Abstract
In this study, we compared the transport of newly synthesized cholesterol with that of influenza virus hemagglutinin (HA) from the endoplasmic reticulum to the plasma membrane. The arrival of cholesterol on the cell surface was monitored by cyclodextrin removal, and HA transport was monitored by surface trypsinization and endoglycosidase H digestion. We found that disassembly of the Golgi complex by brefeldin A treatment resulted in partial inhibition of cholesterol transport while completely blocking HA transport. Further, microtubule depolymerization by nocodazole inhibited cholesterol and HA transport to a similar extent. When the partitioning of cholesterol into lipid rafts was analyzed, we found that newly synthesized cholesterol began to associate with low-density detergent-resistant membranes rapidly after synthesis, before it was detectable on the cell surface, and its raft association increased further upon chasing. When cholesterol transport was blocked by using 15°C incubation, the association of newly synthesized cholesterol with low-density detergent-insoluble membranes was decreased and cholesterol accumulated in a fraction with intermediate density. Our results provide evidence for the partial contribution of the Golgi complex to the transport of newly synthesized cholesterol to the cell surface and suggest that detergent-resistant membranes are involved in the process.
Cholesterol is an essential constituent in the membranes of mammalian cells. These cells obtain cholesterol by two mechanisms: by taking it up from their environment, mostly in the form of low-density lipoproteins (1), or by synthesizing it de novo. The enzymes required for cholesterol biosynthesis reside in the endoplasmic reticulum (ER) (2). From its site of synthesis, cholesterol is transported to other cellular destinations, being most highly enriched in the plasma membrane, where 65–90% of cellular cholesterol resides (3, 4). The cholesterol content of the ER membrane is low, and there is evidence for a cholesterol gradient within the Golgi apparatus, the cis side being relatively sterol-poor and the trans-Golgi network (TGN) more highly sterol-enriched (5, 6). In the late secretory pathway, the higher cholesterol content together with the enrichment of sphingolipids favors the formation of lateral cholesterol/sphingolipid membrane microdomains termed rafts (7, 8). These assemblies have been implicated in membrane trafficking and signal transduction (9).
The subcellular distribution of cholesterol necessitates that the intracellular transfer of newly synthesized cholesterol takes place against a concentration gradient. According to the prevailing view, the carriers responsible for protein transport along the exocytic pathway do not contribute to the flux of biosynthetic cholesterol (for a review, see ref. 10). Instead, cholesterol transport from the ER to the cell surface was shown to be a rapid (t1/2 = 10–20 min), energy- and temperature-dependent process that bypasses the Golgi apparatus and does not require an intact cytoskeleton (10–13). In addition, there is evidence that cytosolic carrier proteins, such as caveolin-1 (14, 15) or sterol carrier protein-2 (16), may play a role in this process. These observations are in contrast to the transport of newly synthesized secretory proteins from the ER to the plasma membrane via vesicular/tubular intermediates that move along cytoskeletal elements and pass through the Golgi complex. However, to compare cholesterol and protein traffic, the two processes should be analyzed in parallel in the same cell system. This has been carried out only in a single report, where biosynthetic labeling of both the protein and cholesterol was performed at a reduced temperature for up to 90 min and the migration of cholesterol to the plasma membrane was assayed by using subcellular fractionation (13).
The objective of the present study was to compare biosynthetic cholesterol transport with biosynthetic transport of a well characterized marker protein, influenza virus hemagglutinin (HA), by using similarly pretreated cells at physiological temperature. The arrival of both the protein and the lipid on the plasma membrane was monitored in intact cells, and nascent cholesterol was resolved by a chromatographic method that separates the intermediates of cholesterol synthesis from the end product. Using such assays, we compared the kinetics and temperature dependence of the two transport processes, and their sensitivity to disruption of the Golgi apparatus or the microtubule network. In addition, we analyzed the partitioning of newly synthesized cholesterol into lipid rafts in normal conditions and when cholesterol transport was inhibited.
Materials and Methods
Materials.
Media and reagents for cell culture were from Life Technologies (Gaithersburg, MD). Influenza stock N virus (A/chick/Germany/49/Hav2Neq1) was grown as described previously (17). [4-14C]Cholesterol (specific activity, 55.0 mCi/mmol), [3H]acetic acid (specific activity, 10.0 Ci/mmol), and Redivue Pro [35S]Met/Cys labeling mix (specific activity, 1,000 Ci/mmol) were from Amersham Pharmacia. Trypsin and soybean trypsin inhibitor were from Worthington. Endoglycosidase H was from Boehringer Mannheim, and brefeldin A (BFA) was from Epicentre Technologies (Madison, WI). Squalestatin was a generous gift from Glaxo, and lovastatin was from Merck Sharp & Dohme. Optiprep was from Nycomed (Oslo). Cycloheximide, mevalonic acid lactone (mevalonate), methyl-β-cyclodextrin, nocodazole (NZ), cholesterol, and other unlabeled lipids were from Sigma. Petroleum ether was from Fischer Scientific, and all other solvents (HPLC-grade) were from Merck.
Cell Culture.
Baby hamster kidney (BHK)-21 clone 13 cells (ATCC CRL8544) were grown as described (18), and HuH7 cells were grown as in ref. 19. The cells were sterol-starved by maintaining in Glasgow's modified Eagle's medium/10 mM Hepes, pH 7.4/2 mM glutamine/100 units/ml penicillin/100 μg/ml streptomycin (experiment medium) supplemented with 5% lipoprotein-deficient serum (LPDS) for 48 h before the labelings. For BHK cells, 1 μM squalestatin was added to the same medium for the last 24 h.
Analysis of HA Transport.
BHK cells grown in LPDS medium on 25-mm-diameter wells were infected with influenza virus (10–20 plaque-forming units/cell) as described (20). At 4 h postinfection the cells were labeled with [35S]Met/Cys (67 μCi/ml) in Met- and Cys-free MEM supplemented with 10 mM Hepes, pH 7.4/0.35 g/liter NaHCO3/2 mM glutamine/100 units/ml penicillin/100 μg/ml streptomycin for 5 min on a 37°C water bath (pulse). The chase was performed in experiment medium containing 10-fold excess of cold Met and Cys and 20 μg/ml cycloheximide. Where indicated, NZ was added to the experiment medium (from a 33-mM stock in DMSO to a final concentration of 33 μM) at 2 h postinfection, and the labeling and chase were performed in the continued presence of the drug. BFA was added only to the chase medium (from a 5-mg/ml stock in ethanol to a final of 5 μg/ml). Control samples received the same amount of DMSO or ethanol, respectively. The amount of HA transported to the plasma membrane was measured by surface trypsinization, and the percentage of HA cleaved was calculated as described (17) [i.e., 2 × HA2/(HA + 2 × HA2) × 100]. HA transport from the ER to the Golgi was measured by endoglycosidase H (endo H) digestion, and the amount of HA transported was calculated as described (18) [i.e., endo H-resistant form/(endo H-resistant form + endo H-sensitive form) × 100].
Analysis of Cholesterol Biosynthesis.
Cells grown on 55-mm dishes in LPDS medium were labeled overnight with [14C]cholesterol (20 nCi/ml) in LPDS medium. The cells were washed with PBS and labeled with [3H]acetate in experiment medium: BHK cells (250 μCi/ml) for 15 min and HuH7 cells (500 μCi/ml) for 5 min at 37°C. The chase was performed in experiment medium containing 10 μM lovastatin and 25 mM mevalonate. The cells then were placed on ice, washed with ice-cold PBS, and scraped into PBS with a rubber “policeman.” After harvesting by centrifugation (2,500 rpm for 5 min), the cells were resuspended in 2% NaCl. Before the lipids were extracted (21), the [14C]cholesterol radioactivity of the cell suspension was determined by liquid-scintillation counting. The extracted lipids were separated by TLC on silica-gel plates by using petroleum ether/diethyl ether/acetic acid (60:40:1) as the solvent. The plates were dried and stained with iodine vapor. The cholesterol band was determined based on the comigration of a cholesterol standard, scraped, and extracted from the silica with chloroform/methanol (1:1). After evaporation of the solvent, the residue was dissolved in a small volume of hexane/acetone (8:2), injected to an Ag+-saturated Nucleosil SA ion-exchange column (250 mm × 4.6 mm, 5-μm particle size; Alltech Associates), and eluted with hexane/acetone (8:2) at a flow rate of 1 ml/min. Fractions of 0.25 ml were collected and [3H]- and [14C]radioactivity was measured by liquid-scintillation counting. Nascent cholesterol was quantified as [3H]radioactivity in the cholesterol peak, corrected for any losses based on the recovery of the [14C]cholesterol internal standard.
Measuring the Arrival of Cholesterol on the Plasma Membrane by Using Cyclodextrin.
Cells were labeled with [14C]cholesterol and were as described above, except that [3H]acetate labeling and chase at 15°C was carried out on a water bath in MEM supplemented with 10 mM Hepes, pH 7.4/0.35 g/liter NaHCO3/2 mM glutamine/100 units/ml penicillin/100 μg/ml streptomycin (air medium). During the last 5 min of the chase the cells were incubated with methyl-β-cyclodextrin (3 mM for BHK and 2 mM for HuH7 cells) in air medium on a shaking-water bath at 37°C. The medium was collected, and the cells were scraped into ice-cold PBS, harvested by centrifugation, and resuspended in 2% NaCl. Aliquots of the medium and the cell suspension were analyzed by liquid-scintillation counting to determine the [14C]cholesterol content. The lipids were extracted from the medium by shaking it three times with an equal volume of hexane, and the combined hexane phases were assayed for [3H]- and [14C]cholesterol with Ag+-HPLC. The cellular fractions were analyzed as described for cholesterol biosynthesis. The arrival of cholesterol on the plasma membrane was defined as the relative portion of [3H]cholesterol radioactivity in the medium of the combined [3H]cholesterol radioactivity in the medium and the cellular fraction at a given time point, corrected with the losses of radioactivity based on the recovery of the [14C]cholesterol. Where indicated, NZ was added to the cells at a 33-μM concentration in experiment medium for 2 h before labeling with [3H]acetate and kept present during the labeling, chase, and cyclodextrin extraction. In the experiments in which BFA was used, it was added to the labeling, chase, and cyclodextrin extraction medium at 5 μg/ml.
Measuring Cholesterol Incorporation in Detergent-Insoluble Membranes.
Detergent insolubility of the cellular membranes was analyzed essentially as described (22). Briefly, the cells were labeled with [14C]cholesterol and [3H]acetate as described above, washed with ice-cold PBS, and harvested by centrifugation. The cell pellets were resuspended in 240 μl TNE (25 mM Tris⋅HCl, pH 7.5/150 mM NaCl/5 mM EDTA), 10% sucrose, 1 mM DTT, protease inhibitor mixture (25 μg/ml each of chymostatin, leupeptin, antipain, and pepstatin A), and 1% Triton X-100 at 4°C. After 10 min on ice, the cell homogenate was mixed with 480 μl of 60% Optiprep and overlaid with 720-μl steps of 35, 30, 25, 20, and 0% Optiprep in TNE/10% sucrose/1% Triton X-100. After a total of 30 min on ice, the gradient was centrifuged at 40,000 rpm for 4 h at 4°C in a SW60 Beckman rotor. Six 720-μl fractions were collected from the top, and lipids were extracted and analyzed as described above.
Other Methods.
LPDS was prepared from FBS by sequential ultracentrifugation by using KBr for density adjustments (23). Cholesterol, triacylglycerol, and phospholipids were determined with enzymatic methods (24). The amount of protein was measured as described (25), and lactate dehydrogenase activity was measured according to ref. 26.
Results
Rate of Cholesterol Synthesis in BHK Cells.
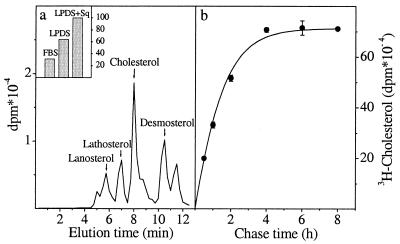
To compare cholesterol and protein transport, we chose fibroblastoid BHK cells as an experimental system because these cells are used frequently to study protein transport and represent peripheral cells engaged in sterol biosynthesis. Newly synthesized cholesterol was detected by pulse labeling the cells with [3H]acetate, extracting the cellular material in organic solvent, and analyzing it by TLC followed by HPLC on a silver-ion column (Ag+-HPLC). This method separates sterols based on the number and position of double bonds (27, 28) and resolves even the immediate precursor desmosterol from cholesterol (Fig. 1a). To up-regulate cholesterol biosynthesis, the cells were cholesterol-starved before labeling by culturing for 48 h in LPDS medium and adding a late-stage inhibitor of cholesterol synthesis, squalestatin (29), for the last 24 h of incubation. The starvation resulted in 3-fold stimulation of cholesterol biosynthesis (Fig. 1a Inset). Under these conditions, reliable detection of nascent cholesterol was achieved by using a 15-min [3H]acetate pulse and 30-min chase (Fig. 1a). At shorter times, the highly abundant precursors masked the cholesterol peak (data not shown). To ensure that the incorporation of radioactivity did not continue during the chase, the chase medium was supplemented with excess unlabeled mevalonate and lovastatin, an inhibitor of hydroxymethylglutaryl-CoA reductase. By using this protocol, we found that only after 4 h of chase was all of the [3H]acetate on the cholesterol biosynthetic pathway in BHK cells converted to cholesterol, with a t1/2 of synthesis of about 1 h (Fig. 1b).
Figure 1.
Cholesterol biosynthesis in BHK cells. (a) BHK cells were incubated in normal culture medium (FBS), in LPDS medium for 48 h (LPDS), or in LPDS medium for 24 h and a further 24 h in the same medium containing squalestatin (LPDS + Sq). Cells were labeled with [3H]acetate for 15 min and chased for 2 h. The relative amount of radioactive cholesterol synthesized in each condition is shown (Inset). Cells cultured as in (LPDS + Sq) were given a 15-min [3H]acetate pulse and chased for 30 min. Lipids were extracted and analyzed by TLC and Ag+-HPLC. The elution times of cholesterol and some of its biosynthetic precursors are indicated. (b) Cells cultured as in (LPDS + Sq) were labeled with [3H]acetate for 15 min and chased for increasing times. Lipids were extracted and analyzed by TLC and Ag+-HPLC, and the radioactivity recovered in the cholesterol peak was plotted as a function of time. Values represent the mean ± SEM of three experiments.
Arrival of Newly Synthesized Cholesterol on the Cell Surface as Detected by Cyclodextrin Extraction.
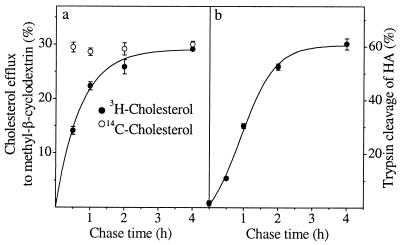
To detect the arrival of newly synthesized cholesterol on the plasma membrane we used cyclodextrin treatment of intact cells. Cyclodextrins are water-soluble cyclic oligosaccharides that have the capacity to sequester cholesterol in their hydrophobic cavity, thereby removing cholesterol from cells rapidly and efficiently (30, 31). With increasing chase times, an increasing fraction of newly synthesized cholesterol should reach the plasma membrane and become accessible to cyclodextrin, whereas steady-state cholesterol label should be removed from the cell surface with similar efficiency at each time point. To minimize the adverse effects that the cholesterol removal may cause, we used a short efflux time (5 min at 37°C) and a low (3 mM) methyl-β-cyclodextrin concentration that resulted in 25–30% efflux of [14C]cholesterol. This treatment was verified not to disrupt the plasma membrane because there was no leakage of cytosolic lactate dehydrogenase from the cells (data not shown). By combining cyclodextrin extraction with the [3H]acetate pulse–chase protocol, we could demonstrate that the percentage of newly synthesized [3H]cholesterol effluxed to cyclodextrin increased during the chase, whereas a constant fraction of the pre-existing [14C]cholesterol was removed from the same cells (Fig. 2a). The earliest time point at which reliably measurable levels of newly synthesized cholesterol could be detected at the surface of BHK cells was after 30 min of chase. At this time point, the extractability of de novo synthesized cholesterol was about half of that of prelabeled [14C]cholesterol (Fig. 2a).
Figure 2.
Transport of newly synthesized cholesterol and HA to the cell surface. Cholesterol-starved BHK cells were prelabeled with [14C]cholesterol overnight, pulsed with [3H]acetate for 15 min, and chased for increasing times. At the end of the chase the cells were treated with methyl-β-cyclodextrin for 5 min. Cells and media were collected, and lipids were extracted and analyzed by TLC and Ag+-HPLC. The percentage of efflux of newly synthesized [3H]cholesterol and prelabeled [14C]cholesterol to cyclodextrin at each time point is indicated (a). Values represent the mean ± SEM of six to nine experiments. To analyze HA transport (b), influenza virus-infected BHK cells were labeled with [35S]Met/Cys for 5 min and chased for increasing times. Arrival of HA on the plasma membrane was detected by surface trypsinization and expressed as a percentage of cleaved HA. Values represent the mean ± SEM of three experiments.
To analyze the transport of newly synthesized HA in the cholesterol-deprived BHK cells, we infected the cells with influenza virus and measured the arrival of pulse-labeled HA to the plasma membrane by surface trypsinization (17, 32). The starvation procedure resulted in about 30% reduction in total cellular cholesterol level. Under these conditions, the transport rate of HA from the ER to the cell surface did not differ significantly from that observed in nondelipidating conditions (17), with a t1/2 of transport of ≈60 min (Fig. 2b). Similar kinetics of HA transport also was obtained when the chase medium was supplemented with lovastatin and mevalonate, indicating that the conditions used for the chase of nascent cholesterol did not affect the surface delivery of HA (data not shown). Although cholesterol depletion has been shown to alter HA transport (18), the reduction in cellular cholesterol level in those experiments was more dramatic (60–70%) than in the present study.
Effect of Temperature on the Transport of Newly Synthesized Cholesterol and HA.
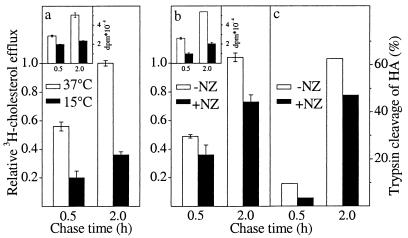
Transport of newly synthesized cholesterol from the ER to the plasma membrane has been shown to be temperature-dependent (12). We therefore tested whether the cyclodextrin extraction assay could be used to detect the inhibitory effect of 15°C incubation on cholesterol transport to the plasma membrane in BHK cells. After a 1-h [3H]acetate pulse at 15°C (the longer pulse was necessary because cholesterol biosynthesis slows down at reduced temperature), a chase of 0.5–2 h was carried out at either 15°C or 37°C. We found that the transport of newly synthesized cholesterol to the plasma membrane was inhibited by 65% when the cells were incubated at the lowered temperature (Fig. 3a). The 15°C block also effectively inhibited biosynthetic protein transport in the same cell system. As demonstrated previously (17), 15°C incubation resulted in the retention of HA in its core glycosylated form and in the lack of the tryptic digestion product HA2 (data not shown).
Figure 3.
Inhibition of transport by reduced temperature or NZ treatment. Transport of nascent [3H]cholesterol was measured as in Fig. 2a after incubating the cells at 15°C or 37°C (a) or in the presence or absence of NZ (b). [3H]cholesterol efflux is expressed as a relative ratio of [14C]cholesterol efflux, which was set at 1.0. The amount of [3H]cholesterol synthesized is shown in Inset. Values represent the mean ± SEM of three experiments. Arrival of HA on the plasma membrane in the presence or absence of NZ (c) was measured by surface trypsinization as in Fig. 2b. Values are the average of duplicate measurements.
Effect of Nocodazole Treatment on the Transport of Newly Synthesized Cholesterol and HA.
Because microtubule depolymerization by NZ has been shown to reduce secretory protein transport (33, 34), we next analyzed the effect of NZ on biosynthetic cholesterol transport. The cells were incubated in the presence of 33 μM NZ for 2 h at 37°C before the [3H]acetate pulse. This treatment results in clear morphological effects of microtubule depolymerization in BHK cells (ref. 35 and data not shown) and allows cholesterol biosynthesis to proceed although at repressed levels (Fig. 3b Inset). The pulse and chase were performed in the continuous presence of the drug. NZ inhibited the transport of newly synthesized cholesterol to the plasma membrane by about 25% (Fig. 3b). Suppression of cholesterol biosynthesis precluded prolonged incubations with NZ or the use of higher drug concentrations. To quantitate the effect of the corresponding NZ treatment on protein transport, we similarly preincubated influenza virus-infected cells for 2 h in the presence of NZ before the [35S]methionine pulse and kept the drug present during the pulse and chase. Under these conditions, the trypsin cleavage of HA was inhibited by about 25% compared with nontreated samples (Fig. 3c).
Effect of BFA Treatment on the Transport of Newly Synthesized Cholesterol and HA.
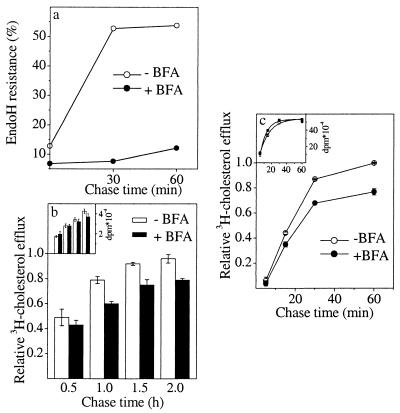
BFA inhibits protein secretion by disassembling the Golgi apparatus and resulting in rapid redistribution of most of the Golgi into the ER (36–38). Accordingly, we found HA transport out of the ER to be inhibited effectively when BFA was present at 5 μg/ml concentration during the chase. Under these conditions, ≈90% of HA remained sensitive to endo H digestion (Fig. 4a) and uncleaved after surface trypsinization (data not shown). When BFA was present at 5 μg/ml during the [3H]acetate pulse and chase, the transport of newly synthesized cholesterol was inhibited by 20%. This inhibition was evident between 1 and 2 h of chase (Fig. 4b). No significant alterations in the rate of cholesterol synthesis were observed in the presence of the drug (Fig. 4b Inset).
Figure 4.
Effect of BFA on the transport of newly synthesized HA and cholesterol. Arrival of HA in the Golgi complex in BHK cells was measured by acquisition of endo H resistance in the presence or absence of BFA and expressed as a percentage of total HA (a). Values are the average of duplicate measurements. Biosynthetic cholesterol transport in BHK cells (b) and HuH7 cells (c) was measured as in Fig. 3a either in the presence or absence of BFA. The amount of [3H]cholesterol synthesized at each time point is shown in Inset. Values represent the mean ± SEM of three experiments.
Because of the inefficiency of [3H]acetate incorporation into cholesterol, it was not possible to reliably analyze the effect of BFA at very early chase times in BHK cells. However, cells of hepatic origin have been shown to synthesize cholesterol from radiolabeled acetate more efficiently (39). We therefore tested the effect of BFA on biosynthetic cholesterol transport by adopting the cyclodextrin extraction assay to a hepatoma cell line, HuH7. In these cells, a 5-min [3H]acetate pulse could be combined with short chase times because of rapid cholesterol synthesis (Fig. 4c Inset). We found that at 5 min of chase, newly synthesized cholesterol could not yet be detected on the cell surface. However, by 15 min of chase, a fraction of newly synthesized cholesterol had reached the surface (about 40% of that found at 60 min of chase), and by 30 min of chase the majority could be found on the plasma membrane. BFA inhibited the transport by 20% already at 15 min of chase, the extent of inhibition also remaining similar at later chase times (Fig. 4c).
Nascent cholesterol recently has been reported to be transported as a cytosolic caveolin–heat shock protein complex to the cell surface (15). Because this mechanism is expected to be BFA-resistant, we tested the effect of cyclosporin A, shown to disrupt the cytosolic chaperone complex and inhibit cholesterol transport (15). Treatment of BHK cells with 1 μM cyclosporin A for 1 h before the [3H]acetate pulse, during the 15-min pulse and the chase of 0.5–2 h did not result in alterations in the transport of newly synthesized cholesterol (data not shown).
Association of Newly Synthesized Cholesterol with Lipid Rafts.
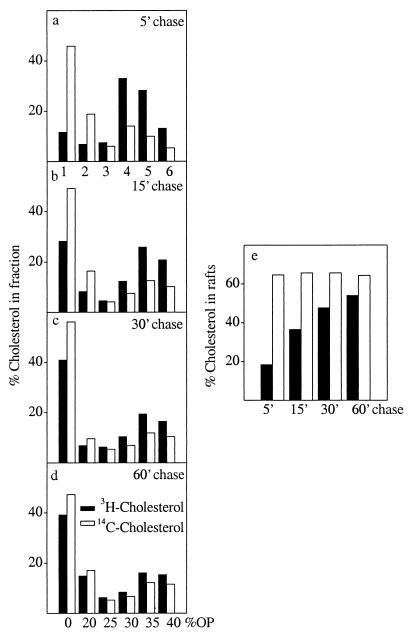
We next analyzed the partitioning of newly synthesized cholesterol in raft microdomains by taking advantage of their resistance to solubilization by Triton X-100 on ice (40). HuH7 cells were pulse-labeled with [3H]acetate for 5 min and chased as in Fig. 4c, the cells were lysed in 1% Triton X-100 on ice, and the lysates were fractionated by an Optiprep density gradient (22). The distribution of newly synthesized [3H]cholesterol and prelabeled [14C]cholesterol between the gradient fractions was measured. Rafts are recovered in the top fractions of the gradient, and this is where the majority of prelabeled [14C]cholesterol was localized (Fig. 5 a– d). At the shortest time point analyzed (5-min chase), about 20% of newly synthesized cholesterol was found in the two top fractions representing rafts (22). With increasing chase time, the raft association of de novo synthesized cholesterol increased, reaching 55% by 60 min of chase, whereas the percentage of [14C]cholesterol in rafts remained constant (65%) (Fig. 5e).
Figure 5.
Detergent solubility of newly synthesized cholesterol. Cholesterol-starved HuH7 cells were prelabeled with [14C]cholesterol overnight, pulsed with [3H]acetate for 5 min, and chased for increasing times. The cells were lysed in ice-cold buffer containing 1% Triton X-100 and fractionated by Optiprep gradient centrifugation. Fractions were collected from the top. Lipids were extracted and analyzed by TLC and Ag+-HPLC. The percentage of newly synthesized and prelabeled [14C]cholesterol in each fraction is indicated (a– d). The percentage of radioactive cholesterol in the two top fractions representing rafts is plotted at each chase time analyzed (e).
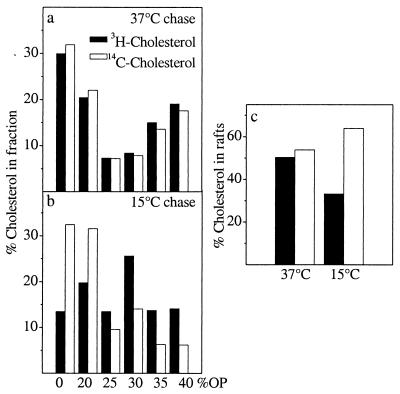
The membranes completely solubilized by the detergent are found in the bottom fractions. However, we noticed that at 5 min of chase a peak of newly synthesized cholesterol was detectable in the middle of the gradient in fraction 4, whereas at longer chase times this was no longer evident (Fig. 5 a vs. b–d). When considered together with the rate of cholesterol surface arrival in HuH7 cells, this raises the possibility that some of the cholesterol in this fraction might represent a pool engaged in the transport process. To further explore this phenomenon, we used the condition found to block cholesterol transport most efficiently, i.e., 15°C incubation (Fig. 3a). After [3H]acetate pulse, BHK cells prelabeled with [14C]cholesterol were incubated either at 15°C or at 37°C for 2 h, extracted with Triton X-100, and analyzed in Optiprep gradients. The 37°C incubation resulted in almost complete cofractionation of newly synthesized [3H]cholesterol with [14C]cholesterol that displayed a clearly biphasic distribution between detergent-insoluble and detergent-soluble membranes (Fig. 6a). However, when the cells were chased at 15°C, a substantial proportion of newly synthesized cholesterol was found to accumulate in the middle of the gradient in fraction 4 (Fig. 6b). There was also a small [14C]cholesterol increment in this fraction that might reflect the continuous routing of cell cholesterol via the ER (41) and its return to the cell surface via the same pathway as newly synthesized cholesterol. The 15°C incubation reduced the percentage of newly synthesized cholesterol recovered in rafts from 50% (observed at 37°C) to 33% whereas the raft association of [14C]cholesterol increased slightly at the reduced temperature (Fig. 6c).
Figure 6.
Effect of 15°C incubation on the detergent solubility of newly synthesized cholesterol. Cholesterol-starved BHK cells were prelabeled with [14C]cholesterol overnight, pulsed with [3H]acetate for 15 min at 37°C, and chased for 2 h at 37°C (a) or 15°C (b). After the chase the cells were treated as in Fig. 5. The percentage of [3H]cholesterol and [14C]cholesterol in each fraction is indicated. The percentage of radioactive cholesterol in the two top fractions representing rafts is plotted at both temperatures (c).
Discussion
In this study, we developed a high-resolution cholesterol transport assay based on cyclodextrin extraction of the plasma membrane and separation of sterols by HPLC analysis. Much of the work done on biosynthetic cholesterol delivery has relied solely on the use of TLC to detect newly synthesized cholesterol. It is obvious from our experiments that it is very difficult to resolve the biosynthetic products deriving from acetate with TLC-based methods only. This is true especially for the other C27 sterols, i.e., immediate precursors of cholesterol, and in accordance with the report of Ruan et al. (27). The transport of biosynthetic sterol intermediates is known to differ from that of the end product (39), and pooling of the sterol precursors together with cholesterol therefore will disturb the analysis. This is manifested especially at very early chase times, when a significant portion of the label is not yet incorporated into cholesterol. In fact, we observed that desmosterol is effluxed from cells more readily than cholesterol, in accordance with Phillips et al. (42) (unpublished results). Further, the detection of cholesterol arrival on the plasma membrane traditionally has relied either on subcellular fractionation or enzymatic oxidation of cholesterol. However, it is difficult to obtain plasma membrane fractions entirely devoid of contaminating membranes. Also, the latter technique is problematic because, with some cholesterol oxidation protocols, intracellular cholesterol pools are not protected from oxidation (ref. 10 and unpublished observations).
The cholesterol transport assay was applied successfully in two different cell types. This revealed that not only cholesterol production but also its biosynthetic transport rate varied between cell types. In hepatoma cells, the half-time of cholesterol synthesis was only about one-sixth of that in fibroblastoid cells (compare Fig. 1b and Fig. 4c Inset). Moreover, whereas the half-time of transport was estimated to be about 30 min in fibroblastoid cells (Fig. 2a), it was closer to the previously reported values of 10–20 min in hepatoma cells (Fig. 4c). The improved methodology used in this work enabled the identification of a partial sensitivity of cholesterol transport to NZ and BFA treatments. These data are consistent with the idea that, in contrast to previous results, the Golgi complex and the microtubule network are not excluded from the machinery responsible for the anterograde movement of newly synthesized cholesterol. Nevertheless, the complete inhibition of HA transport by BFA in comparison with the 20% inhibition of nascent cholesterol transport argues that the majority of newly synthesized cholesterol uses transport route(s) that HA is unable to access, presumably bypassing the Golgi complex, to reach the cell surface. The analysis of transport in the presence or absence of BFA in the hepatoma cells failed to reveal two kinetically different components. This suggests that the minor BFA-sensitive and the major BFA-insensitive transport mechanisms operate in parallel and with roughly similar rates. The inhibition by BFA at early chase times implies also that the BFA-sensitive transport component in HuH7 cells is considerably fast. The rapid rate does not contradict with the involvement of the Golgi complex, because various cargo molecules have been shown to travel through the secretory pathway with markedly different kinetics (43, 44).
Our data show that the association of newly synthesized cholesterol with rafts begins intracellularly, before its delivery to the cell surface. This was evident at the shortest chase time analyzed (5 min), at which there was no newly synthesized cholesterol captured by cyclodextrin but yet about 20% of nascent cholesterol floated to the top fractions in the Optiprep–Triton gradient. We envisage that the raft assembly may be initiated already in the ER. Although in mammalian cells the incorporation of proteins into rafts seems to take place at the level of the Golgi complex (45), where cholesterol and glycosphingolipids coincide, the initial building blocks of rafts already may form in the ER. Interestingly, recent findings demonstrate that rafts are present in the ER in yeast (46) and Triton-insoluble fractions with associated GPI-anchor intermediates also could be isolated from the ER of mammalian cells (47). We detected, however, a major increase in the proportion of raft-associated nascent cholesterol roughly paralleling its appearance on the cell surface. This suggests that the partitioning of cholesterol into rafts becomes more extensive and is stabilized upon biosynthetic delivery, potentially because of association of nascent cholesterol with glycosphingolipids on the plasma membrane or in late transport intermediates.
After a short chase time, a transient peak of nascent cholesterol could be pinpointed in the Optiprep gradient in an intermediate region between the raft-associated detergent-insoluble and the completely detergent-soluble pools. A similar peak was evident when transport was inhibited by reduced temperature. The nature of cholesterol interactions with other lipids or proteins resulting in this behavior remains to be elucidated. However, one can envisage that the intermediate fraction might represent a subdomain of the ER where nascent cholesterol accumulates at the reduced temperature or correspond to hypothetical carrier vesicles or tubules detached from the ER. The introduced cyclodextrin-based cholesterol transport assay coupled with monitoring the increasing partitioning of nascent cholesterol in lipid rafts provide a basis for further delineation of the biosynthetic cholesterol transport machinery.
Acknowledgments
We thank Matti Jauhiainen for help with LPDS preparation, Arja Puolasmaa from Merck Sharp & Dohme for providing lovastatin; Marjukka Lovio from Glaxo Wellcome for squalestatin, Birgitta Rantala, and Ritva Keva for expert technical assistance. This work was supported by the Academy of Finland (Grants 37661, 43184 to E.I., and 43668; Grants 42163, 36282, and 45817 to V.M.O.), the Sigrid Juselius Foundation, and the Päivikki and Sakari Sohlberg Foundation.
Abbreviations
- BFA
brefeldin A
- BHK
baby hamster kidney
- ER
endoplasmic reticulum
- HA
hemagglutinin
- LPDS
lipoprotein-deficient serum, NZ, nocodazole
- TGN
trans-Golgi network
- endo H
endoglycosidase H
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140218797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140218797
References
- 1.Brown M S, Goldstein J L. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart M P, Billheimer J T, Faust J R, Gaylor J L. J Biol Chem. 1987;262:9649–9655. [PubMed] [Google Scholar]
- 3.Lange Y, Swaisgood M H, Ramos B V, Steck T L. J Biol Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- 4.Warnock D E, Roberts C, Lutz M S, Blackburn W A, Young W W, Jr, Baenziger J U. J Biol Chem. 1993;268:10145–10153. [PubMed] [Google Scholar]
- 5.Orci L, Montesano R, Meda P, Malaisse-Lagae F, Brown D, Perrelet A, Vassalli P. Proc Natl Acad Sci USA. 1981;78:293–297. doi: 10.1073/pnas.78.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bretscher M S, Munro S. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 7.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 8.Brown D A, London E. Annu Rev Cell Dev Biol. 1998;14:111–36. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 9.Kurzchalia T V, Parton R G. Curr Opin Cell Biol. 1999;11:424–431. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 10.Liscum L, Munn N J. Biochim Biophys Acta. 1999;1438:19–37. doi: 10.1016/s1388-1981(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 11.DeGrella R F, Simoni R D. J Biol Chem. 1982;257:14256–14262. [PubMed] [Google Scholar]
- 12.Kaplan M R, Simoni R D. J Cell Biol. 1985;101:446–453. doi: 10.1083/jcb.101.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbani L, Simoni R D. J Biol Chem. 1990;265:1919–1923. [PubMed] [Google Scholar]
- 14.Smart E J, Ying Y, Donzell W C, Anderson R G. J Biol Chem. 1996;271:29427–29435. doi: 10.1074/jbc.271.46.29427. [DOI] [PubMed] [Google Scholar]
- 15.Uittenbogaard A, Ying Y, Smart E J. J Biol Chem. 1998;273:6525–6532. doi: 10.1074/jbc.273.11.6525. [DOI] [PubMed] [Google Scholar]
- 16.Puglielli L, Rigotti A, Greco A V, Santos M J, Nervi F. J Biol Chem. 1995;270:18723–18726. doi: 10.1074/jbc.270.32.18723. [DOI] [PubMed] [Google Scholar]
- 17.Matlin K S, Simons K. Cell. 1983;34:233–243. doi: 10.1016/0092-8674(83)90154-x. [DOI] [PubMed] [Google Scholar]
- 18.Keller P, Simons K. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 20.Yoshimori T, Keller P, Roth M G, Simons K. J Cell Biol. 1996;133:247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bligh E G, Dryer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 22.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 24.Bergmeyer H U. In: Methods of Enzymatic Analysis. Bergmeyer J, Grassl M, editors. New York: VCH; 1985. pp. 1–160. [Google Scholar]
- 25.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Absolom D R. Methods Enzymol. 1986;132:95–180. doi: 10.1016/s0076-6879(86)32005-6. [DOI] [PubMed] [Google Scholar]
- 27.Ruan B, Shey J, Gerst N, Wilson W K, Schroepfer G J., Jr Proc Natl Acad Sci USA. 1996;93:11603–11608. doi: 10.1073/pnas.93.21.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan B, Gerst N, Emmons G T, Shey J, Schroepfer G J., Jr J Lipid Res. 1997;38:2615–2626. [PubMed] [Google Scholar]
- 29.Thelin A, Peterson E, Hutson J L, McCarthy A D, Ericsson J, Dallner G. Biochim Biophys Acta. 1994;1215:245–249. doi: 10.1016/0005-2760(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 30.Neufeld E B, Cooney A M, Pitha J, Dawidowicz E A, Dwyer N K, Pentchev P G, Blanchette-Mackie E J. J Biol Chem. 1996;271:21604–21613. doi: 10.1074/jbc.271.35.21604. [DOI] [PubMed] [Google Scholar]
- 31.Yancey P G, Rodrigueza W V, Kilsdonk E P C, Stoudt G W, Johnson W J, Phillips M C, Rothblat G H. J Biol Chem. 1996;271:16026–16034. doi: 10.1074/jbc.271.27.16026. [DOI] [PubMed] [Google Scholar]
- 32.Matlin K S, Simons K. J Cell Biol. 1984;99:2131–2139. doi: 10.1083/jcb.99.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogalski A A, Singer S J. J Cell Biol. 1984;99:1092–1100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschberg K, Miller C M, Ellenberg J, Presley J F, Siggia E D, Phair R D, Lippincott-Schwartz J. J Cell Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Annaert W G, Becker B, Kistner U, Reth M, Jahn R. J Cell Biol. 1997;139:1397–1410. doi: 10.1083/jcb.139.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y. J Biol Chem. 1986;261:11398–11403. [PubMed] [Google Scholar]
- 37.Doms R W, Russ G, Yewdell J W. J Cell Biol. 1989;109:61–72. doi: 10.1083/jcb.109.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson W J, Fischer R T, Phillips M C, Rothblat G H. J Biol Chem. 1995;270:25037–25046. doi: 10.1074/jbc.270.42.25037. [DOI] [PubMed] [Google Scholar]
- 40.Brown D A, Rose J K. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 41.Lange Y, Ye J, Rigney M, Steck T L. J Lipid Res. 1999;40:2264–2270. [PubMed] [Google Scholar]
- 42.Phillips J E, Rodrigueza W V, Johnson W J. J Lipid Res. 1998;39:2459–2470. [PubMed] [Google Scholar]
- 43.Lodish H F, Kong N, Snider M, Strous G J A M. Nature (London) 1983;304:80–83. doi: 10.1038/304080a0. [DOI] [PubMed] [Google Scholar]
- 44.Wieland F T, Gleason M L, Serafini T A, Rothman J E. Cell. 1987;50:289–300. doi: 10.1016/0092-8674(87)90224-8. [DOI] [PubMed] [Google Scholar]
- 45.Brown D A, London E. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 46.Bagnat M, Keränen S, Shevchenko A, Shevchenko A, Simons K. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sevlever D, Pickett S, Mann K J, Sambamurti K, Medof M E, Rosenberry T L. Biochem J. 1999;343:627–635. [PMC free article] [PubMed] [Google Scholar]