type calcium channels. Nature. 1999;399:155–159. doi: 10.1038/20194. [DOI] [PubMed] [Google Scholar]
- 21.Beitz E, Liu K, Ikeda M, Guggino WB, Agre P, Yasui M. Determinants of AQP6 trafficking to intracellular sites versus the plasma membrane in transfected mammalian cells. Biol Cell. 2006;98:101–109. doi: 10.1042/BC20050025. [DOI] [PubMed] [Google Scholar]