Summary
Understanding the biological potential of fetal stem/progenitor cells will help define mechanisms in liver development and homeostasis. We isolated epithelial fetal human liver cells and established phenotype-specific changes in gene expression during continuous culture conditions. Fetal human liver epithelial cells displayed stem cell properties with multilineage gene expression, extensive proliferation and generation of mesenchymal lineage cells, although the initial epithelial phenotype was rapidly supplanted by mesoendodermal phenotype in culture. This meso-endodermal phenotype was genetically regulated through cytokine signaling, including transforming growth factor β, bone morphogenetic protein, fibroblast growth factor and other signaling pathways. Reactivation of HNF3α (FOXA1) transcription factor, a driver of hepatic specification in the primitive endoderm, indicated that the meso-endodermal phenotype represented an earlier developmental stage of cells. We found that fetal liver epithelial cells formed mature hepatocytes in vivo, including after genetic manipulation using lentiviral vectors, offering convenient assays for analysis of further cell differentiation and fate. Taken together, these studies demonstrate plasticity in fetal liver epithelial stem cells, offer paradigms for defining mechanisms regulating lineage switching in stem cells, and provide potential avenues for regulating cell phenotypes for applications of stem cells, such as for cell therapy.
Keywords: Fetal, Epithelial, Liver, Mesenchymal, Stem cells, Differentiation
Introduction
The fetal human liver is enriched in stem/progenitor cells suitable for clinical applications and basic studies of stem cell biology. In recent studies, stem cells isolated from the fetal human liver replicated extensively, withstood genetic manipulations and, under specific circumstances recapitulated aspects of additional lineages – the pancreatic β-cell lineage with insulin expression and correction of hyperglycemia in animals (Malhi et al., 2002; Lazaro et al., 2003; Wege et al., 2003; Zalzman et al., 2003; Dan et al., 2006). These properties, which are distinct from those of adult hepatocytes, should offer insight into the basis of stemness in fetal liver stem cells, including the study of lineage advancement mechanisms.
Recently, the possibility of epithelial-mesenchymal transition (EMT) in endodermal cells from the liver or pancreas, generated interest in epithelial plasticity as a way to expand cells for cell therapy (Gershengorn et al., 2004). Although the phenomenon of EMT has been well studied – for example, in oncogenesis (Pagan et al., 1995; Hugo et al., 2007) – the molecular pathways regulating EMT in stem cells are incompletely understood.
To obtain further insight into the potential of epithelial fetal liver stem cells that undergo expansion in vitro, we considered that further study of cells expressing epithelial cell adhesion molecule (EpCAM) would be appropriate. This molecule is selectively displayed by epithelial cells and characterizes subsets of hepatic stem cells (Winter et al., 2003; Inada et al., 2007). From 7 to 24 weeks of fetal liver development, EpCAM-positive cells are in a state of extensive proliferation, with coexpression of hepatic markers, such as albumin (Alb) and α-fetoprotein (AFP), and biliary markers, such as cytokeratin (CK)-19 and γ-glutamyltranspeptidase (GGT). Here, we demonstrate by multiple assays that fetal liver stem cells develop unique phenotype-specific changes during continuous cell culture, where the initial endodermal phenotype was supplanted by a conjoint meso-endodermal phenotype. In this situation, cells coexpressed epithelial and mesenchymal properties with evidence of regression toward more-primitive developmental stages. Analysis of genome-wide gene expression identified coordinated alteration in the expression of multiple genes, including cytokine-specific signaling networks. We developed promoter-based assays to demonstrate retention of hepatic properties under both in vitro and in vivo conditions following transplantation of genetically modified fetal liver cells, which proved capable of generating mature hepatocytes in the liver of immunodeficient mice without fusion with native liver cells. These findings will be relevant for understanding how lineage development in stem cells either requires or benefits from this conjoint meso-endodermal stage equivalent to that of fetal liver cells.
Results
Fetal cells display stem-cell properties
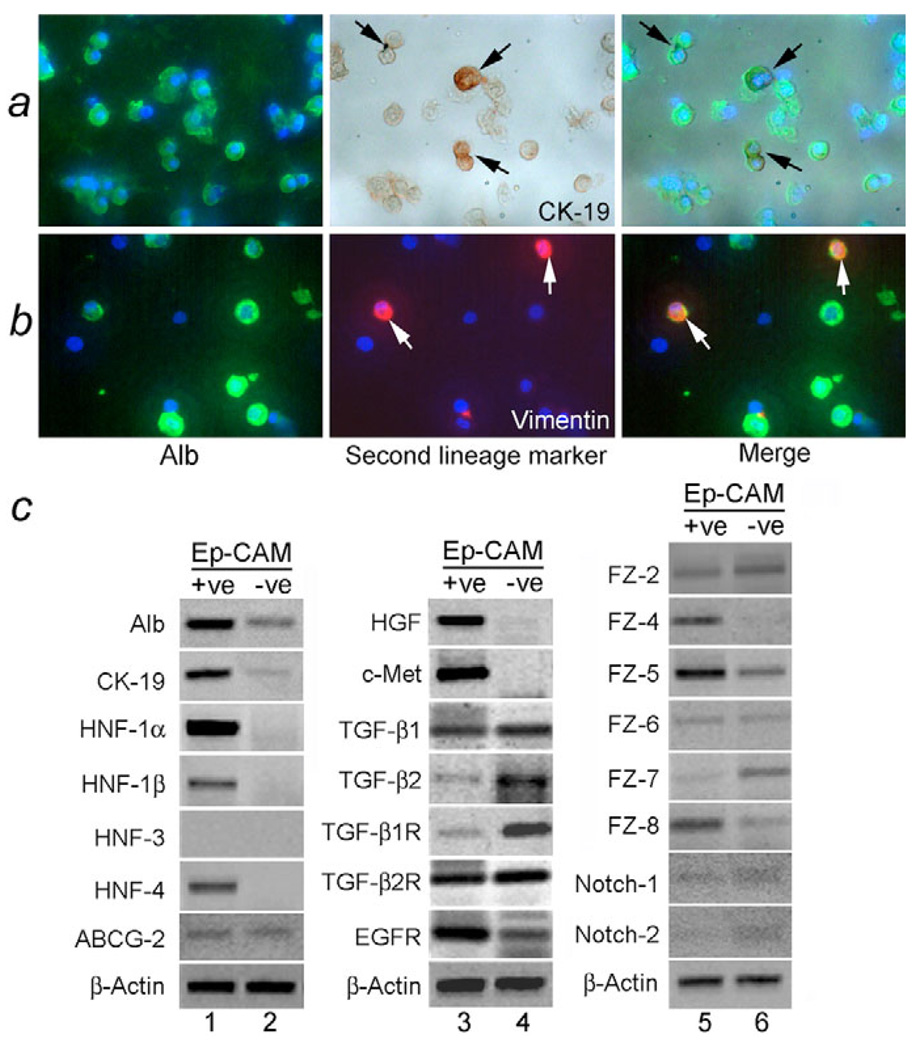
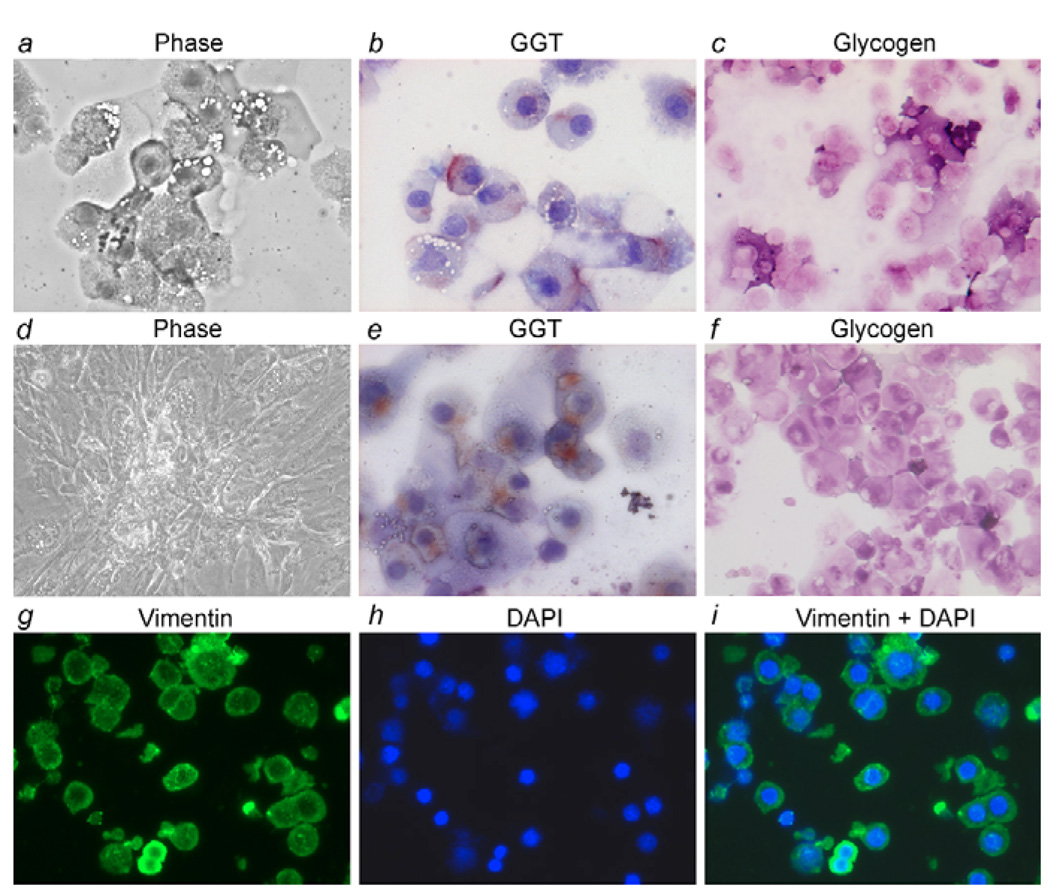
Dissociation of 19- to 24-week-old fetal livers (n=19) with collagenase yielded 1.0±0.4×109 nucleated cells with a viability of 88±8%. Immunomagnetic cell sorting for EpCAM provided 6.6±2×107 epithelial cells per liver. Secondary immunostaining verified EpCAM-positivity in sorted cells (supplementary material Fig. S1). In all sorted cells, immunohistochemical assays identified the presence of one or more hepatobiliary markers, such as glycogen, α1 antitrypsin, GGT, DPPIV or CK-19, indicating separation of only epithelial cells. Some of these cells simultaneously expressed vimentin or α-SMA, although it was noteworthy that no EpCAM-positive cells stained for mesenchymal markers without a hepatobiliary marker. This indicated that isolated cells had not been contaminated with mesenchymal cells. Moreover, coexpression of the hepatobiliary markers, Alb and CK-19 or glycogen and CK-19, was observed in 15±0.4% and 15±1% of EpCAM-positive cells, respectively. We observed coexpression of Alb and vimentin in 6±3% cells (Fig. 1; supplementary material Fig. S2 and Table S1). These findings indicated that freshly isolated fetal liver epithelial cells were characterized by multilineage gene expression.
Fig. 1.
Gene expression in primary fetal liver cells following MACS. (a,b) Gene expression with two-color immunostaining in EpCAM-positive cells to demonstrate coexpression of Alb and CK-19 or vimentin (arrows). DAPI nuclear counterstain, blue color. Magnification: ×200. (c) Gene expression by RT-PCR amplification in EpCAM-positive or -negative cell fractions, including hepatobiliary markers (Alb, CK-19), liver transcription factors (hepatic nuclear factors, HNFs) (lanes 1, 2), growth factors and receptors (lanes 3, 4) and cell-signaling molecules Wnt receptors, frizzled (FZ) and Notch (lanes 5, 6).
Analysis of the repertoire of gene expression in EpCAM-positive cells demonstrated expression of transcription factors, growth factors and intracellular signaling factors, which were not observed in EpCAM-negative cells (Fig. 1). Of note was ABCG2, which is expressed in stem cells (Zhou et al., 2001), Notch members, which help direct cell differentiation (Morrison et al., 2000), and cell signaling members, such as those involved in Wnt signaling (Ober et al., 2006). Differences in the expression of stem-cell-related genes in EpCAM-positive and EpCAM-negative cell fractions probably reflected different cell types in the fractions.
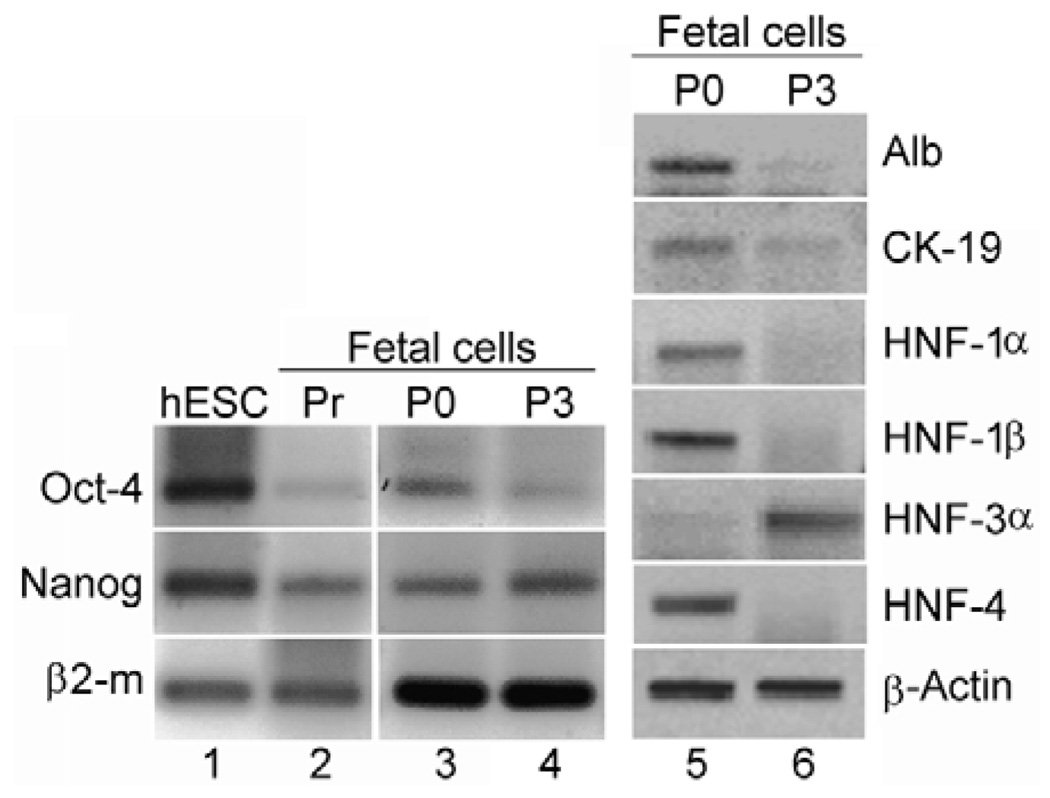
EpCAM-positive cells efficiently attached to culture dishes and showed extensive proliferation capacity with Ki67 expression in 63±9%, 60±10% and 40±6% cells in P0 cultures and cells passaged four (P4) and seven (P7) times, respectively (supplementary material Fig. S3). Moreover, EpCAM-positive fetal cells shared expression of Nanog, Oct4 and alkaline phosphatase with undifferentiated human embryonic stem cells (hESCs) (Fig. 2). By contrast, expression of hepatobiliary genes, including Alb, AFP, CK-19 and hepatic nuclear factors (HNFs) declined in subpassaged fetal liver cells, with the exception of HNF-3α. Surprisingly, most cultured fetal cells began to display the mesenchymal marker, vimentin. The onset of HNF-3α expression was noteworthy because this transcription factor is expressed very early during embryonic hepatic specification, at least in the mouse (Gualdi et al., 1996), and suggested regression of differentiation in cultured fetal cells. Although Wnt pathway genes and regulatory interactions between Nanog, Oct4 and Sox2 transcription factors play seminal roles in maintaining hESCs in pluripotential states (Sato et al., 2004; Boyer et al., 2005), the functions of these and other genes in fetal cells are unclear.
Fig. 2.
RT-PCR analysis of gene expression in Ep-CAM-positive cells. Gene expression in hESCs cultured under undifferentiating conditions (lane 1), EpCAM-positive fetal liver cells before cell culture (Pr, lane 2), during primary cell culture (P0, lanes 3, 5) and during the third passage in cell culture (P3, lanes 4, 6).
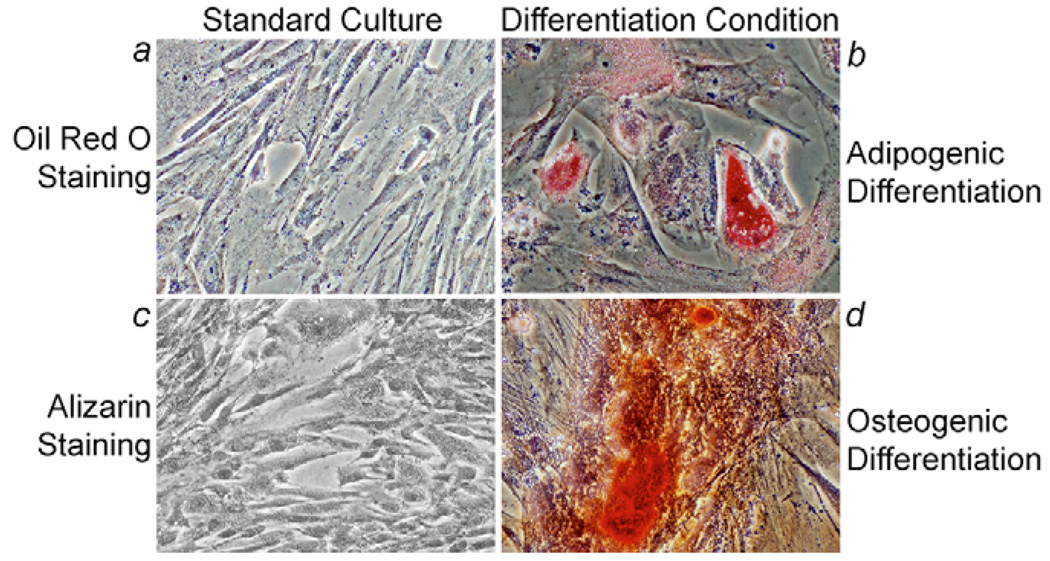
To determine whether fetal liver cells undergoing phenotypic alterations with coexpression of epithelial and mesenchymal markers would generate additional lineages, we cultured P2 cells with established protocols for inducing osteogenic or adipogenic differentiation in mesenchymal stem cells generated from hESCs (Olivier et al., 2006). We cultured cells for 3 weeks under these differentiation conditions followed by analysis of osteogenic (Alizarin Red staining for calcium) or adiopocytic (Oil Red O staining for lipid) differentiation. These studies demonstrated that cultured fetal liver epithelial cells that had acquired additional mesenchymal properties were capable of generating osteocytes and adiopocytes (Fig. 3), which was in agreement with cellular multipotency.
Fig. 3.
Generation of mesenchymal lineages from cultured fetal liver cells. P2 passage cells cultured under conditions to induce adipogenic and osteogenic differentiation. Cells cultured under standard conditions did not show staining for Oil Red O (a) or Alizarin (c). Under differentiation induction conditions, cells began to accumulate lipid droplets and acquired a foamy appearance (b), which is typically seen in adipocytes, or exhibited calcium deposition (d), which is typically seen in osteocytes. Magnification: ×200.
Cultured fetal liver cells retain hepatic properties
We considered that for assessing the extent of phenotypic changes in fetal liver cells, studies of promoter regulation would be appropriate, because promoter activity is governed by dominant negative and positive lineage-specific cellular factors, even in liver cells (Ott et al., 1999a). As lentivirus vectors can transduce fetal liver cells permanently, we used lentivirus vector constructs to analyze expression of the green fluorescent protein (GFP) reporter under control of either the ubiquitously active phosphoglycerate kinase (PGK) promoter or the Alb and transthyretin (TTR) promoters, which strongly favor hepatocytes.
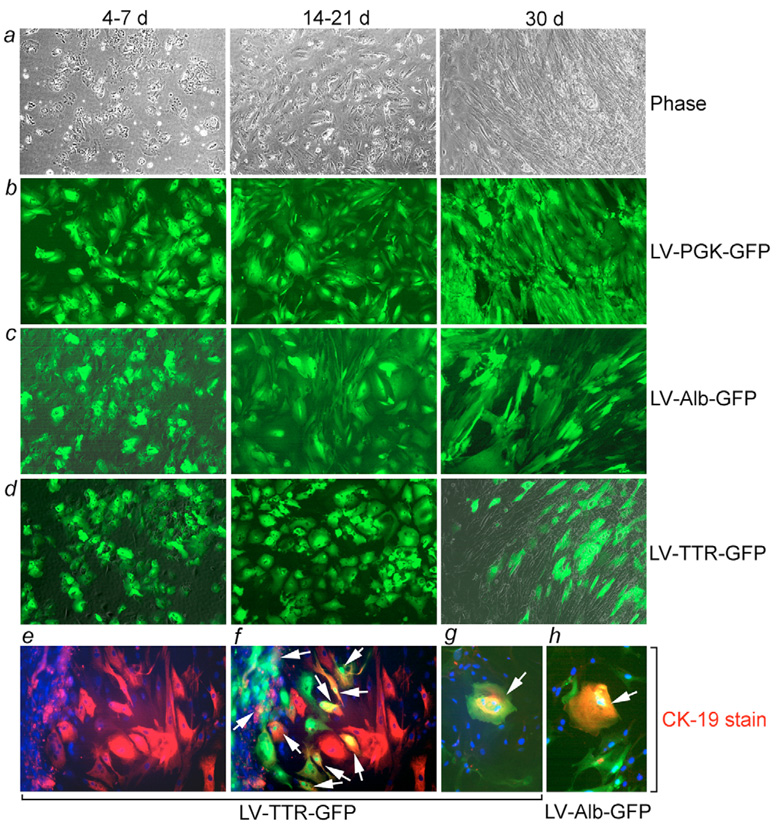
In primary culture (P0), EpCAM-positive fetal liver cells demonstrated characteristic hepatocyte-like morphology with a compact and rounded appearance, large nucleus, prominent nucleoli and complex cytoplasmic structure (Fig. 4a). When cells were transduced with lentivirus vectors in titers to transduce virtually 100% cells, for example with the PGK-GFP construct (Fig. 4b), we observed Alb and TTR promoter activity in virtually all P0 fetal cells (Fig. 4c,d), which indicated a hepatic-predominant phenotype. In these studies, the extent of cell transduction was verified by colocalization of cytoplasmic GFP and cell nuclei with 4′-6-diamidino-2-phenylindole (DAPI) staining (supplementary material Fig. S4). In continuous culture conditions for 1–2 weeks, cells retaining Alb and TTR promoter activity started to change their hepatocyte-like morphology to assume a flattened shape, although the Alb promoter remained active in these cells (Fig. 4d). We verified the extent of promoter activity by flow cytometry in cells cultured continuously over two passages. PGK-GFP was expressed in 92%, Alb-GFP in 84% and TTR-GFP in 53% of these P2 cells. Mean GFP intensity indicated that promoter strength was in the order of PGK>Alb>TTR, with relative intensity, 1.0:0.2:0.1. The cellular phenotype was verified by staining for hepatobiliary markers, which showed that despite altered morphology in 30-day P0 cultures, 60%, 10% and 20% cells expressed Alb, AFP and CK-19, respectively. Of cells with CK-19 expression, 10% coexpressed GFP under control of the TTR promoter. Moreover, cells showed histochemical staining for glycogen and GGT. These findings of liver gene expression were again consistent with the multipotent phenotype of cells and cells expressing CK-19 and TTR or Alb promoter activities at high levels were present in cultures over 7 weeks after multiple cell passages (Fig. 4e,f).
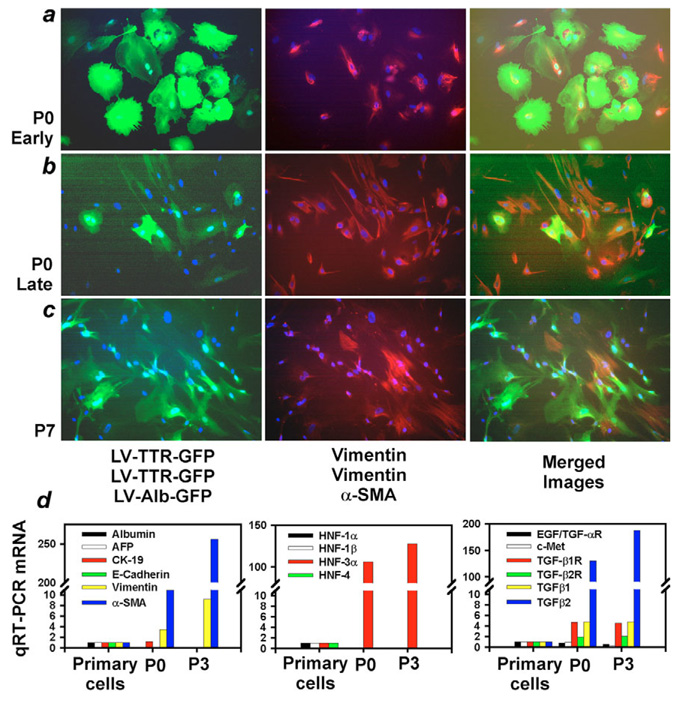
Fig. 4.
Promoter activity in EpCAM-positive cells. P0 cells in early (4–7 day) intermediate (14–21 day) and late (30 day) cultures. (a) Cells with advancement along fibroblast-like morphology. (b-d) GFP expression in P0 cells transduced with lentiviral vector using PGK, Alb and TTR promoters. (e,f) Late P0 cultures with CK-19 staining (e, red color) and merged image showing coexpression of GFP under TTR promoter (f, green color=GFP; arrows indicate CK-19-GFP coexpressing cells). (g,h) Epithelial cells in P3 cultures with merged images of CK-19 immunostaining and GFP under either TTR (g) or Alb promoter (h) (arrows). DAPI nuclear counterstain (blue). Magnification: ×200.
To further verify results from mass cultures, we studied cells from three additional fetal livers and stained freshly isolated cells in cytospun preparations for glycogen alone, GGT alone, or both glycogen and GGT. This demonstrated staining of EpCAM-sorted cells for GGT, as well as glycogen in 100% cells (Fig. 5a-c). Next, we established single-cell colonies for analyzing the properties of clonally derived cells. The resultant cell colonies were monitored and selected with cloning rings for further expansion. The colonies formed by these cells initially showed a characteristic epithelial morphology. Subsequently, cell morphology changed (Fig. 5d), although all clonal cultured cells remained positive for glycogen. When clonal cells were released from late P0 and P1 cultures after several weeks, cytospun preparations were stained for hepatobiliary markers, and showed that many cells were positive for GGT and that all cells were positive for glycogen (Fig. 5e,f). Moreover, virtually all clonal cells expressed vimentin (Fig. 5g,h), along with epithelial markers, as indicated by costaining for glycogen and vimentin (not shown). RT-PCR assays verified that clonal fetal cells expressed both epithelial (albumin, CK-19) and mesenchymal markers (vimentin, alpha-SMA). These studies verified that data obtained in mass cultures of EpCAM-positive cells were representative of clonal cultured cells. Therefore, we considered it was appropriate to continue studies with non-clonal fetal cells.
Fig. 5.
Analysis of clonal EpCAM-positive cells. (a-c) Freshly isolated cells under phase (a), after GGT staining, red color, with hematoxylin counterstaining, blue color (b) and after glycogen staining (c). (d) An expanded colony of clonal P1 cells after culture for 6 weeks. (e-i) Cytospun preparations of cells released from late P1 culture with GGT (e) glycogen (f), as well as vimentin (g-i) staining in cells. The data verified that clonal cultured cells maintained epithelial characteristics while acquiring additional mesenchymal properties. Magnification: ×200.
Further analysis of phenotypic alteration and mesoendodermal features
Within one week in mass P0 culture, the majority of EpCAM positive fetal cells with characteristic hepatocytic morphology acquired filamentous forms of the mesenchymal marker, vimentin, while retaining epithelial properties (Fig. 6). This remarkable change persisted throughout P0 culture and after continuous cell culture during serial passages, except that vimentin expression became more prominent and α-SMA was also expressed in the presence of TTR promoter or Alb promoter activity (Fig. 6b,c). It was noteworthy that not all cells expressed vimentin and α-SMA simultaneously (supplementary material Fig. S5), which was in agreement with well-established differences in the way expression of intermediate filaments and α-SMA is regulated in mesenchymal cells (hepatic stellate cells) during injury responses or myofibroblast transformation in vivo (Benten et al., 2005b). Retention of Alb and TTR promoter activity indicated that cells maintained hepatocyte like intracellular transcriptional context. However, cellular gene expression was profoundly altered during the acquisition of this meso-endodermal phenotype (Fig. 6d). For instance, quantitative RT-PCR showed downregulation of liver epithelial genes, including Alb, AFP, CK-19, and the ubiquitous epithelial gene, E-cadherin, while mesenchymal genes, vimentin and α-SMA, were highly expressed. Particularly noteworthy was the altered expression of regulatory transcription factors, e.g. HNF1α, HNF1β and HNF4, which were suppressed, whereas HNF3α (FOXA1) was newly expressed, since its expression was undetectable in EpCAM positive cells immediately after cell isolation (see Fig. 1). We observed changes in regulatory cytokines, such that expression of hepatocyte growth factor (HGF) and its receptor, c-Met, declined, while expression of TGFβ1, TGFβ2 and corresponding TGFβ receptors increased significantly in cultured fetal cells (Fig. 6d).
Fig. 6.
Meso-endodermal phenotype in fetal liver cells. (a) Immunostaining of early P0 culture (7 days) with GFP lentiviral vector expression under TTR promoter (left, green color), filamentous vimentin (middle, red color) and merged image (on right) showing coexpression in cells of GFP and vimentin. (b) Similar findings in late P0 culture, with coexpression of GFP lentiviral vector under Alb promoter and filamentous vimentin, except for morphological alterations. (c) P7 culture with GFP expression under Alb promoter, filamentous α-SMA expression (middle) and merged image showing cells with either or both properties (right). DAPI nuclear counterstain. Magnification: ×200. (d) Summary of gene expression switches with quantitative RT-PCR showing mean fold change in mRNA abundance in freshly isolated fetal liver cells (primary), early P0 culture (7 d) and multi-week P3 cultures. Alb, AFP, CK-19, E-cadherin, and HNF1 and HNF4 were expressed less in P0 and P3 cells, while HNF3α (FOXA1) expression reappeared. By contrast, expression of vimentin, α-SMA, TGFβ1 and TGFβ2 and receptors, increased. Data were normalized against β2 microglobulin. An arbitrary baseline was assigned to HNF3α.
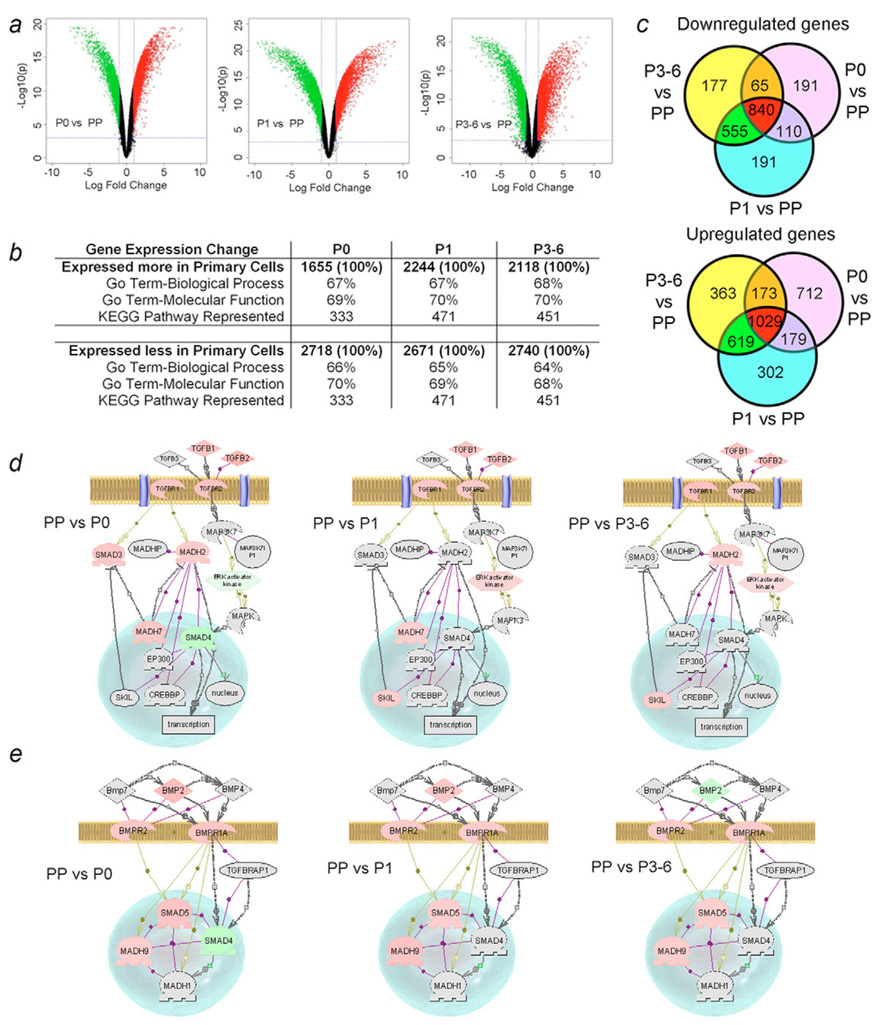
To analyze the extent of genetic reprogramming in cells undergoing the meso-endodermal phenotype, we used Affymetrix microarrays that span the entire human genome (Fig. 7). Cellular gene expression changed rapidly when freshly isolated primary (PP) cells were in P0 culture with further changes in P1 and P3 or P6 culture conditions (Fig. 7a-c). Cumulative analysis indicated that significant gene fractions were up- or downregulated more than twofold in PP cells compared with cultured cells. The differentially regulated genes represented multiple genetically defined cellular pathways. As changes in PP vs. P3 or PP vs. P6 conditions were essentially similar, to segregate uniquely regulated genes we subtracted commonly perturbed genes in P0, P1 and combined P3 P6 conditions (Fig. 7c). This comparison established that <2% of all genes expressed in cells under various conditions were either uniquely up- or downregulated in P0, P1 or P3-P6 culture. Analysis of selected cytokine-signaling networks [TGFβ, bone morphogenetic proteins (BMPs), HGF, EGF/TGFα, fibroblast growth factors (FGFs) and platelet derived growth factor (PDGF)] and regulatory pathways (Notch, Wnt) produced noteworthy findings. For instance, TGFβ signaling was inhibited in PP, whereas TGFβ inhibitory controls were progressively lost (Fig. 7d), and BMP2 became more active in P3-P6 cells (Fig. 7e). In PP cells, HGF signaling was intact with c-Met expression; in later cultures, HGF signaling was perturbed and c-Met was downregulated, with complex changes in the signaling pathways (supplementary material Fig. S6). Although TGFα signaling was intact in PP cells, in cultured cells, TGFα expression was upregulated, along with that of STAT-3, the transcriptional regulator. PDGF expression and signaling were largely unaffected. Similarly, FGFR1 expression and signaling remained intact throughout, whereas FGFR2 and FGFR3 were more highly expressed in cultured cells (supplementary material Fig. S7). Finally, expression of Notch and β-catenin, as well as Wnt signaling, was altered, although these pathways were incompletely activated in fetal liver cells (not shown).
Fig. 7.
Microarray analysis of gene expression in fetal liver cells. (a) Volcano plots of gene expression in freshly isolated cells (PP) vs P0, P1 or P3-P6 cultures. Upper right and left regions show >twofold and statistically significant differences in gene expression (green=down, red=up). (b) Overall distribution of differentially expressed genes and fractions representing two gene ontology groups and KEGG pathways. (c) Identification of genes expressed in various cell populations. The data were from total gene sequences considered to have been expressed, range, 44,256 to 50,319. Compared with PP cells, 191 genes were uniquely downregulated in P0 and P1 and 177 genes were uniquely downregulated in P3-P6. By contrast, 712, 302 and 363 genes were upregulated in P0, P1 and P3-P6, respectively. Changes in TGFβ signaling (d) and BMP signaling (e) in cultured cells compared with PP cells. Genes with higher expression in PP are in pink, those expressed less well in PP are in green, and those not represented are in grey. In PP cells, Smad2 (MADH2) and Smad3 expression was consistent with their positive roles in endoderm development and hepatic differentiation, while Smad7 (MADH7, the negative regulator of Smad2 and Smad3) was expressed simultaneously, and Smad4 and ERK activator kinase were less active. By contrast, during subsequent cell culture, Smad7 inhibition was removed, along with downregulation of ERK activator kinase and Ski-like (SKIL), which inhibit the Smad signaling regulating endodermal differentiation. Analysis of BMP signaling (e) indicated that Smad4 was expressed more highly in P0 cells and BMP2 was expressed more highly in P3-P6 cells. Depending on the context of Smad4 activity, BMP2 signaling can favor mesoderm or endoderm induction. Taken together, perturbations in TGFβ and BMP signaling were in agreement with the fetal cell phenotypes observed.
Reversible elements in mesoendodermal phenotype switch
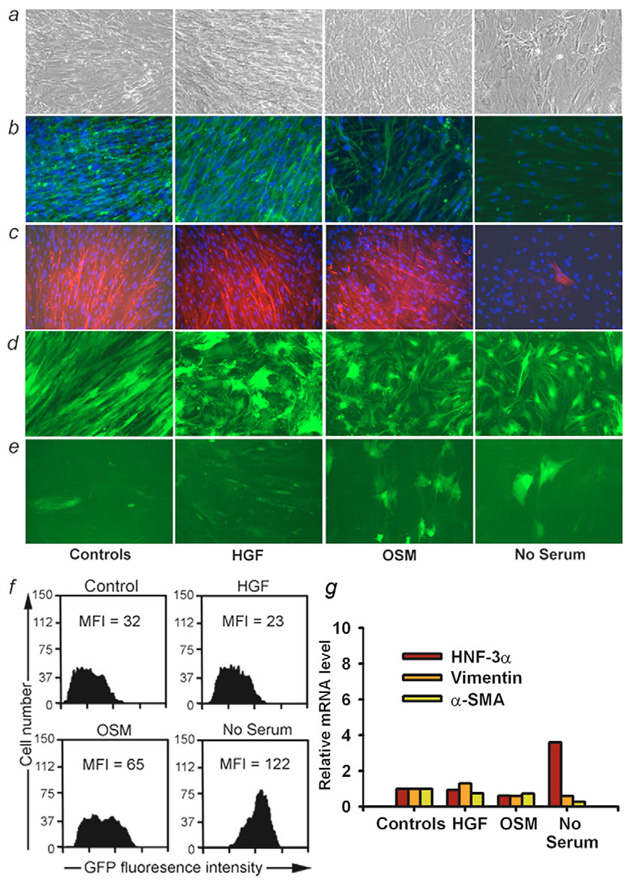
To study the possibility of manipulating cellular phenotype, which will be helpful in many situations, we transduced P2-P6 fetal liver cells with GFP lentivirus vector using Alb, TTR and PGK promoters and cultured cells for up to 2 weeks in the presence or absence of fetal bovine serum (FBS), hHGF and oncostatin M (OSM) to regulate cell growth and differentiation (Schwartz et al., 2002; Shahdadfar et al., 2005; Okaya et al., 2005). Control cells cultured with FBS again showed flattened morphologies, although culture with OSM or without serum produced less spindle-like and more rounded morphology (Fig. 8a). When P3 cells were maintained with OSM and, especially with no serum, vimentin and α-SMA expression declined (Fig. 8b,c), whereas Alb promoter activity was enhanced in some cells (Fig. 8d,e). This change was quantitated by flow cytometric measurement of mean fluorescence intensity of the GFP reporter and showed a fourfold increase in cells cultured without serum compared with control cells (Fig. 8f). Of note, in P3 cells cultured with no serum, expression of HNF3α (FOXA1), AFP and Alb mRNAs increased, whereas expression of vimentin and α-SMA mRNA decreased (Fig. 8g). Similarly, expression of HNF1β and HNF3β (FOXA2) mRNAs increased slightly, although expression of HNF4 mRNA remained downregulated. These results were not simply accounted for by cell cycle perturbations, as shown by flow cytometry (supplementary material Fig. S8), and indicated that P3 cells were responsive to some extent following perturbation of intracellular signaling.
Fig. 8.
Regulation of meso-endodermal phenotype. Perturbations in P3 fetal liver cells cultured with FBS (control) or hHGF, OSM and no serum. (a) Cell morphology under phase contrast with more compact cells in OSM or no serum. (b) Vimentin staining (green color). (c) α-SMA staining (red color) with characteristic filamentous pattern seen in mesenchymal cells and less pronounced expression in cells cultured with no serum (panel on extreme right). (d,e) Cells transduced with GFP lentiviral vector under PGK or Alb promoter, respectively. While PGK expression remained unaltered, Alb promoter was more active in cells cultured with OSM or no serum. Magnification: 200×. (f) Flow cytometry analysis indicating twofold and fourfold greater mean fluorescence intensity (MFI) of GFP expression under Alb promoter in cells cultured with OSM or no serum. (g) Changes in gene expression in P3 cells with qRT-PCR using β2-microglobulin as internal control. Vimentin expression declined to 60% of controls and α-SMA expression declined to 72% and 26% of controls in cells cultured with OSM or no serum, respectively. In cells without serum, HNF-3α (FOXA1) expression increased.
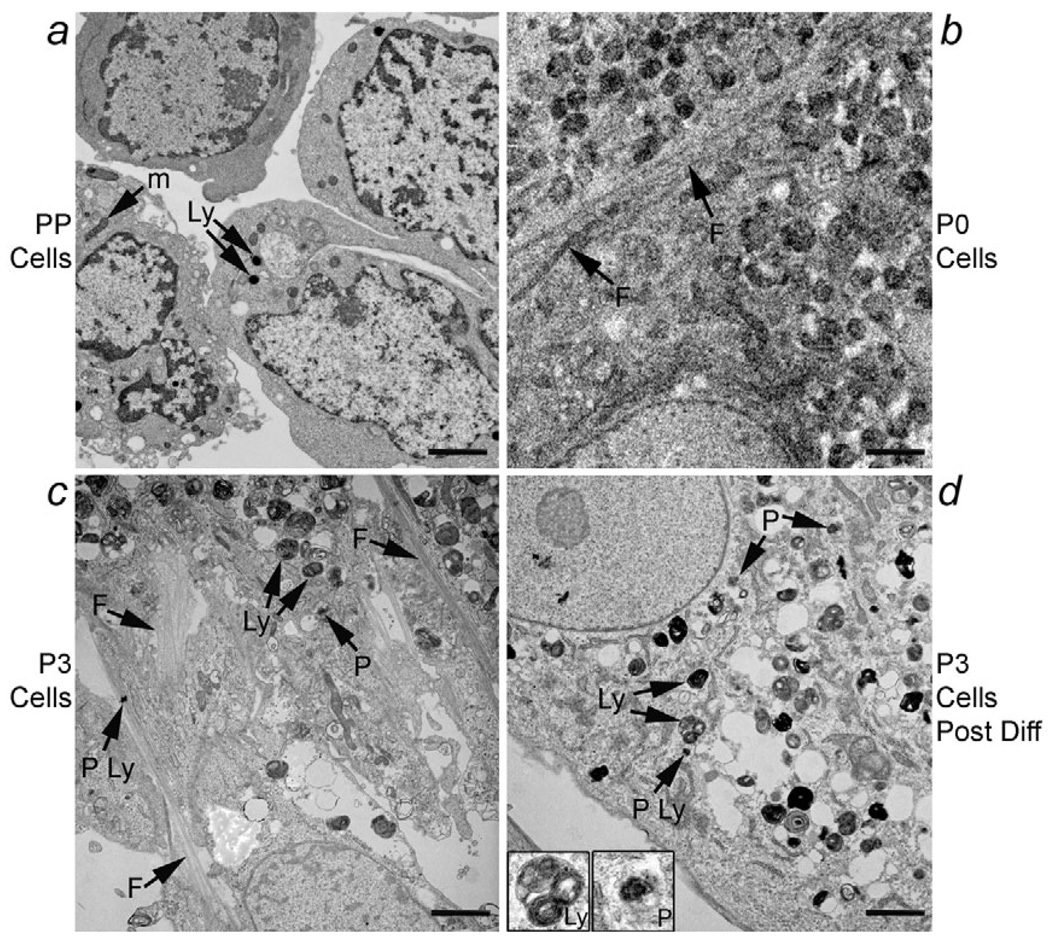
To further demonstrate changes in cellular phenotype, we performed ultrastructural analysis with transmission electron microscopy of freshly isolated EpCAM-positive PP cells, P0 cells after three weeks in culture, P2 cells, P3 cells, as well as P3 cells that had been cultured without serum for inducing phenotype alterations. These studies demonstrated that the initial epithelial phenotype of freshly isolated PP cells changed rapidly in P0 and subsequent passages with the appearance of intermediate filaments while aspects of the epithelial phenotype were present (Fig. 9a,b,c). After culture in the absence of serum, fewer cells showed intermediate filaments (>90% in P3 versus <25% in P3 without serum condition), although the epithelial phenotype was not restored to that of primary PP cells (Fig. 9d). These findings were in agreement with the rapid onset of the conjoint meso-endodermal phenotype after cell culture and the requirement of further cell differentiation to restore specific phenotypes in cells.
Fig. 9.
EM analysis of cellular ultrastructure. (a) Freshly isolated PP cells with large nuclei, prominent nucleoli and relatively sparse cytoplasm with scattered mitochondria (m) and secondary lysosomes (Ly). P0 cells (b) and P3 cells (c) that had been in culture for three weeks and seven weeks, respectively. These cells showed intermediate filaments (F, arrows) as well as complex cytoplasmic appearance with multiple mitochondria, vacuoles, occasional primary lysosomes (P Ly) as well as secondary lysosomes and microperoxisomes (P) consistent with alterations in cell morphology and presence of both mesenchymal and epithelial features. (d) P3 cells cultured without serum. Under this condition, most cells no longer contained intermediate filaments, although the epithelial phenotype did not resemble that of freshly isolated PP cells, indicating the incompleteness of phenotypic reversion. These cells again contained primary and secondary lysosomes as well as microperoxisomes, which are additionally shown in insets. Scale bars: 2 µm.
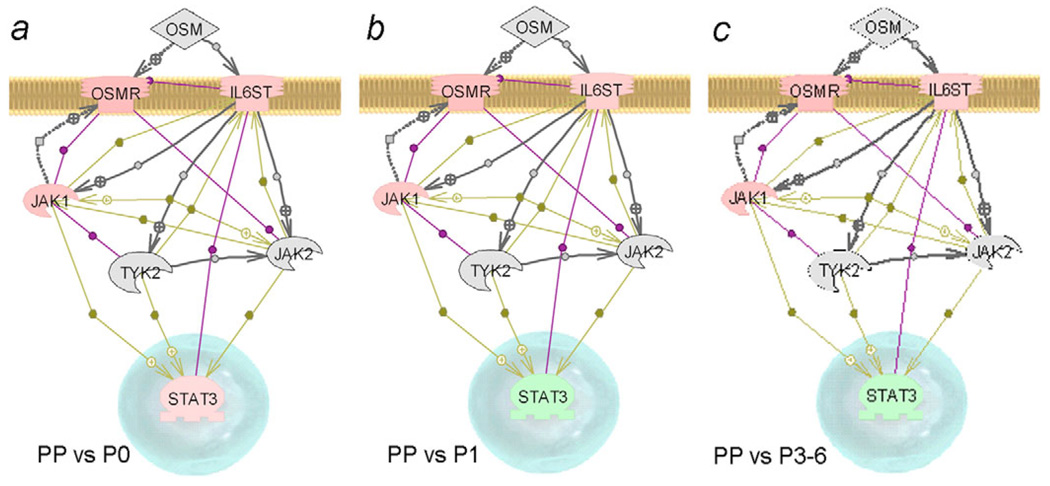
To determine whether intracellular signaling pathways relevant to cell culture manipulations were appropriately maintained, we interrogated microarray gene expression data for OSM signaling (Fig. 10). In cultured cells, the OSM signaling pathway was intact, with expression of the OSM receptor, as well as interleukin-6 signal transducer (IL6ST), albeit at lower levels than in freshly isolated PP cells. These findings were in agreement with our results showing some responsiveness of cultured cells to OSM. Moreover, the JAK1-STAT3 pathway was active in these cells rather than the JAK2-STAT3 pathway, although understanding the significance of this will require more work.
Fig. 10.
Changes in OSM signaling. (a-c) Microarray data analysis of OSM signaling in PP cells versus cultured cells. Genes with higher expression in PP cells are in pink, those expressed less well in PP cells are in green, those not expressed are in grey (IL6ST, interleukin-6 signal transducer). The OSM signaling pathway was intact, although STAT3 was upregulated in cultured cells, probably explaining the responsiveness of P3 cells to added OSM.
In vivo fate of fetal liver cells
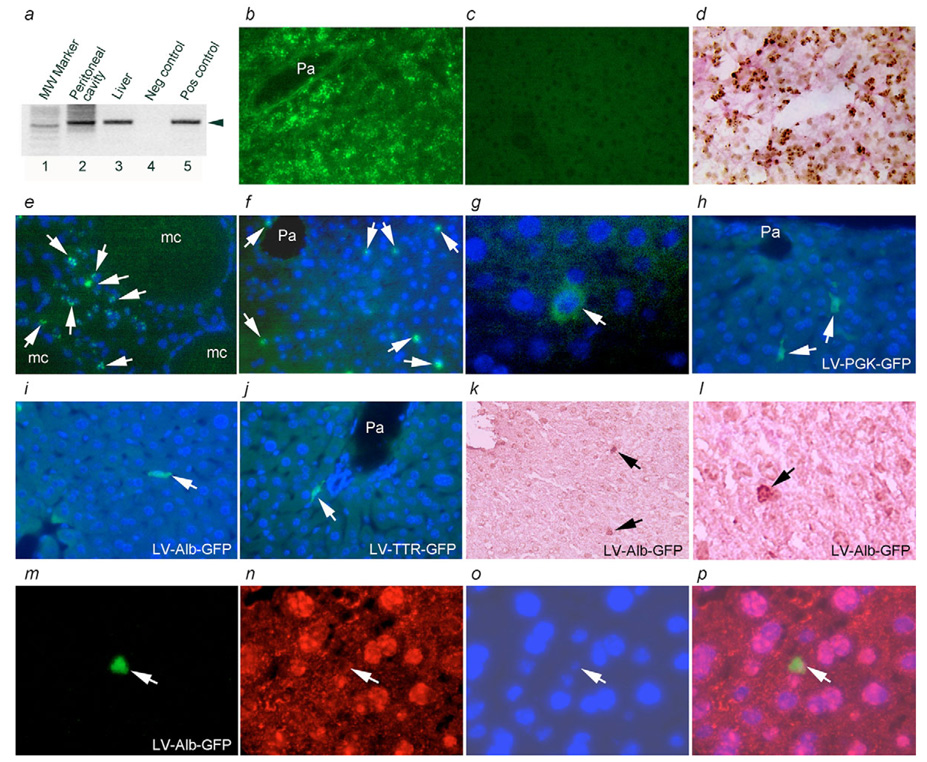
To determine whether the cell differentiation program can be completed in vivo, including after cells had been cultured and manipulated to restore given phenotypes, analysis of cell fate in vivo will be critical. Therefore, we examined whether lentivirus transduced cells would engraft in immunodeficient SCID mice. We transplanted 1×106 EpCAM-positive freshly isolated PP cells in multiple animals, with or without prior cell transduction with a GFP lentivirus vector containing one of PGK, Alb or TTR promoters. Tissue analysis 5 days after cell transplantation identified transplanted cells in an extrahepatic site, the peritoneal cavity, as well as in the liver itself (Fig. 11a). In situ hybridization using a human-specific probe showed that transplanted cells were within the expected locations in these sites (Fig. 11b-i), including next to microcarriers in the peritoneal cavity (Fig. 11e) as well as in the liver parenchyma after injection of cells into the portal vein (Fig. 11f). Transplanted cells contained immunodetectable albumin (Fig. 11g), which was in agreement with their capacity to maintain an epithelial function in vivo. Analysis of multiple cell recipients showed one to three transplanted cells adjacent to portal areas, which was similar to transplanted cell numbers in immunodeficient mouse recipients of adult human hepatocytes in a previous study (Cho et al., 2004). Moreover, we observed engraftment of cells transduced with lentivirus-GFP using PGK, Alb or TTR promoters (Fig. 11h-j). Transplanted cells retained Alb and TTR promoter activity consistent with hepatic epithelial function. Histochemical staining along with in situ hybridization verified that transplanted cells also contained glycogen (Fig. 11k,l), which was again in agreement with their epithelial nature. To exclude the possibility of fusion between transplanted and native mouse cells, we combined in situ hybridization with human and mouse pancentromere probes. Analysis of 15–20 consecutive transplanted cells in multiple tissues samples from two recipients showed absence of fusion in transplanted and mouse cells (Fig. 11m-p).
Fig. 11.
Fate of fetal cells in vivo. (a) Transplanted cells in the peritoneal cavity or liver (lanes 2, 3) of immunodeficient mice with PCR probe identifying CMT1A human sequences. (b) In situ hybridization signals with human pancentromeric probe using FITC-conjugated anti-digoxigenin in human liver. (c) No hybridization signals with this probe were seen in mouse liver. (d) Histochemical staining of human liver for glycogen (pink color in cytoplasm) followed by in situ hybridization with human pancentromeric probe using peroxidase-conjugated anti-digoxigenin and development with diaminobenzidine (dark brown color in nuclei). (e,f) Transplanted fetal human cells (arrows) in the peritoneal cavity or liver (f) using human in situ hybridization probe. mc, microcarriers; Pa, portal area. (g) Immunostaining for human albumin in a transplanted cell (arrow) in the liver. (h-j) Transplanted cells (arrows) transduced with GFP lentiviral vector containing PGK, Alb or TTR promoters, with GFP immunostaining (green) and DAPI nuclear stain (blue). (k,l) Histochemical staining for glycogen (pink) followed by in situ hybridization using human pancentromeric probe to verify that lentiviral-vector-transduced cells (brown nuclear signals, arrows) were hepatocytes. (m-p) The same liver area verifying absence of fusion in lentiviral-vector-transduced human cells and native mouse cells with in situ hybridization using human plus mouse pancentromeric probes. A transplanted cell (arrow, m) hybridizes with the human probe. Hybridization signals with the mouse probe in cells other than the human cell (n), DAPI staining of nuclei (o) and merged view of m-o (p) to indicate that mouse signals were absent in the transplanted human cell and vice versa. Magnification: ×400 (b-f, h-k); ×600 (g,l, m-p).
Discussion
Our findings indicated that EpCAM-positive fetal liver cells possess the capacity for multilineage gene expression, including expression of Oct4, Nanog and alkaline phosphatase, which are markers of pluripotency in hESCs, and extensive proliferation in vitro, while acquiring a meso-endodermal phenotype with some reversible elements. We verified this meso-endodermal phenotype by ultrastructural analysis of fetal liver cells and demonstrated that after regression to a more primitive phenotype, fetal liver cells were capable of generating mesenchymal lineage cells in vitro, while possessing the capacity to generate mature hepatocytes in vivo at an early stage. This context-specific phenotype in fetal liver stem cells was compatible in general with their plasticity and advances insights into their biological potential beyond a recent summary (Schmelzer et al., 2006). Moreover, identification of phenotypic changes in fetal liver cells will be particularly helpful in understanding lineage transitions during induction of differentiation in various stem cells. Also, the ability of cells to engraft in the liver, to integrate in the liver parenchyma and to acquire features of mature hepatocytes, including after genetic modification with lentivirus vectors, was in agreement with properties desirable for future cell and gene therapies, and other applications.
Our analysis of promoter regulation demonstrated that fetal cells with mesenchymal properties did not simply represent mesenchymal cells that developed from epithelial cells, but that cells were in a novel meso-endodermal state, supported by the activity of Alb and TTR promoters in cells. The permissivity of cells to hepatic promoter activity indicated that positive transcriptional regulators took precedence over their negative regulatory counterparts. The known dominance of stem cell phenotypes in somatic cell hybrids suggested that cellular transcriptional programs would preferentially maintain the stem cell state rather than the differentiated state (Cowan et al., 2005), which was in further agreement with our studies establishing this novel phenotype in fetal stem cells.
During embryonic development, as well as in disease, bi-directional epithelialmesenchymal and mesenchymal-epithelial cell transitions have been recognized (Hay and Zuk, 1995; Hugo et al., 2007). Also, cells emanating from adult pancreatic islets were recently observed to develop mesenchymal properties (Gershengorn et al., 2004). Our data favor the possibility that epithelial fetal liver cells reverted in culture to a more primitive phenotype that included mesenchymal elements. This notion was strengthened by the findings of downregulated expression of epithelial genes, Alb, AFP and CK-19, the ubiquitous epithelial marker, E-cadherin, as well as the transcription factors, HNF1 and HNF4, in cultured cells. However, HNF3α, which helps specify the endoderm at the earliest stages of liver and pancreas development (Gualdi et al., 1996; Lee et al., 2005), was newly expressed. The recent demonstration that hepatic mesenchymal cells can express hepatic genes raises intriguing further possibilities concerning the origin of various liver cell types and the role of a mesoendodermal fetal stage in that process (Sicklick et al., 2006).
We acknowledge that enzymatic dissociation of epithelial cells from the fetal liver, especially during the stages of active acinar reorganization, should perturb cells in multiple ways. Loss of cell-cell adhesion and/or polarity and cytoskeletal remodeling required for attachment of cells to extracellular matrix components should produce further outside-in perturbations in cell signaling. The ability of fetal liver cells to express soluble signals during these perturbations, including those capable of inducing EMT, such as TGFβ, EGF, HGF, FGFs and Wnt ligands (Thiery, 2002), should be relevant in mechanisms capable of producing phenotypic alterations. For outside-in signaling, many extracellular factors require specific receptors or cascades of secondary and tertiary messengers, such as Smad protein family members (e.g. TGFβ engages Smad2, Smad3, Smad4 and Smad7, HGF engages Smad3, and EGF engages Smad7), and glycogen synthase kinase (GSK)-3, which participates in Wnt signaling (Mishra et al., 2005). Among extracellular signaling molecules, TGFβs play significant roles in mesenchymal activations, including through multiple cellular pathways, such as those involving activated Ras, extracellular signal-regulated kinase (ERK) or mitogen-activated protein kinase (MAPK), Notch, Wnt–GSK3–β-catenin, nuclear factor (NF)-κB, and phosphatidylinositol 3-kinase (Zavadil and Bottinger, 2005). Many nuclear transcription factors serve as downstream mediators in these pathways. However, the master transcriptional factors, Nanog, Oct4 and Sox2 directly regulate key components of intracellular signaling, including TGFβ, Wnt and other pathways (Boyer et al., 2005). Therefore, although we identified critical changes in the expression of TGFβ1, TGFβ2, TGFα, FGFs and their receptors and regulators, phenotypic changes in fetal liver cells were probably multifuctional and complex, as also emphasized by our microarray data analysis.
Currently much interest is focused on induction of endodermal (hepatic, pancreatic) differentiation in pluripotent cells. The mesoendodermal reversion of lineage-committed fetal liver stem cells offers novel directions in such pursuits. Our consideration that mesoendodermal conversion represents a more primitive state of fetal liver cells should be similar to embryonic gastrulation and differentiation of hESCs along the endoderm (D’Amour et al., 2005). In this context, derepression of HNF3α (FOXA1) should be in agreement with cellular reversion to a more primitive state, because HNF3α (FOXA1) is expressed initially during endodermal reorganization, including in the vicinity of the primitive mesoderm and ectoderm in invertebrates and Drosophila, as well as in mammals (Gualdi et al., 1996; Fritzenwanker et al., 2004). Similarly, HNF3-transduced hESCs were more amenable to producing endodermal cells (Ishizaka et al., 2002). Therefore, identification of a more primitive meso-endodermal intermediate in fetal liver cells should suggest potential additional mechanisms to explore differentiation in stem cells that have advanced along the mesenchymal lineage (Barberi et al., 2005).
Such an ability to alter phenotypes in fetal liver cells should offer opportunities for developing experiments involving the use of agonists and/or antagonists, similarly to endodermal lineage differentiation in bone-marrow-derived multipotent mesenchymal cells (Schwartz et al., 2002). Epithelial fetal human liver cells were recently found capable of generating mesenchymal cells, such as endothelial cells and adipocytes (Dan et al., 2006). Similarly, we found that cultured fetal liver cells were responsive to soluble factors and generated adipocytes, as well as osteocytes, which was in agreement with their reversion to a multipotent phenotype. Finally, despite reversion to a more primitive phenotype in cell culture, the ability of fetal liver cells to engraft and maintain an epithelial phenotype in intact mice will help to demonstrate further mechanisms in cell differentiation. Moreover, the ability of fetal liver cells to engraft and proliferate in the liver will advance efforts to generate chimeric animal models and to develop cell and gene therapy applications, although this will require further analysis of primary and cultured fetal liver cells.
Materials and Methods
Tissues
The Fetal Tissue Repository at Albert Einstein College of Medicine provided 19- to 24-week fetal livers with approval from the Committee on Clinical Investigations (Institutional Review Board at Einstein).
Cells
Livers were dissociated in Leffert's buffer with 0.03% collagenase (Cat. no. 2139, Worthington Biochemical Corp., Lakewood, NJ), 5 mM CaCl2 and 500 U/ml DNase (D-5025, Sigma Chemical Co., St. Louis, MO). Dispersed cells were passed through 80 µm dacron, pelleted at 350 g for 5 minutes at 4°C, and cryopreserved in University of Wisconsin solution, as previously described (Malhi et al., 2002). Red blood cells were lysed in 155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA for 7 minutes on ice. Epithelial cells were isolated by EpCAM immunomagnetic sorting (Miltenyi Biotec, Bergisch Gladbach, Germany) by resuspending 1×107 cells per 60 µl Buffer B (phosphate-buffered saline, pH 7.2, 2 mM EDTA, 0.5% bovine serum albumin and DNase) (Sigma). FCR blocking agent (20 µl per 107 cells) was added (Cat. no. 130-059-901, Miltenyi Biotec) followed by 20 µl per 1×107 cells of Ep-CAM antibodyconjugated magnetic particles (130-061-101, Miltenyi Biotec) for 30 minutes at 4°C. Cells were washed in Buffer B, pelleted at 350 g for 5 minutes at 4°C and applied in Buffer B to separation columns (#130-041-407, Miltenyi Biotec). Cells were resuspended in Dulbecco's Minimal Essential Medium (DMEM) and pelleted at 350 g for 5 minutes at 4°C. Cell viability was determined with Trypan Blue exclusion.
Cell culture
Cells were resuspended in DMEM with 10% FBS (Atlanta Biologicals, Norcross, GA), 5 µg/ml insulin, 5 µM hydrocortisone, 100 U/ml penicillin, 100 µg/ml streptomycin and 250 ng/ml amphotericin B (DMEM; Life Technologies, Rockville, MD). 0.1–1×105 cells/cm2 were plated in plastic culture dishes in 5% CO2. Medium was changed after 1 day and then every 3 days. Cells were subpassaged 1:3 from near-confluent cultures with trypsin-EDTA for 3 minutes at 37°C. In growth factor studies, cells were cultured with 25 ng/ml hepatocyte growth factor (HGF) (H9661, Sigma) or 10 ng/ml oncostatin M (O9635, Sigma). All experiments were carried out in triplicate and were repeated for reproducibility.
For analysis of EpCAM-sorted cells under clonal conditions, cryopreserved cells were sorted and cytospins were prepared for histochemical staining of glycogen and/or GGT and for immunostaining of CK-19, as described previously (Malhi et al., 2002). Simultaneously, cells were cultured as above under low-density conditions to obtain single cell colonies under intermittent microscopic observation. Resultant cell colonies were fixed in situ for staining, and individual cell colonies were isolated after 4–6 weeks under P0 and then again in P1 conditions using cloning rings to obtain cytospun preparations for glycogen, GGT and CK-19 staining, as well as for RT-PCR analysis of epithelial and mesenchymal markers (see below).
To obtain mRNA from hESCs, WA-01 cells (WiCell Research Institute, Madison, WI) after ~50 passages, were cultured in the hESC Core Facility at Albert Einstein College of Medicine. Undifferentiated cells were maintained on mouse embryonic fibroblasts (MEFs) (irradiated to 80cGy) in DMEM-F12 medium (Invitrogen) supplemented with 20% Knockout Serum Replacer (Invitrogen), 1% MEM nonessential amino acids (Invitrogen), 1 mM L-glutamine, 4 ng/ml bFGF (ProSpecTany TechnoGene, Rehovot, Israel) and 1% penicillin-streptomycin according to published protocols (Thomson et al., 1998). Medium was changed daily and cells passaged weekly by dissociation with 1 mg/ml collagenase IV (Invitrogen). RNA was extracted by Trizol Reagent (Invitrogen).
Analysis of cell proliferation
Cells were immunostained for Ki-67 (see supplementary material Table S2). For cell cycle analysis with flow cytometry, cells were detached by trypsin-EDTA, washed with PBS, fixed in 100% ethanol for 10 minutes at room temperature and stained with 3 µM propidium iodide (P-1304, Molecular Probes, Eugene, OR). FACScan was used to collect at least 10,000 events per sample and CellQuest software was used for data analysis (BD Biosciences Pharmingen, San Diego, CA).
Osteogenic and adipogenic differentiation of fetal cells
To induce osteogenic differentiation, cells were cultured in DMEM containing 10% FBS, 100 nm dexamethasone, 50 µm ascorbic acid-2-phosphate and 10 mM β-glycerophosphate (Sigma) for 3 weeks with fresh medium every 3–4 days. To examine differentiation, cells were washed twice with PBS, fixed in 70% cold ethanol for 1 hour and incubated for 10 minutes with 40 mM Alizarin Red (Sigma). Adipogenic differentiation was induced by culturing cells with DMEM containing 10% FBS, 1 µM dexamethasone, 0.2 mM indomethacin, 10 µg/ml insulin, 0.5 mM 3-isobutyl-1 xanthine (Sigma) for 3 weeks with fresh medium every 3–4 days. To demonstrate differentiation, cells were washed with PBS, fixed in 10% formalin for 1–2 hours, and incubated for 15 minutes with fresh Oil Red-O solution (Sigma) (Olivier et al., 2006).
Gene expression
Immunohistochemistry and reverse transcription-polymerase chain reactions (RT PCR) were carried out as summarized in supplementary material Table S2 and Table S3. Total RNA was extracted by Trizol Reagent (Invitrogen) or RNeasy Mini Kit (Qiagen, Valencia, CA) and treated with DNase (Qiagen). cDNA was prepared from 2–4 µg total cellular RNA using the Omniscript RT Kit (Qiagen). Equal amounts of cDNAs were subjected to PCR with Platinum PCR Supermix (Invitrogen) using 35 cycles with denaturation at 94°C for 30 seconds, annealing at Tm selected for each primer pair for 30 seconds and final extension at 72°C for 2 minutes (supplementary material Table S3).
For quantitative analysis, qRT-PCR was performed in a total volume of 20 µl with SYBRgreen RT-PCR Kit (Qiagen) using 1 µg RNAs and 1 µM of each primer (supplementary material Table S3). Reactions were carried out in a LightCycler (Roche, Indianapolis, IN) with reverse transcription at 50°C for 20 minutes and initial PCR activation at 95°C for 15 minutes. Thermal cycles were repeated 40 times with denaturation at 94°C for 15 seconds, annealing at Tm selected for each primer pair for 25 seconds and primer extension at 72°C for 30 seconds. Gene expression levels in samples were determined by comparison against the level of β2-microglobulin expression. This comparison utilized the 2(–ΔΔCT) method of Livak and Schmittgen (Livak and Schmittgen, 2001). This method permitted analysis of differences across multiple conditions of cells. Data were obtained in replicate conditions with normalization using β2-microglobulin.
Electron microscopy
Freshly isolated EpCAM+ cells were pelleted immediately for examination or cultured as described above to obtain samples from P0, P2 and P3 conditions. In addition, P3 cells were cultured under serum-free conditions to alter cellular phenotype. Cells were fixed in 2.5% glutaraldehyde in cacodylate buffer, postfixed in osmium tetroxide and stained with 1% uranyl acetate before embedding in plastic. Ultrathin sections were examined under a JEOL 1200 transmission electron microscope with examination of 25–50 consecutive cells in each condition to determine differences in phenotype.
Microarray studies
EpCAM-positive cells were isolated from three fetal livers (19 weeks, male; 20 weeks, male; 23 weeks, female). Cells were cultured in parallel to obtain P0 (one week cell culture), P1 (one passage), P3 (three passages) and P6 (six passages) in triplicate. Total cellular RNA was extracted from freshly isolated primary (PP) and cultured cells (RNeasy Mini Kit; Qiagen). RNAs were processed individually for 15 samples in the Molecular Biology Core of the Feinstein Institute for Medical Research, North Shore-Long Island Jewish Health System, Manhasset, New York, and individually hybridized in U133 2.0 Plus human oligonucleotide arrays (Affymetrix, Santa Clara, CA).
The probe level intensities were adjusted by quantile normalization and gene expression measures were summarized by the robust multi-array analysis (RMA) approach (Irizarry et al., 2003), which is considered superior to MAS and dChip (Bolstad et al., 2003). We used Bioconductor Software for probe level data analyses. To identify differentially expressed genes among groups, empirical Bayes-moderated t-tests were performed for each gene (Bolstad et al., 2003). Statistical analysis of gene expression data used the LIMMA package for R. To account for potential inflation in type-1 error rates due to multiple testing, we calculated adjusted P values for each gene as described (Reiner et al., 2003). Genes meriting investigation were identified by adjusted P values ≤0.01 (controlling expected false-discovery rates to not more than 1%) and fold difference in mean expression ≥2.0 to compare PP cells with other cell samples. To visualize differentially expressed genes, volcano plots were prepared, which summarize fold change in gene expression and Bayes-moderated t -test results using scatter plot of −log10 transformed adjusted P values against log2 fold change. Significant genes were in the upper left or upper right portions of the volcano plot; upregulated genes were highlighted in red and downregulated genes in green. Differentially expressed genes in PP versus cultured cells were compared to identify uniquely expressed genes with SAS software algorithms (SAS Institute, Cary, NC) and Venn diagrams were prepared. The differentially expressed genes were annotated and categorized with the assistance of Database for Annotation, Visualization and Integrated Discovery (DAVID) and the related Expression Analysis Systematic Explorer (EASE) package (National Institute of Allergy and Infectious Disease, Bethesda, MD) (Dennis et al., 2003). This permitted identification of gene ontology groupings, as well as pathways, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG). To demonstrate representation of specific pathways in lists of differentially expressed genes, datasets were analyzed with PathwayStudio software (Ariadne Genomics, Rockville, MD). Expression data involving differentially expressed genes were utilized to generate biological association networks and these were queried with curated pathways in PathwayStudio. Gene expression profiles in freshly isolated cells (PP) were used as denominators to establish changes in P0, P1, P3 and P6 cultured cells. For final data analysis, we pooled data from P3 and P6 samples because of similarities in their gene expression profiles. The data were shown in a pictorial format, according to PathwayStudio definitions concerning gene expression, interactions and regulations.
Lentivirus gene transfer
Vesicular stomatitis virus (VSV)-pseudotyped lentivirus stocks were obtained by cotransfecting pCCL.PPT.hPGK.GFP.Wpre, pCCL.PPT.mALB.GFP.Wpre and pCCL.PPT.TTR.GFP.Wpre transfer constructs, the third-generation packaging constructs pMDLg/pRRE and pRSV-REV and pMD2.G envelope into 293T cells as previously described (Follenzi et al., 2002). Lentiviral vectors (LV) expressed GFP under control of the ubiquitously active PGK promoter, the liver-specific mouse albumin enhancer-promoter or the liver-specific synthetic TTR promoter (Follenzi et al., 2002; Vigna et al., 2005). Vectors were titered on HepG2 cells and showed titers of 1–3×107 TU (titration units) per ml. Cultured cells were incubated with 0.1 to 1×107 TU LV per 1×106 cells for 16–18 hours at 37°C followed by analysis of GFP expression under fluorescence microscopy, which was initially 4 days after vector transduction. For analyzing gene expression in cells in vivo, primary fetal human cells were sorted by MACS and sorted cells were incubated with 1×107 TU LVs for 1 hour at 37°C, washed and transplanted into animals.
In vivo studies using SCID mice
We transplanted 0.6–0.9 ×106 freshly isolated or cultured fetal human liver cells with or without LV transduction into SCID mice in the Balb/c background. Typically, cells were transplanted into 3 mice per group into either portal vein or peritoneal cavity. For the latter, cells were incubated with 1 ml Cytodex 3™ microcarrier beads (Amersham Biosciences, Piscataway, NJ) for 30 minutes at 37°C. Animals were sacrificed 5 days after cell transplantation to analyze cell engraftment and gene expression (see supplement).
Identification of transplanted cells in tissues
Transplanted cells were identified with in situ hybridization using 7-µm-thick cryosections fixed in 4% paraformaldehyde in PBS as described previously (Benten et al., 2005a). Sections were incubated in 2× sodium chloride and sodium citrate buffer (SSC) for 30 minutes at 37°C, denatured in 70% formamide for 2 minutes at 80°C and hybridized overnight with a digoxigenin-labeled pancentromeric probe at 37°C followed by washes in 50% formamide and 2× SSC. To visualize hybridization signals, sections were incubated with FITC-conjugated anti-digoxigenin Fab fragments (1 207 741, Enzo Life Sciences, Farmingdale, NY) for 1 hour at room temperature.
To demonstrate GFP expression, liver samples were fixed in 4% paraformaldehyde in PBS (PAF), equilibrated in 20% sucrose and frozen in methylbutane at −80°C. Immunostaining used 5-µm-thick cryostat sections postfixed in 4% paraformaldehyde. Cells were directly fixed in PAF. Blocking was with 5% goat serum, 1% BSA in PBS containing 0.1% Triton X-100 (PBS-T) for 1 hour at room temperature followed by incubation with rabbit anti-GFP in PBS-T (1:300, Molecular Probes). Sections were incubated with FITC-conjugated goat anti-rabbit IgG with counterstaining using DAPI-Antifade.
To reveal additional hepatic markers in cells, we performed glycogen staining followed by in situ hybridization using the human pancentromeric probe, according to our previously published protocols (Ott et al., 1999b). To reveal albumin, cryosections were used for immunostaining according to the conditions in supplementary material Table S2.
To determine whether transplanted fetal human liver cells engrafted in the mouse liver without undergoing genetic fusion with native cells, we performed in situ hybridization using our pancentromeric human probe, along with a mouse pancentromeric probe from Cambio (1697; Cambridge, UK). Tissue sections were prepared and processed as described above and then hybridized with human and mouse probes together followed by post-hybridization washes as described above. For visualizing hybridization signals, sections were incubated with FITC-conjugated antidigoxigenin Fab fragments (1:50, Enzo Life Sciences) plus mouse avidin-Texas-Red and biotinylated anti-avidin (1:200, Cambio) in 1% bovine serum albumin, 4× SSC, 0.1% Tween 20 for 90 minutes at 37°C. Sections were washed with PBS three times for 5 minutes each at room temperature, counterstained with DAPI (4′,6-diamidino 2-phenylindole)-Antifade (Molecular Probes) and examined under epifluorescence.
Statistical methods and analysis
Data are shown as means ± s.d. where appropriate. The significance of differences was analyzed by the Student’s t-test, chi-square test or ANOVA. P values <0.05 were considered significant.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/121/7/1002/DC1
Acknowledgments
Prof. L. Naldini provided the TTR promoter construct. The authors have no conflicts of interest to report. Funding Support: NIH grants R01 DK46952, P01 DK52956, P20 M075037, P30 DK41296 and P30 CA13330.
References
- Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PloS Med. 2005;2:e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benten D, Cheng K, Gupta S. Identification of transplanted human cells in animal tissues. In: Derby IA, Hewitson TD, editors. In Situ Hybridization Protocols (Methods in Molecular Biology, Vol. 326) Totowa, NJ: Humana Press; 2005a. pp. 189–201. [DOI] [PubMed] [Google Scholar]
- Benten D, Kumaran V, Joseph B, Schattenberg J, Popov Y, Schuppan D, Gupta S. Hepatocyte transplantation activates hepatic stellate cells with beneficial modulation of cell engraftment in the rat. Hepatology. 2005b;42:1072–1081. doi: 10.1002/hep.20889. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Joseph B, Sappal BS, Giri RK, Wang R, Ludlow J, Susick R, Gupta S. Analysis of the functional integrity of cryopreserved human liver cells including xenografting in immunodeficient mice to address suitability for clinical applications. Liver Int. 2004;24:361–370. doi: 10.1111/j.1478-3231.2004.0938.x. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1491–1492. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Dan YY, Riehle KJ, Lazaro C, Teoh N, Haque J, Campbell JS, Fausto N. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc. Natl. Acad. Sci. USA. 2006;103:9912–9917. doi: 10.1073/pnas.0603824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Follenzi A, Sabatino G, Lombardo A, Boccacio C, Naldini L. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum. Gene Ther. 2002;13:243–260. doi: 10.1089/10430340252769770. [DOI] [PubMed] [Google Scholar]
- Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev. Biol. 2004;15:389–402. doi: 10.1016/j.ydbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Gershengorn MC, Hardikar AA, Wei C, Geras-Raaka E, Marcus-Samuels B, Raaka BM. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science. 2004;306:2261–2264. doi: 10.1126/science.1101968. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Hay ED, Zuk A. Transformations between epithelium and mesenchyme: normal, pathological, and experimentally induced. Am. J. Kidney Dis. 1995;26:678–690. doi: 10.1016/0272-6386(95)90610-x. [DOI] [PubMed] [Google Scholar]
- Heremans Y, Van De Casteele M, in’t Veld P, Gradwohl G, Serup P, Madsen O, Pipeleers D, Heimberg H. Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J. Cell Biol. 2002;159:303–312. doi: 10.1083/jcb.200203074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J. Cell. Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- Inada M, Benten D, Cheng K, Joseph B, Berishvili E, Badve S, Logdberg L, Dabeva M, Gupta S. Stage-specific regulation of adhesion molecule expression segregates epithelial stem/progenitor cells in fetal and adult human livers. Hepatol. Int. 2007 doi: 10.1007/s12072-007-9023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishizaka S, Shiroi A, Kanda S, Yoshikawa M, Tsujinoue H, Kuriyama S, Hasuma T, Nakatani K, Takahashi K. Development of hepatocytes from ES cells after transfection with the HNF-3beta gene. FASEB. J. 2002;16:1444–1446. doi: 10.1096/fj.01-0806fje. [DOI] [PubMed] [Google Scholar]
- Lazaro CA, Croager EJ, Mitchell C, Campbell JS, Yu C, Foraker J, Rhim JA, Yeoh GC, Fausto N. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095–1106. doi: 10.1053/jhep.2003.50448. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lemm I, Lingott A, Pogge v Strandmann E, Zoidl C, Bulman MP, Hattersley AT, Schulz WA, Ebert T, Ryffel GU. Loss of HNF1alpha function in human renal cell carcinoma: frequent mutations in the VHL gene but not the HNF1alpha gene. Mol. Carcinog. 1999;24:305–314. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malhi H, Irani AN, Gagandeep S, Gupta S. Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes. J. Cell Sci. 2002;115:2679–2688. doi: 10.1242/jcs.115.13.2679. [DOI] [PubMed] [Google Scholar]
- Mishra L, Derynck R, Mishra B. Transforming growth factor-β signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DYR. Mesodermal wnt2b signaling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Okaya A, Kitanaka J, Kitanaka N, Satake M, Kim Y, Terada K, Sugiyama T, Takemura M, Fujimoto J, Terada N, et al. Oncostatin M inhibits proliferation of rat oval cells, OC15–5, inducing differentiation into hepatocytes. Am. J. Pathol. 2005;166:709–719. doi: 10.1016/S0002-9440(10)62292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier EN, Rybicki AC, Bouhassira EE. Differentiation of human embryonic stem cells into bipotent mesenchymal stem cells. Stem Cells. 2006;24:1914–1922. doi: 10.1634/stemcells.2005-0648. [DOI] [PubMed] [Google Scholar]
- Ott M, Ma Q, Li B, Gagandeep S, Rogler LE, Gupta S. Regulation of hepatitis B virus expression in progenitor and differentiated cell-types: evidence for negative transcriptional control in nonpermissive cells. Gene Expr. 1999a;8:175–186. [PMC free article] [PubMed] [Google Scholar]
- Ott M, Rajvanshi P, Sokhi R, Alpini G, Aragona E, Dabeva M, Shafritz DA, Gupta S. Differentiation-specific regulation of transgene expression in a diploid epithelial cell line derived from the normal F344 rat liver. J. Pathol. 1999b;187:365–373. doi: 10.1002/(SICI)1096-9896(199902)187:3<365::AID-PATH237>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Pagan R, Llobera M, Vilaro S. Epithelial-mesenchymal transition in cultured neonatal hepatocytes. Hepatology. 1995;21:820–831. [PubMed] [Google Scholar]
- Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99:507–512. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- Schmelzer E, Wauthier E, Reid LM. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Reyes M, Koodie L, Jiang Y, Blackstad M, Lund T, Lenvik T, Johnson S, Hu WS, Verfaillie CM. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J. Clin. Invest. 2002;109:1291–1302. doi: 10.1172/JCI15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahdadfar A, Fronsdal K, Haug T, Reinholt FP, Brinchmann JE. In vitro expansion of human mesenchymal stem cells: choice of serum is a determinant of cell proliferation, differentiation, gene expression, and transcriptome stability. Stem Cells. 2005;23:1357–1366. doi: 10.1634/stemcells.2005-0094. [DOI] [PubMed] [Google Scholar]
- Sicklick JK, Choi SS, Bustamante M, McCall SJ, Perez EH, Huang J, Li YX, Rojkind M, Diehl AM. Evidence for epithelial-mesenchymal transitions in adult liver cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;291:G575–G583. doi: 10.1152/ajpgi.00102.2006. [DOI] [PubMed] [Google Scholar]
- Suaud L, Joseph B, Formstecher P, Laine B. mRNA expression of HNF-4 isoforms and of HNF-1alpha/HNF-1beta variants and differentiation of human cell lines that mimic highly specialized phenotypes of intestinal epithelium. Biochem. Biophys. Res. Commun. 1997;235:820–825. doi: 10.1006/bbrc.1997.6888. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vigna E, Amendola M, Benedicenti F, Simmons AD, Follenzi A, Naldini L. Efficient Tet-dependent expression of human factor IX in vivo by a new self regulating lentiviral vector. Mol. Ther. 2005;11:763–775. doi: 10.1016/j.ymthe.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Walsh J, Andrews PW. Expression of Wnt and Notch pathway genes in a pluripotent human embryonal carcinoma cell line and embryonic stem cell. APMIS. 2003;111:197–210. doi: 10.1034/j.1600-0463.2003.1110124.x. [DOI] [PubMed] [Google Scholar]
- Wege H, Le HT, Chui MS, Liu L, Wu J, Giri RK, Malhi H, Sappal BS, Kumaran V, Gupta S, et al. Telomerase reconstitution immortalizes human fetal hepatocytes without disrupting their differentiation potential. Gastroenterology. 2003;124:432–444. doi: 10.1053/gast.2003.50064. [DOI] [PubMed] [Google Scholar]
- Winter MJ, Nagtegaal ID, van Krieken JH, Litvinov SV. The epithelial cell adhesion molecule (Ep-CAM) as a morphoregulatory molecule is a tool in surgical pathology. Am. J. Pathol. 2003;163:2139–2148. doi: 10.1016/S0002-9440(10)63570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulbrand U, Wied M, Zöfel P, Göke B, Arnold R, Fehmann H. Growth factor receptor expression in human gastroenteropancreatic neuroendocrine tumours. Eur. J. Clin. Invest. 1998;28:1038–1049. doi: 10.1046/j.1365-2362.1998.00397.x. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Gupta S, Giri RK, Berkovich I, Sappal BS, Karnieli O, Zern MA, Fleischer N, Efrat S. Reversal of hyperglycemia in mice using human expandable insulin-producing cells differentiated from fetal liver progenitor cells. Proc. Natl. Acad. Sci. USA. 2003;100:7253–7258. doi: 10.1073/pnas.1136854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/121/7/1002/DC1