Abstract
All living cells routinely expel Na+ ions, maintaining lower concentration of Na+ in the cytoplasm than in the surrounding milieu. In the vast majority of bacteria, as well as in mitochondria and chloroplasts, export of Na+ occurs at the expense of the proton-motive force. Some bacteria, however, possess primary generators of the transmembrane electrochemical gradient of Na+ (sodium-motive force). These primary Na+ pumps have been traditionally seen as adaptations to high external pH or to high temperature. Subsequent studies revealed, however, the mechanisms for primary sodium pumping in a variety of non-extremophiles, such as marine bacteria and certain bacterial pathogens. Further, many alkaliphiles and hyperthermophiles were shown to rely on H+, not Na+, as the coupling ion. We review here the recent progress in understanding the role of sodium-motive force, including (i) the conclusion on evolutionary primacy of the sodium-motive force as energy intermediate, (ii) the mechanisms, evolutionary advantages and limitations of switching from Na+ to H+ as the coupling ion, and (iii) the possible reasons why certain pathogenic bacteria still rely on the sodium-motive force.
1. Introduction
The transmembrane electrochemical gradient of H+ ions (proton-motive force, or PMF) is the source of energy for a variety of cellular processes in most bacteria, mitochondria, and chloroplasts. A typical H+ cycle includes the generation of PMF by primary transport systems (H+ pumps) and its utilization for ATP synthesis, solute transport, motility, reverse electron transport, etc [1, 2]. In bacteria, protons are expelled out of the cell, so that their concentration inside is larger than outside. However, maintaining high levels of PMF becomes increasingly difficult for the bacteria living in alkaline environments, where the external concentration of H+ ions is low [1, 3–6]. To reconcile the life at alkaline pH values with the chemiosmotic principle, Skulachev proposed that alkaliphilic bacteria could use Na+ as a coupling ion instead of or in addition to H+ and coined the term “Sodium World” [1]. Another reason for a switch from H+ to Na+ was suggested by Konings and colleagues, who observed a rapid increase of proton leakage through the membrane at elevated temperatures [7] and reasoned that hyperthermophilic bacteria would benefit from relying on the sodium-motive force (SMF) instead of PMF [8, 9]. Similarly to the H+ cycle, the Na+ cycle would include generators of SMF (primary Na+ pumps) and SMF consumers, such as Na+-translocating membrane ATP synthase, Na+-dependent membrane transporters for nutrient uptake and/or a Na+-dependent flagellar motor for motility.
The idea that SMF might substitute for PMF has been verified by studies of such anaerobic bacteria as Propionigenium modestum, Malonomonas rubra, and Clostridium (renamed Caloramator) fervidus, which rely exclusively on Na+ ions for their energy metabolism [10–12]. However, none of these bacteria was either an alkaliphile or a hyperthemophile (C. fervidus is a moderate thermophile growing optimally at 60°C). Further, recent studies as well as analyses of the genomic data have questioned the very premise that alkaliphiles and/or hyperthermophiles must rely on the SMF to overcome their energetic difficulties. Several alkaliphiles and hyperthermophiles had no (known) primary Na+ pumps encoded in their genomes [13–17]. In contrast, Na+ cycling has been inferred for a number of mesophilic bacteria, including various marine strains (reviewed in [18]) and several important human pathogens [19, 20]. As a result, the reasons why certain bacteria and archaea depend on the SMF or PMF for their energy metabolism became even more obscure than ever before.
We review here the recent progress in understanding the role of sodium-motive force, including the conclusion that SMF was the initial form of membrane energy intermediate, the mechanisms and evolutionary advantages of switching from Na+ to H+ as the coupling ion, and the possible reasons why certain pathogenic bacteria still rely on the sodium-motive force. We try to minimize the overlap with several excellent reviews of the Na+-dependent systems published in the past several years [21–27]. Instead, based on own attempts to understand the different facets of the Sodium World [20, 28–30], we try to provide here a holistic picture of this world in its evolutionary development.
2. Structural organization of the F-type and V-type ATPases and similarity of their Na+-binding sites
While the PMF and SMF can be generated by several different enzymes (see below), the only enzyme capable of catalyzing PMF- or SMF-energized ATP synthesis is the membrane ATP synthase. Therefore the coupling ion specificity of the ATP-synthase defines whether the organism relies on a Na+- or H+-type membrane energetics.
As shown in Fig. 1, ATP synthases are rotary molecular machines that exist in two distinct types, namely, the F-type that is present in bacteria, some archaea, and eukaryotic organelles [30–35] and the V-type, represented in archaea and some bacteria [30, 36–41]. Prokaryotic V-type ATP synthases are evolutionarily related to eukaryotic V-type ATPases, which hydrolyze ATP to acidify certain cellular compartments, in particular, the vacuoles [37, 40, 42]. Some authors classify the simpler, prokaryotic V-type ATPases into a separate subgroup of A-type (from archaeal) ATPases/ATP synthases [39, 43, 44]. Others, instead, prefer to speak about prokaryotic and eukaryotic V-type ATPases [30, 37, 38, 40, 41].
Figure 1. Similar and distinct features in the organization of prokaryotic F- and V-type ATPases.
Homologous subunits in the two types of ATPases are indicated by identical colors and shapes, whereas analogous by evolutionarily unrelated subunits of the central stalk are shown by different colors and shapes. Subunits that show structural analogy but do not appear to be homologous are shown by different but similar colors. The dual notation of some V-ATPase subunits (e.g., c/K) reflects their designations in eukaryotic and prokaryotic V-ATPases, respectively. For further details, see ref. [53].
All F/V-ATPases have a mushroom-like structure, with the hexameric head part (F1 or V1, respectively) protruding ~100 Å from the membrane and carrying ATP/ADP-binding catalytic sites (see Fig. 1 and refs. [39–41, 45, 46]). The F/V–type ATPases are rotary dynamo machines in which the central stalk together with the membrane oligomeric ring is thought to slide along the interface with the stator membrane subunits bound via the peripheral stalk to the catalytic hexamer (see Fig. 1 and [31, 34, 47, 48]). This sliding movement is coupled to the transmembrane ion transfer [1, 49–52].
While sharing a common overall scaffold (Fig. 1), F-type and V-type ATPases differ in many structural and functional features (see Fig. 1 and [40, 42, 46, 53]). In phylogenetic trees, bacterial, archaeal and eukaryotic V-type ATP synthases/ATPases invariably cluster together and separately from the F-type ATPases [54–56].
Proton-translocating and Na+-translocating forms are found among both F- and V-type ATPases. In the absence of Na+ ions, Na+-translocating ATPases of F- and V-types could translocate protons [57, 58]. In contrast, H+-translocating ATPases are apparently incapable of translocating Na+ ions [59]. This asymmetry is most likely due to the higher coordination number of Na+, which requires six ligands to draw it from water and to keep it in the non-polar inner space of the membrane [60]. Comparative studies of the membrane rotor subunits c of the Na+-translocating and H+-translocating ATPases identified several residues that were involved in Na+ binding and served as principal determinants of the coupling ion specificity [59, 61–63]. However, the exact modes of Na+ ion binding in the F- and V-ATPases remained obscure until the structures of the membrane-spanning, rotating oligomers of the Na+-translocating ATP synthases of the F- and V-type were resolved [64, 65]. These structures revealed the same overall configuration of the Na+-binding sites, including conservation of all key ligands (Fig. 2). The minor difference between the two binding sites turned out to be the presence of a Gln residue that could bind the Na+ ion directly in the c/K subunit of a V-type ATPase [65], whereas the corresponding Thr residue in the c subunit of the F-type ATPase apparently coordinates the Na+ ion via an additional water molecule (Thomas Meier, personal communication). As seen in Fig. 2, the structural conservation even extends to the non-ligating Tyr residue that is important for stabilization of the principal, Na+-binding Glu residue [58].
Figure 2. Similar structural organization of membrane rotor subunits and their Na+-binding sites in the F- and V-type Na+-translocating ATP synthases.
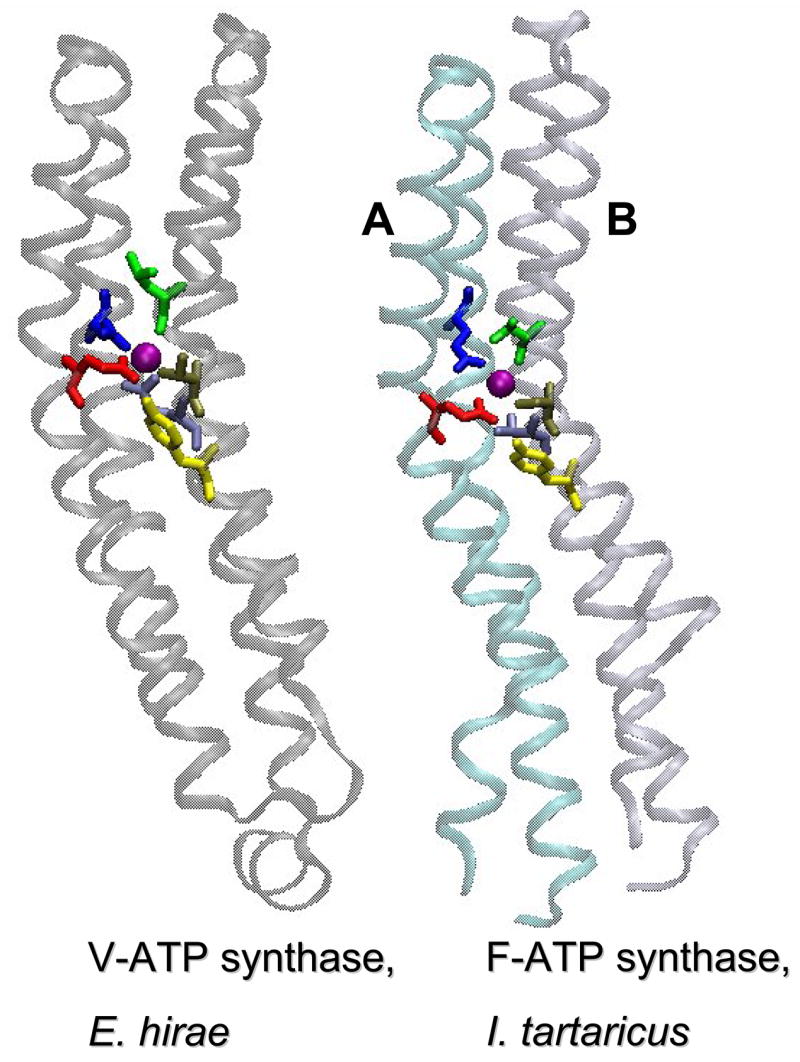
The structures were drawn by using the VMD software package [123]. Left panel, a single K subunit of the Na+-translocating V-type ATP synthase of Enterococcus chirae (Protein Data Bank entry 2BL2 [65]); right panel, two c subunits, A (cyan) and B (ice-blue), of the Na+-translocating F-type ATP synthase of Ilyobacter tartaricus (Protein Data Bank entry 1YCE [64]). In both structures, the Na+ ion is shown as a purple ball, amino acid residues that coordinate the Na+ ion are shown in stick representation and colored. The principal Na+-coordinating Glu residue (Glu65A in I. tartaricus and Glu139 in E. hirae) is colored red; other Na+ ligands are colored as follows: Gln32A in I. tartaricus and Gln110 in E. hirae, blue; Ser66B in I. tartaricus and Thr64 in E. hirae, tan; Val63B in I. tartaricus and Leu61 in E. hirae (which coordinate Na+ with their backbone carbonyls), green. One more bond is provided by a grey-colored Gln65 in E. hirae and, via a water molecule, by a Thr67B in I. tartaricus (T. Meier, personal communication). The remaining sixth bond is provided, most likely, by the unseen water molecules as discussed in more detail elsewhere [30]. The Tyr residue (Tyr70B in I. tartaricus and Tyr68 in E. hirae) that is important for stabilization of the principal Na+-binding Glu residue [58] is colored yellow.
3. Ancestral status of the Na+ bioenergetics
Deciphering of binding modes of Na+ ions in the F-type and V-type ATPases made it possible to address the question of the cation specificity of the common ancestor of these two types of membrane ATPase. The nearly identical arrangement of the Na+-binding sites in F-type and V-type c subunits (Fig. 2) strongly suggests that the common ancestor of V-type and F-type ATPases also was a Na+-translocating enzyme. In the phylogenetic tree of the F/V ATPases, as shown elsewhere [30], Na+-translocating ATPases did not form a clade within either the F or the V branch but instead comprised three distinct lineages among V-ATPases and at least three lineages among F-ATPases with the same set of Na+ ligands conserved in each of these clades. Independent emergence of nearly identical sets of Na+ ligands in several distinct lineages does not look plausible, leading to the conclusion that the common ancestor of V- and F-ATPases had a Na+-binding site [30]. This would mean that the ability to translocate Na+ was independently lost in multiple lineages of F-type and V-type ATPases, yielding H+-translocating enzymes. Multiple instances of ligand loss in the absence of purifying selection are far more plausible than multiple instances of ligand gain and are consistent with the conserved sequence patterns in the vicinity of the H+-binding Glu residue in the c subunits of various bacterial phyla [30]. As discussed above, this also indicates that the primordial organism that harbored this ancestral enzyme possessed a Na+-based membrane energetics.
The possibility that sodium bioenergetics antedated proton bioenergetics has been considered previously [1, 20, 66] but has not drawn much attention, and proton-dependent energy metabolism is widely believed to be ancestral [67]. The primacy of proton bioenergetics is attractive considering the intrinsic chemical coupling between protonation/deprotonation events and redox reactions, in particular, those of water and diverse quinones [68–70]. However, the complexity of the proton-tight membranes serves as a powerful argument against proton energetics being the ancestral state. Indeed, the transition to proton energetics could not have happened until the cellular membranes became impermeable for protons. As argued elsewhere [30, 71, 72] and discussed in some detail below, organization of such membranes is anything but trivial and their origin must have involved several distinct evolutionary steps.
4. Emergence of membrane bioenergetics in the Sodium World
It is conceivable that the functional evolution of the F/V-ATPases was determined by the evolution of the membranes and not the other way around [30], so that the tentative evolutionary scenario has to consider parallel, coupled changes of membrane enzymes and membranes per se. One such evolutionary reconstruction suggested that F/V ATPases evolved from ATP-dependent RNA/protein translocases that functioned within primordial membranes, which were permeable for both H+ and Na+ but not for RNA or protein [53]. The ancient translocase could have used a ring of small membrane subunits to form a membrane channel and could employ Na+ ions to crosslink and stabilize them (as in Ilyobacter tartaricus [64], see Fig. 2), preventing eventual destruction of the channel by the translocated polymer. This suggestion is supported by experiments demonstrating a dramatic decrease in the stability of the c-oligomers of Na+-translocating F-ATPases from I. tartaricus and Propionigenium modestum in the absence of Na+ [73]. Thus, even if primordial membranes were leaky for both Na+ and H+, there could have been a mechanistic demand for Na+-binding and, accordingly, selection for the corresponding set of amino acid ligands.
The next stage of evolution could be envisaged as selection for tighter membranes that would maintain the ionic homeostasis of the evolving cells, and concomitantly, would create the opportunity for the utilization of ion gradients. As suggested by Skulachev [1], sodium-tight membranes could precede proton-tight membranes as structurally less demanding (see also below). With sodium-impermeable membranes, a mechanism for pumping Na+ out of the cell, in response to the increasing salinity of the primordial ocean, would provide a clear evolutionary advantage, driving the transition from an ATP-dependent protein translocase to an ion-translocating membrane ATPase (see [53] for details). The common ancestor of the F- and V-ATPases, thanks to its rotating scaffold, would then be able to translocate Na+ ions in both directions, depending on the magnitude of the SMF. Upon further increase in the external salinity, reversal of the rotation could result in the Na+-driven synthesis of ATP by this primordial rotary machine.
The transition from an ATPase to an ATP synthase could only have happened if other primary Na+ pumps were available for maintaining the Na+ gradient. Several classes of such pumps have been characterized. The first one includes Na+-transporting oxaloacetate decarboxylase, described in 1980 by Dimroth [74], and similar biotin-dependent membrane-bound enzymes that couple export of Na+ ions to the decarboxylation of malonate, methylmalonyl-CoA, or glutaconyl-CoA (reviewed in [27]). The second class of primary Na+ pumps is represented by the Na+-translocating N5-methyltetrahydromethanopterin:coenzyme M methyltransferase [75], detected so far only in methanogenic archaea (reviewed in [76]). One more class of primary Na+ pumps includes Na+-transporting ATPases that belong to the ABC (ATP binding cassette)-type or P-type ATPase families. An ABC-type Na+-transporting ATPase has been described in Bacillus subtilis [77], while a P-type Na+-transporting ATPase has been reported in another member of Bacillaceae, the alkaliphilic bacterium Exiguobacterium aurantiacum [78]. Close homologs of these enzymes are found in a variety of other bacteria, suggesting that ATP-dependent export of Na+ might be widespread in the microbial world. The next class of primary Na+ pumps is represented by the Na+-translocating NADH:ubiquinone oxidoreductase (NaNQR), a respiratory Na+ pump, first described in a marine bacterium Vibrio alginolyticus by Tokuda and Unemoto [79], and a closely related RNF enzyme, originally identified by its role in Rhodobacter nitrogen fixation [80] and proposed to function as a Na+-translocating ferredoxin:NAD oxidoreductase [23]1.
A recent work by Malinen and coauthors [90] identified an entirely new, fifth, class of primary Na+ pumps. The membrane-bound inorganic pyrophosphatase (PPase) has been long known to be an energy-linked enzyme, capable of coupling pyrophosphate hydrolysis to the transfer of H+ ions across the membrane. Previous studies of the membrane-bound PPase from Thermotoga maritima showed that its activity was Na+-dependent [91], suggesting that this enzyme might translocate Na+ ions. A direct study of ion translocation by T. maritima PPase expressed in E. coli and characterized in sub-bacterial membrane vesicles proved it to be an electrogenic Na+ pump extruding sodium ions from the cytoplasm at the expense of pyrophosphate hydrolysis. The ability to translocate Na+ ions was also demonstrated for the closely related PPase enzymes from the moderately thermophilic bacterium Moorella thermoacetica and the mesophilic archaeon Methanosarcina mazei [90]. Given that close homologs of these PPases are found in a wide variety of bacteria and archaea, generation of the SMF at the expense of pyrophosphate may turn out to be a very common mechanism of membrane energy transformation.
Out of the five classes of primary sodium pumps, at least two, namely the Na+-pumps related to the Na+-transporting decarboxylases [27, 57] and the Na+-transporting pyrophosphatase [90], are present both in bacteria and archaea and thus appear to antedate the divergence of the three domains of life. It would be reasonable to assume that the plethora of Na+-coupled secondary membrane transporters, encoded in various bacteria and archaea, also originate from the Sodium World. Many of these Na+-dependent transporters do not have proton-driven counterparts, so substrate transport into the cell essentially depends on a system of Na+/H+ exchangers transmitting part of the PMF into SMF even in the organisms with proton energetics (reviewed in [92, 93]).
Taken together, ancient Na+ pumps, the Na+-driven ATP synthase, and Na+-driven secondary transporters could complete the first, sodium-dependent energy cycle in the primitive cell membrane, marking the onset of the membrane bioenergetics.
5. Evolutionary advantages and constraints of switching from Na+ to H+ energetics
As argued by Haines, proton impermeability of the membrane is achieved by tighter packing of atoms in the middle of the bilayer, between its two leaflets [71]. Representatives of the three domains of life utilize distinct solutions to make their membranes tighter to protons [26, 30, 71, 72], which additionally buttresses the suggestion on the independent transition from the sodium to proton energetics in different lineages. Anyway, the majority of bacteria, archaea and eukaryotes all found ways to make the membrane proton-tight (with a notable exception of animal plasma membranes that remained “sodium membranes” [1]).
As briefly noted above, proton energetics is chemically more advantageous. The use of H+ as a coupling ion offers the benefit of direct mechanistic linkage of scalar redox reactions to vectorial translocation of proton across the membrane, resulting in the generation of the PMF. Thus, a plethora of primary redox-driven H+ translocators with different characteristic mid-point potentials became available for the organization of orderly electron-transfer chains with multiple coupling sites, sequentially covering the redox span of about 1.2 eV from organic substrates to oxygen [1, 2]. In contrast, the (known) redox-driven Na+ pumps operate over a much smaller redox gap, namely from NADH to quinone in case of the NQR and from ferredoxin to NAD+ in case of RNF. Besides, the number of required coordinating ligands for the proton is either one (for H+) or three (if H3O+ is translocated [94, 95]), compared to as many as six for the Na+ ion [60]. Not surprisingly, after membranes became proton-tight, the more robust and versatile proton-transporting devices spread all over the microbial world. Why then proton energetics did not completely replace the sodium energetics? Why do some organisms still rely upon sodium energetics?
In the historical perspective, Na+ ion cycle was originally proposed as an adaptive mechanism to solve the bioenergetic problems arising in organisms populating the habitats where generation and maintenance of sufficiently high PMF seems to be impossible, most importantly, in extreme alkaliphiles [1, 3–5]. In addition, Na+ ion cycle has been suggested as a solution for extreme thermophiles. Indeed, prokaryotic growth at high temperatures is limited by the increased permeability of the cytoplasmic membrane for ions resulting in low levels of the total PMF [7]. Remarkably, membrane permeability for Na+ ions is several orders of magnitude lower than for protons [7, 26], due to the fundamentally different mechanisms of H+ and Na+ transfer across the membrane [30, 71, 96–99].
An important consequence of the structural characterization of the Na+-binding sites in the c subunits of F-type and V-type ATPases [64, 65] was that it finally allowed unequivocal assignment of the cation specificity for the membrane ATPases encoded in numerous sequenced genomes [30] and, accordingly, a large-scale check of the correlations between the growth conditions of various organisms and the types of membrane energetics they employ. These assignments, while confirming many earlier observations, brought some surprising results.
The original concept of the Na+ ion cycle as an adaptation to alkaline environments proved to be correct only for a limited number of organisms. While some alkaliphiles have a fully functioning Na+ cycle, i.e. encode one or more primary Na+ pumps and have a Na+-dependent For V-type ATP synthase, others can grow at pH 10 and above using H+ as the coupling ion (Table 1). These data clearly show that Na+ cycle is not necessary for survival in alkaline environments, confirming previous results from the Krulwich group (reviewed in [24, 100–102]), as well as reports by others [17, 103]2.
Table 1.
Utilization of Na+ and H+ cycle by alkaliphilic bacteria and archaea
| Organism | Growth at pH
|
Na+ pumpsa | ATPase ion specificityb | |
|---|---|---|---|---|
| opt | max | |||
| Archaea | ||||
| Natronomonas pharaonis | 8.5 | 11.0 | – | H+ |
| Bacteria | ||||
| Alkalilimnicola ehrlichei | 9.3 | 10.2 | – | H+ |
| Alkaliphilus metalliredigens | 9.6 | 11.0 | OAD (2x) PP, RNF | Na+ |
| Bacillus halodurans | 9.0 | 10.8 | – | H+ |
| Bacillus clausii | 9.0 | 10.5 | – | H+ |
| Clostridium paradoxum | 9.3 | 10.2 | nd | Na+ |
| Vibrio cholerae | 7.6 | 9.6 | NQR, OAD, RNF | H+ |
| Vibrio parahaemolyticus | 7.8–8.6 | 11.0 | NQR, OAD, RNF | H+ |
| Vibrio vulnificus | 7.8 | 10.0 | NQR, OAD, RNF | H+ |
| Yersinia enterocolitica | 7.4 | 10.0 | NQR, RNF | H+ |
- The Na+ pumps encoded in completely sequenced genomes are abbreviated as follows: NQR, Na+-translocating NADH:quinone oxidoreductase; OAD, Na+-translocating oxaloacetate decarboxylase (A. metalliredigens carries two oad operons); PP, Na+-translocating pyrophosphatase; RNF - putative Na+-translocating ferredoxin:NAD+ oxidoreductase. A dash indicates absence of the encoded Na+ pumps, nd - no data (Clostridium paradoxum genome has not been sequenced).
- Cation specificity of the respective ion-translocating ATPases has been predicted based on the presence or absence of the complete set of Na+ ligands (see Fig. 1). Experimentally characterized ion selectivity is indicated by bold typeface.
How can alkaliphilic, proton-dependent prokaryotes manage to energize ATP synthesis in the conditions of high pH? The first reports of proton energetics in extreme alkaliphiles already asked for consideration of so-called local coupling mechanisms [6]. One group of such hypothetical mechanisms implies that the effective concentration of protons on the external surface of the membrane is higher than that in the bulk; a rapid lateral H+ movement between the respiratory H+ pump and the ATP synthase could then be able to support ATP synthesis [6, 105–107]. This mechanism might rely on a particular trait of the membrane/water interface: the negatively charged membrane surface is separated from the bulk water phase by an electrostatic barrier. For protons, its height could be estimated as about 0.12 eV, which is high enough to keep the pH value at the external surface of metabolizing alkaliphiles neutral even if the surrounding medium is strongly alkaline [29, 108, 109]. Structural analysis of prokaryotic proton pumps has revealed that their periplasmic surface is covered by Asp and Glu residues that should facilitate proton transfer along the membrane/water interface [29]. Further on, a direct intramembrane transfer of H+ from respiratory complex to FO portion of the ATP synthase was suggested to occur via protein-protein interaction [6], a mechanism that echoes the original hypothesis by R.J.P. Williams [105, 110]. A recent paper from the Krulwich group shows a direct interaction between cytochrome caa3 and the F-type ATP synthase in liposomes made from the alkaliphile Bacillus pseudofirmus [111]. The two mechanisms of local coupling, in fact, are not contradictory. The already noted structural analysis of prokaryotic proton pumps has also revealed a buried plexus of Arg and Lys residues beyond the surface layer of acidic side chains. These buried bases should operate as proton buffers – proton sponges (see [29] and references therein). Proton transfer between the sponges of two neighboring enzymes is well imaginable. The potential importance of such proton transfer is supported by the finding that replacement of a single Lys residue in the FO part of the alkaliphilic ATP synthase of B. pseudofirmus by Gly leads to the loss of the non-fermentative, respiration-driven growth at high pH [112].
As follows from Table 2, the idea that sodium cycle was indispensable for growth at high temperatures also did not prove to be valid. Certain bacterial and archaeal hyperthermophiles (defined as organisms that grow optimally at or above 77°C) encode primary Na+ pumps and have a Na+-dependent ATP synthase, indicating that they do rely on the Na+ ion cycle for their energy metabolism (see also [113]). Nevertheless, a significant number of hyperthermophilic bacteria and archaea do not encode any (known) primary Na+ pumps and have H+-translocating ATP synthases (Table 2). Thus, a functional Na+ cycle is not necessary for survival at high temperature either. How then would bacterial cells overcome massive transmembrane proton leakage at high temperatures? Or, rather, which factors define whether a particular group of organisms uses H+ or Na+ as the coupling ion?
Table 2.
Utilization of Na+ and H+ cycle by hyperthermophilic bacteria and archaea
| Archaea | T°opt | Na+ pumpsa | ATPase ion specificitya | O2 tolerance, e− acceptor |
|---|---|---|---|---|
| Aeropyrum pernix | 95°C | – | H+ | Aerobe |
| Archaeoglobus fulgidus | 85°C | OAD | H+ | Anaerobe, sulfate |
| Hyperthermus butylicus | 106°C | – | H+ | Anaerobe, sulfur |
| Ignicoccus hospitalis | 90°C | – | H+ | Anaerobe, sulfur |
| Methanocaldococcus jannaschii | 85°C | MTase | Na+ | Anaerobe, CO2 |
| Methanopyrus kandleri | 98°C | MTase | Na+ | Anaerobe, CO2 |
| Nanoarchaeum equitans | 90°C | – | Na+ | Anaerobe |
| Pyrobaculum aerophilum | 100°C | PP | H+ | Aerobe, O2 or nitrate |
| Pyrobaculum arsenaticum | 95°C | PP | H+ | Anaerobe, arsenate |
| Pyrobaculum calidifontis | 95°C | PP | H+ | Aerobe, O2 or nitrate |
| Pyrobaculum islandicum | 100°C | PP | H+ | Anaerobe, sulfite |
| Pyrococcus abyssi | 96°C | MCD | Na+ | Anaerobe, sulfur |
| Pyrococcus furiosus | 100°C | MCD | Na+ | Anaerobe, sulfur |
| Pyrococcus horikoshii | 98°C | MCD | Na+ | Anaerobe, sulfur |
| Sulfolobus solfataricus | 87°C | – | H+ | Aerobe |
| Sulfolobus tokodaii | 80°C | – | H+ | Aerobe |
| Thermococcus kodakarensis | 95°C | MCD | Na+ | Anaerobe, sulfur |
| Thermofilum pendens | 90°C | – | H+ | Anaerobe, sulfur |
| Bacteria | ||||
| Aquifex aeolicus | 95°C | – | H+ | Aerobe |
| Thermoanaerobacter tengcongensis | 75°C | MCD, PP | H+ | Anaerobe, sulfur, thiosulfate |
| Thermotoga maritima | 80°C | OAD, PP, RNF | Na+ | Anaerobe, sulfur |
| Thermotoga petrophila | 80°C | OAD, PP, RNF | Na+ | Anaerobe, sulfur |
| Thermus thermophilus | 75°C | – | H+ | Aerobe |
- Abbreviations and symbols as in Table 1; MCD, Na+-tranlocating methylmalonyl-CoA decarboxylase; MTase, Na+-tranlocating N5-methyltetrahydromethanopterin:coenzyme M methyltransferase.
Comparative analysis of microbial genomes clearly demonstrates that the distribution of the Na+ cycle in hyperthermophilic species is rather patchy and does not correlate with the optimal growth temperature (Table 2). However, there is a clear correlation between the H+ cycle and utilization of oxygen: every archaeal and bacterial hyperthermophile capable of growing in aerobic or microaerophilic conditions has an H+-translocating ATP synthase, even if it encodes a potential Na+ pump and uses SMF to energize substrate symports and/or motility. Hyperthermophilic anaerobes that utilize such high-potential alternative electron acceptors as nitrate, sulfate, or sulfite also tend to have an H+-translocating ATP synthase (Table 2). The Na+-translocating ATP synthase, and, accordingly, the full Na+ cycle are found primarily in anaerobic hyperthermophiles that grow by fermentation. We think that this trend, as well as the distribution of the Na+ cycle in analyzed genomes in general, could be explained by the following simple considerations:
The H+ cycle represents a relatively recent evolutionary acquisition that spread only after proton-tight membranes appeared, as discussed in detail previously [30]. Importantly, the proton-tightness of modern membranes is not absolute: even with all the improvements their conductivity to protons is by orders of magnitude higher than to Na+ ions [71, 96–99]. Accordingly, the organisms that rely on proton energetics have to cope with a notable leakage of H+ ions that should dramatically increase with temperature [7, 26] and with the rise in the external pH. Although psychrophilic and mesophilic bacteria, as well as archaea, are able to adjust the lipid composition of their membranes in order to limit the H+ permeability (so-called “homeo-proton permeability adaptation”), the capacity of such adjustment is limited [26, 71, 114, 115].
As discussed above, the H+ energetics is, generally, lucrative; that is why it is employed by mitochondria, chloroplasts, and the vast majority of modern microorganisms. Using a respiratory chain and the H+ cycle, it becomes possible to gain plenty of free energy upon oxidation of external substrates - but only if the high-potential electron acceptors, such as oxygen, nitrate, sulfate, or sulfite, and respective oxidoreductases are available. Otherwise, the energy gain from proton energetics becomes no better than that from sodium energetics, which appears to operate over small redox gaps, as discussed above.
Therefore, whether an organism relies on Na+ or H+ as the coupling ion, seems to depend on a trade-off between amount of potentially available free energy and the intensity of ion leakage across the coupling membrane. Under mesophilic conditions, when proton leaks appear to be moderate, prokaryotes routinely select the more efficient proton energetics. Only in some obligate anaerobes, whose energy budget is tight and cannot cover the losses caused by proton leaks, Na+ energetics may become the favored one. At high temperature and/or high pH, when proton leaks are expected to be large, the sodium energetics becomes more advantageous than under mesophilic conditions, so that obligate anaerobes routinely exploit the sodium cycle. Those anaerobic organisms that, in addition, are capable of reducing sulfur (or polysulfide) make a threshold case – some of them use sodium energetics while Thermofilum pendens and possibly others have an H+-ATPase (see Table 2). This distribution reflects the fact that the ability to reduce sulfur seems to give only a marginal advantage as compared to fermentation alone. Indeed, the energy gain from sulfur reduction by hydrogen has been estimated as 30 kJ/mol at pH 7.0 as compared to >150 kJ/mol for the reduction of sulfate, sulfite or thiosulfate [116], not to mention oxygen. Accordingly, extremophiles that have ample supply of powerful electron acceptors, be it oxygen, nitrate or sulfate, are less energy-limited and can rely on proton energetics even while paying a heavy price of elevated proton leakage at high temperature or at high pH (see Table 2). The need to “pay” for increased proton leaks might explain a sharp increase in the H+-extruding respiration in some thermophiles in response to the elevated temperatures [117], and in the three-fold increase of the cytochrome oxidase content in B. pseudofirmus grown at pH 10.5 [118].
6. Why certain pathogenic bacteria still rely on the sodium-motive force?
Existence of pathogenic microorganisms that fully depend on sodium energetics [20] finds its explanation if we take into account the anaerobic conditions inside animal body and the absence of alternative electron acceptors such as nitrate, sulfate, or sulfite. However, many bacterial pathogens encode primary Na+ pumps, such as NQR, RNF, and/or oxaloacetate decarboxylase, even if their ATP synthases are H+-dependent [19, 20, 63, 119]. This intriguing property could be rationalized by taking into account the bioenergetic challenges confronting pathogenic bacteria at certain stages of their life cycles.
For example, in the case of human pathogen Vibrio cholerae, preservation of primary Na+ pumps, together with extended system of Na+/H+ antiport and a plethora of SMF consumers, could be related to the estuarine environment that V. cholerae inhabits during the free-living part of its life cycle. Estuaries show large fluctuations in physical parameters including pH (reaching up to 9.5) and salinity [120]. Na+-based energetics could be critical for survival of this pathogen during periods of alkaline swings that occur in estuaria. Remarkably, secretion of the cholera toxin during the colonization of the human gut by V. cholerae results in elevated levels of Na+ in alkaline intestinal lumen, almost mimicking the estuarian conditions. Cholera toxin-mediated sodium enrichment of the lumen environment is probably an adaptive mechanism that boosts the efficiency of Na+ circulation in V. cholerae colonizing the host [121].
Obligate intracellular parasites with a complete Na+ cycle, such as the Chlamydia species that possess all key elements of the Na+-cycle, including a respiratory Na+ pump (NaNQR), Na+-specific ATP synthase, a Na+/H+ antiporter, and Na+-substrate symporters [20, 63], might have additional reasons for reliance on the SMF. These organisms proliferate in the homeostatic, nutrient-rich cytoplasm of an infected cell that has relatively low Na+ concentration and near-neutral pH, next to the mitochondria and chloroplasts that exclusively utilize the H+ gradient. A recently suggested model presents a possible mechanism by which chlamydia manipulate ion homeostasis of the host cell in such a way that the Na+ cycle could become important to their survival [122]. It posits that, after exhausting the energy resources of the host cell, proliferating chlamydia switch to amino acid fermentation, which results in chlamydial microenvironment becoming relatively alkaline and sodium-rich. Under these conditions, the SMF becomes the preferred source of energy for nutrient uptake by chlamydial cells (see [122] for detailed discussion).
7. Conclusions
Recent results, coming from structural characterization of membrane ATPases, experimental studies, and comparative genome analyses, provide strong evidence that, rather than being an exotic adaptation to extreme conditions, the sodium-motive force was an ancestral, albeit not very efficient, mode of membrane energy metabolism. In most ecological niches, the availability of usable terminal electron acceptors, such as oxygen, nitrate, or sulfate, led to gradual replacement of the Na+ ion cycle with the far more efficient proton cycle. In some sense, the Na+ cycle is an evolutionary relic that has been preserved by natural selection only in a limited set of fermentative organisms, which, incidentally, includes some important human pathogens. A much wider group of organisms retained one or more primary Na+ pumps that are utilized under conditions of lowered PMF (e.g., anaerobiosis) or high salinity (e.g., in marine bacteria). The traces of Na+-based energetics are still seen in the universal distribution of Na+ gradients and Na+-dependent systems of solute transport in virtually all known cell types.
Acknowledgments
We thank Drs. A.A. Baykov, A.V. Bogachev, W. Junge, E.V. Koonin, T.H. Haines, K.S. Makarova, T. Meier, V.P. Skulachev and Y.I. Wolf for helpful discussions. This work was supported in part by grants from Deutsche Forschungsgemeinschaft (Mu-1285/10-1) and the Volkswagen Foundation (AYM), the Natural Sciences and Engineering Research Council of Canada (PD), and by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health (MYG).
Abbreviations
- PMF
proton-motive force
- SMF
sodium-motive force
- NaNQR
Na+-translocating NADH:ubiquinone oxidoreductase
Footnotes
There have been reports of Na+-translocating terminal oxidases in a variety of bacteria, including Escherichia coli, Vibrio alginolyticus, Bacillus halodurans, and Vitreoscilla sp. [81–85]. However, most of those studies were conducted on whole cells and the ability of any terminal oxidase to transport Na+ ions still remains to be confirmed with a purified enzyme preparation. One more enzyme, formyl-methanofuran dehydrogenase of Methanothermobacter thermautotrophicus and other methanogenic archaea, can translocate sodium ions [86] but normally works in the reverse direction, using the PMF or SMF to generate an electron donor for CO2 reduction [87–89]
It should be noted that no genome sequences of obligate alkaliphilic bacteria (defined by Horikoshi as organisms that require pH 9 or more for their growth and have an optimal growth pH of around 10 [104]) are available at this time.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Skulachev VP. Membrane Bioenergetics. Springer-Verlag; Berlin: 1988. [Google Scholar]
- 2.Cramer WA, Knaff DB. Energy Transduction in Biological Membranes: A Textbook of Bioenergetics. Springer-Verlag; 1990. [Google Scholar]
- 3.Skulachev VP. Membrane-linked energy transductions. Bioenergetic functions of sodium: H+ is not unique as a coupling ion. Eur J Biochem. 1985;151:199–208. doi: 10.1111/j.1432-1033.1985.tb09088.x. [DOI] [PubMed] [Google Scholar]
- 4.Skulachev VP. The sodium cycle: a novel type of bacterial energetics. J Bioenerg Biomembr. 1989;21:635–647. doi: 10.1007/BF00762683. [DOI] [PubMed] [Google Scholar]
- 5.Dimroth P. Bacterial energy transductions coupled to sodium ions. Res Microbiol. 1990;141:332–336. doi: 10.1016/0923-2508(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 6.Krulwich TA. Alkaliphiles: ‘basic’ molecular problems of pH tolerance and bioenergetics. Mol Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 7.van de Vossenberg JLCM, Ubbink-Kok T, Elferink MG, Driessen AJM, Konings WN. Ion permeability of the cytoplasmic membrane limits the maximum growth temperature of bacteria and archaea. Mol Microbiol. 1995;18:925–932. doi: 10.1111/j.1365-2958.1995.18050925.x. [DOI] [PubMed] [Google Scholar]
- 8.Lolkema JS, Speelmans G, Konings WN. Na+-coupled versus H+-coupled energy transduction in bacteria. Biochim Biophys Acta. 1994;1187:211–215. doi: 10.1016/0005-2728(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 9.Albers SV, van de Vossenberg JLCM, Driessen AJM, Konings WN. Bioenergetics and solute uptake under extreme conditions. Extremophiles. 2001;5:285–294. doi: 10.1007/s007920100214. [DOI] [PubMed] [Google Scholar]
- 10.Hilpert W, Schink B, Dimroth P. Life by a new decarboxylation-dependent energy conservation mechanism with sodium as coupling ion. EMBO J. 1984;3:1665–1670. doi: 10.1002/j.1460-2075.1984.tb02030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speelmans G, Poolman B, Abee T, Konings WN. Energy transduction in the thermophilic anaerobic bacterium Clostridium fervidus is exclusively coupled to sodium ions. Proc Natl Acad Sci USA. 1993;90:7975–7979. doi: 10.1073/pnas.90.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimroth P, Hilbi H. Enzymic and genetic basis for bacterial growth on malonate. Mol Microbiol. 1997;25:3–10. doi: 10.1046/j.1365-2958.1997.4611824.x. [DOI] [PubMed] [Google Scholar]
- 13.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Otsuka R, Nakazawa H, Takamiya M, Kato Y, Yoshizawa T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Masuda S, Yanagii M, Nishimura M, Yamagishi A, Oshima T, Kikuchi H. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 2001;8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- 17.Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 2005;15:1336–1343. doi: 10.1101/gr.3952905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kogure K. Bioenergetics of marine bacteria. Curr Opin Biotechnol. 1998;9:278–282. doi: 10.1016/s0958-1669(98)80059-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, Bertsova YV, Feng B, Tsatsos P, Verkhovskaya ML, Gennis RB, Bogachev AV, Barquera B. Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry. 1999;38:16246–16252. doi: 10.1021/bi991664s. [DOI] [PubMed] [Google Scholar]
- 20.Häse CC, Fedorova ND, Galperin MY, Dibrov PA. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev. 2001;65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimroth P, Cook GM. Bacterial Na+- or H+-coupled ATP synthases operating at low electrochemical potential. Adv Microb Physiol. 2004;49:175–218. doi: 10.1016/S0065-2911(04)49004-3. [DOI] [PubMed] [Google Scholar]
- 22.Bogachev AV, Verkhovsky MI. Na+-translocating NADH:quinone oxidoreductase: progress achieved and prospects of investigations. Biochemistry (Mosc) 2005;70:143–149. doi: 10.1007/s10541-005-0093-4. [DOI] [PubMed] [Google Scholar]
- 23.Boiangiu CD, Jayamani E, Brugel D, Herrmann G, Kim J, Forzi L, Hedderich R, Vgenopoulou I, Pierik AJ, Steuber J, Buckel W. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J Mol Microbiol Biotechnol. 2005;10:105–119. doi: 10.1159/000091558. [DOI] [PubMed] [Google Scholar]
- 24.Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: new insights. Biochim Biophys Acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimroth P, von Ballmoos C, Meier T. Catalytic and mechanical cycles in F-ATP synthases. EMBO Rep. 2006;7:276–282. doi: 10.1038/sj.embor.7400646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konings WN. Microbial transport: adaptations to natural environments. Antonie van Leeuwenhoek. 2006;90:325–342. doi: 10.1007/s10482-006-9089-3. [DOI] [PubMed] [Google Scholar]
- 27.Dimroth P, von Ballmoos C. ATP synthesis by decarboxylation phosphorylation. Results Probl Cell Differ. 2007 doi: 10.1007/400_2007_045. [DOI] [PubMed] [Google Scholar]
- 28.Dibrov P. The sodium cycle in vibrio cholerae: riddles in the dark. Biochemistry (Mosc) 2005;70:150–153. doi: 10.1007/s10541-005-0094-3. [DOI] [PubMed] [Google Scholar]
- 29.Mulkidjanian AY, Heberle J, Cherepanov DA. Protons @ interfaces: implications for biological energy conversion. Biochim Biophys Acta. 2006;1757:913–930. doi: 10.1016/j.bbabio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Mulkidjanian AY, Galperin MY, Makarova KS, Wolf YI, Koonin EV. Evolutionary primacy of sodium bioenergetics. Biol Direct. 2008;3:13. doi: 10.1186/1745-6150-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 32.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 33.Capaldi RA, Aggeler R. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem Sci. 2002;27:154–160. doi: 10.1016/s0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- 34.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 35.Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006;25:5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukohata Y, Yoshida M. The H+-translocating ATP synthase in Halobacterium halobium differs from F0F1-ATPase/synthase. J Biochem. 1987;102:797–802. doi: 10.1093/oxfordjournals.jbchem.a122118. [DOI] [PubMed] [Google Scholar]
- 37.Perzov N, Padler-Karavani V, Nelson H, Nelson N. Features of V-ATPases that distinguish them from F-ATPases. FEBS Lett. 2001;504:223–228. doi: 10.1016/s0014-5793(01)02709-0. [DOI] [PubMed] [Google Scholar]
- 38.Nishi T, Forgac M. The vacuolar H+-ATPases - Nature’s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 39.Müller V, Gruber G. ATP synthases: structure, function and evolution of unique energy converters. Cell Mol Life Sci. 2003;60:474–494. doi: 10.1007/s000180300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drory O, Nelson N. The emerging structure of vacuolar ATPases. Physiology (Bethesda) 2006;21:317–325. doi: 10.1152/physiol.00017.2006. [DOI] [PubMed] [Google Scholar]
- 41.Yokoyama K, Imamura H. Rotation, structure, and classification of prokaryotic V-ATPase. J Bioenerg Biomembr. 2005;37:405–410. doi: 10.1007/s10863-005-9480-1. [DOI] [PubMed] [Google Scholar]
- 42.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 43.Ihara K, Abe T, Sugimura KI, Mukohata Y. Halobacterial A-ATP synthase in relation to V-ATPase. J Exp Biol. 1992;172:475–485. doi: 10.1242/jeb.172.1.475. [DOI] [PubMed] [Google Scholar]
- 44.Schäfer G, Meyering-Vos M. F-type or V-type? The chimeric nature of the archaebacterial ATP synthase. Biochim Biophys Acta. 1992;1101:232–235. doi: 10.1016/0005-2728(92)90233-r. [DOI] [PubMed] [Google Scholar]
- 45.Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 46.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 47.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 48.Panke O, Gumbiowski K, Junge W, Engelbrecht S. F-ATPase: specific observation of the rotating c subunit oligomer of EFoEF1, FEBS Lett. 2000;472:34–38. doi: 10.1016/s0014-5793(00)01436-8. [DOI] [PubMed] [Google Scholar]
- 49.Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 50.Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 51.Cherepanov DA, Mulkidjanian AY, Junge W. Transient accumulation of elastic energy in proton translocating ATP synthase. FEBS Lett. 1999;449:1–6. doi: 10.1016/s0014-5793(99)00386-5. [DOI] [PubMed] [Google Scholar]
- 52.Mulkidjanian AY. Proton in the well and through the desolvation barrier. Biochim Biophys Acta. 2006;1757:415–427. doi: 10.1016/j.bbabio.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 53.Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat Rev Microbiol. 2007;5:892–899. doi: 10.1038/nrmicro1767. [DOI] [PubMed] [Google Scholar]
- 54.Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, Konishi J, Denda K, Yoshida M. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilario E, Gogarten JP. Horizontal transfer of ATPase genes--the tree of life becomes a net of life. Biosystems. 1993;31:111–119. doi: 10.1016/0303-2647(93)90038-e. [DOI] [PubMed] [Google Scholar]
- 56.Hilario E, Gogarten JP. The prokaryote-to-eukaryote transition reflected in the evolution of the V/F/A-ATPase catalytic and proteolipid subunits. J Mol Evol. 1998;46:703–715. doi: 10.1007/pl00006351. [DOI] [PubMed] [Google Scholar]
- 57.Dimroth P. Primary sodium ion translocating enzymes. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 58.von Ballmoos C, Dimroth P. Two distinct proton binding sites in the ATP synthase family. Biochemistry. 2007;46:11800–11809. doi: 10.1021/bi701083v. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Fillingame RH. Changing the ion binding specificity of the Escherichia coli H+-transporting ATP synthase by directed mutagenesis of subunit c. J Biol Chem. 1995;270:87–93. doi: 10.1074/jbc.270.1.87. [DOI] [PubMed] [Google Scholar]
- 60.Frausto da Silva JJR, Williams RJP. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Clarendon Press; Oxford: 1991. [Google Scholar]
- 61.Rahlfs S, Müller V. Sequence of subunit c of the Na+-translocating F1F0 ATPase of Acetobacterium woodii: proposal for determinants of Na+ specificity as revealed by sequence comparisons. FEBS Lett. 1997;404:269–271. doi: 10.1016/s0014-5793(97)00088-4. [DOI] [PubMed] [Google Scholar]
- 62.Kaim G, Wehrle F, Gerike U, Dimroth P. Molecular basis for the coupling ion selectivity of F1F0 ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry. 1997;36:9185–9194. doi: 10.1021/bi970831q. [DOI] [PubMed] [Google Scholar]
- 63.Dzioba J, Hase CC, Gosink K, Galperin MY, Dibrov P. Experimental verification of a sequence-based prediction: F1F0-type ATPase of Vibrio cholerae transports protons, not Na+ ions. J Bacteriol. 2003;185:674–678. doi: 10.1128/JB.185.2.674-678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 65.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 66.Dibrov PA. The role of sodium ion transport in Escherichia coli energetics. Biochim Biophys Acta. 1991;1056:209–224. doi: 10.1016/s0005-2728(05)80052-0. [DOI] [PubMed] [Google Scholar]
- 67.Deamer DW. The first living systems: a bioenergetic perspective. Microbiol Mol Biol Rev. 1997;61:239–261. doi: 10.1128/mmbr.61.2.239-261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Junge W, Haumann M, Ahlbrink R, Mulkidjanian A, Clausen J. Electrostatics and proton transfer in photosynthetic water oxidation. Philos Trans R Soc B Biol Sci. 2002;357:1407–1417. doi: 10.1098/rstb.2002.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faxen K, Gilderson G, Adelroth P, Brzezinski P. A mechanistic principle for proton pumping by cytochrome c oxidase. Nature. 2005;437:286–289. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- 70.Mulkidjanian AY. Proton translocation by the cytochrome bc1 complexes of phototrophic bacteria: introducing the activated Q-cycle. Photochem Photobiol Sci. 2007;6:19–34. doi: 10.1039/b517522d. [DOI] [PubMed] [Google Scholar]
- 71.Haines TH. Do sterols reduce proton and sodium leaks through lipid bilayers? Prog Lipid Res. 2001;40:299–324. doi: 10.1016/s0163-7827(01)00009-1. [DOI] [PubMed] [Google Scholar]
- 72.Konings WN, Albers SV, Koning S, Driessen AJM. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie van Leeuwenhoek. 2002;81:61–72. doi: 10.1023/a:1020573408652. [DOI] [PubMed] [Google Scholar]
- 73.Meier T, Dimroth P. Intersubunit bridging by Na+ ions as a rationale for the unusual stability of the c-rings of Na+-translocating F1F0 ATP synthases. EMBO Rep. 2002;3:1094–1098. doi: 10.1093/embo-reports/kvf216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dimroth P. A new sodium-transport system energized by the decarboxylation of oxaloacetate. FEBS Lett. 1980;122:234–236. doi: 10.1016/0014-5793(80)80446-7. [DOI] [PubMed] [Google Scholar]
- 75.Lienard T, Becher B, Marschall M, Bowien S, Gottschalk G. Sodium ion translocation by N5-methyltetrahydromethanopterin: coenzyme M methyltransferase from Methanosarcina mazei Gö1 reconstituted in ether lipid liposomes. Eur J Biochem. 1996;239:857–864. doi: 10.1111/j.1432-1033.1996.0857u.x. [DOI] [PubMed] [Google Scholar]
- 76.Gottschalk G, Thauer RK. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim Biophys Acta. 2001;1505:28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 77.Cheng J, Guffanti AA, Krulwich TA. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki Y, Ueno S, Ohnuma R, Koyama N. Cloning, sequencing and functional expression in Escherichia coli of the gene for a P-type Na+-ATPase of a facultatively anaerobic alkaliphile, Exiguobacterium aurantiacum. Biochim Biophys Acta. 2005;1727:162–168. doi: 10.1016/j.bbaexp.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Tokuda H, Unemoto T. Na+ is translocated at NADH:quinone oxidoreductase segment in the respiratory chain of Vibrio alginolyticus. J Biol Chem. 1984;259:7785–7790. [PubMed] [Google Scholar]
- 80.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 81.Dibrov PA, Lazarova RL, Skulachev VP, Verkhovskaya ML. The sodium cycle. II. Na+-coupled oxidative phosphorylation in Vibrio alginolyticus cells. Biochim Biophys Acta. 1986;850:458–465. doi: 10.1016/0005-2728(86)90114-3. [DOI] [PubMed] [Google Scholar]
- 82.Semeykina AL, Skulachev VP, Verkhovskaya ML, Bulygina E, Chumakov KM. The Na+-motive terminal oxidase activity in an alkalo- and halo-tolerant Bacillus. Eur J Biochem. 1989;183:671–678. doi: 10.1111/j.1432-1033.1989.tb21097.x. [DOI] [PubMed] [Google Scholar]
- 83.Avetisyan AV, Dibrov PA, Semeykina AL, Skulachev VP, Sokolov MV. Adaptation of Bacillus FTU and Escherichia coli to alkaline conditions: the Na+-motive respiration. Biochim Biophys Acta. 1991;1098:95–104. [PubMed] [Google Scholar]
- 84.Kostyrko VA, Semeykina AL, Skulachev VP, Smirnova IA, Vaghina ML, Verkhovskaya ML. The H+-motive and Na+-motive respiratory chains in Bacillus FTU subcellular vesicles. Eur J Biochem. 1991;198:527–534. doi: 10.1111/j.1432-1033.1991.tb16046.x. [DOI] [PubMed] [Google Scholar]
- 85.Park C, Moon JY, Cokic P, Webster DA. Na+-translocating cytochrome bo terminal oxidase from Vitreoscilla: some parameters of its Na+ pumping and orientation in synthetic vesicles. Biochemistry. 1996;35:11895–11900. doi: 10.1021/bi9530503. [DOI] [PubMed] [Google Scholar]
- 86.Kaesler B, Schönheit P. The role of sodium ions in methanogenesis. Formaldehyde oxidation to CO2 and 2H2 in methanogenic bacteria is coupled with primary electrogenic Na+ translocation at a stoichiometry of 2–3 Na+/CO2. Eur J Biochem. 1989;184:223–232. doi: 10.1111/j.1432-1033.1989.tb15010.x. [DOI] [PubMed] [Google Scholar]
- 87.Kaesler B, Schönheit P. The sodium cycle in methanogenesis. CO2 reduction to the formaldehyde level in methanogenic bacteria is driven by a primary electrochemical potential of Na+ generated by formaldehyde reduction to CH4. Eur J Biochem. 1989;186:309–316. doi: 10.1111/j.1432-1033.1989.tb15210.x. [DOI] [PubMed] [Google Scholar]
- 88.de Poorter LM, Geerts WG, Theuvenet AP, Keltjens JT. Bioenergetics of the formyl-methanofuran dehydrogenase and heterodisulfide reductase reactions in Methanothermobacter thermautotrophicus. Eur J Biochem. 2003;270:66–75. doi: 10.1046/j.1432-1033.2003.03362.x. [DOI] [PubMed] [Google Scholar]
- 89.Stojanowic A, Hedderich R. CO2 reduction to the level of formylmethanofuran in Methanosarcina barkeri is non-energy driven when CO is the electron donor. FEMS Microbiol Lett. 2004;235:163–167. doi: 10.1016/j.femsle.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 90.Malinen AM, Belogurov GA, Baykov AA, Lahti R. Na+-pyrophosphatase: a novel primary sodium pump. Biochemistry. 2007;46:8872–8878. doi: 10.1021/bi700564b. [DOI] [PubMed] [Google Scholar]
- 91.Belogurov GA, Malinen AM, Turkina MV, Jalonen U, Rytkonen K, Baykov AA, Lahti R. Membrane-bound pyrophosphatase of Thermotoga maritima requires sodium for activity. Biochemistry. 2005;44:2088–2096. doi: 10.1021/bi048429g. [DOI] [PubMed] [Google Scholar]
- 92.Saier MH., Jr Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- 93.Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boyer PD. Bioenergetic coupling to protonmotive force: should we be considering hydronium ion coordination and not group protonation? Trends Biochem Sci. 1988;13:5–7. doi: 10.1016/0968-0004(88)90005-9. [DOI] [PubMed] [Google Scholar]
- 95.Bühl M, Ludwig R, Schurhammer R, Wipff G. Hydronium ion complex of 18-crown-6: Theory confirms three "normal" linear hydrogen bonds. J Phys Chem A. 2004;108:11463–11468. [Google Scholar]
- 96.Deamer DW. Proton permeation of lipid bilayers. J Bioenerg Biomembr. 1987;19:457–479. doi: 10.1007/BF00770030. [DOI] [PubMed] [Google Scholar]
- 97.Nagle JF. Theory of passive proton conductance in lipid bilayers. J Bioenerg Biomembr. 1987;19:413–426. doi: 10.1007/BF00770027. [DOI] [PubMed] [Google Scholar]
- 98.Paula S, Volkov AG, Van Hoek AN, Haines TH, Deamer DW. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys J. 1996;70:339–348. doi: 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tepper HL, Voth GA. Mechanisms of passive ion permeation through lipid bilayers: insights from simulations. J Phys Chem B. 2006;110:21327–21337. doi: 10.1021/jp064192h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krulwich TA, Guffanti AA. The Na+ cycle of extreme alkalophiles: a secondary Na+/H+ antiporter and Na+/solute symporters. J Bioenerg Biomembr. 1989;21:663–677. doi: 10.1007/BF00762685. [DOI] [PubMed] [Google Scholar]
- 101.Krulwich TA, Ito M, Gilmour R, Hicks DB, Guffanti AA. Energetics of alkaliphilic Bacillus species: physiology and molecules. Adv Microb Physiol. 1998;40:401–438. doi: 10.1016/s0065-2911(08)60136-8. [DOI] [PubMed] [Google Scholar]
- 102.Krulwich TA, Ito M, Guffanti AA. The Na+-dependence of alkaliphily in Bacillus. Biochim Biophys Acta. 2001;1505:158–168. doi: 10.1016/s0005-2728(00)00285-1. [DOI] [PubMed] [Google Scholar]
- 103.Hoffmann A, Dimroth P. The electrochemical proton potential of Bacillus alcalophilus. Eur J Biochem. 1991;201:467–473. doi: 10.1111/j.1432-1033.1991.tb16304.x. [DOI] [PubMed] [Google Scholar]
- 104.Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Williams RJP. The multifarious couplings of energy transduction. Biochim Biophys Acta. 1978;505:1–44. doi: 10.1016/0304-4173(78)90007-1. [DOI] [PubMed] [Google Scholar]
- 106.Rottenberg H. Phase transitions and coupling in energy transducing membranes. FEBS Lett. 1978;94:295–297. doi: 10.1016/0014-5793(78)80960-0. [DOI] [PubMed] [Google Scholar]
- 107.Kell DB. On the functional proton current pathway of electron transport phosphorylation. An electrodic view. Biochim Biophys Acta. 1979;549:55–99. doi: 10.1016/0304-4173(79)90018-1. [DOI] [PubMed] [Google Scholar]
- 108.Cherepanov DA, Feniouk BA, Junge W, Mulkidjanian AY. Low dielectric permittivity of water at the membrane interface: effect on the energy coupling mechanism in biological membranes. Biophys J. 2003;85:1307–1316. doi: 10.1016/S0006-3495(03)74565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cherepanov DA, Junge W, Mulkidjanian AY. Proton transfer dynamics at the membrane/water interface: dependence on the fixed and mobile pH buffers, on the size and form of membrane particles, and on the interfacial potential barrier. Biophys J. 2004;86:665–680. doi: 10.1016/S0006-3495(04)74146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams RJP. Possible functions of chains of catalysts. J Theor Biol. 1961;1:1–17. doi: 10.1016/0022-5193(61)90023-6. [DOI] [PubMed] [Google Scholar]
- 111.Liu X, Gong X, Hicks DB, Krulwich TA, Yu L, Yu CA. Interaction between cytochrome caa3 and F1F0-ATP synthase of alkaliphilic Bacillus pseudofirmus OF4 is demonstrated by saturation transfer electron paramagnetic resonance and differential scanning calorimetry assays. Biochemistry. 2007;46:306–313. doi: 10.1021/bi0619167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Z, Hicks DB, Guffanti AA, Baldwin K, Krulwich TA. Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and non-fermentative growth at pH 10.5. J Biol Chem. 2004;279:26546–26554. doi: 10.1074/jbc.M401206200. [DOI] [PubMed] [Google Scholar]
- 113.Pisa KY, Huber H, Thomm M, Müller V. A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 2007;274:3928–3938. doi: 10.1111/j.1742-4658.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- 114.van de Vossenberg JLCM, Driessen AJM, da Costa MS, Konings WN. Homeostasis of the membrane proton permeability in Bacillus subtilis grown at different temperatures. Biochim Biophys Acta. 1999;1419:97–104. doi: 10.1016/s0005-2736(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 115.Albers SV, van de Vossenberg JLCM, Driessen AJM, Konings WN. Adaptations of the archaeal cell membrane to heat stress. Front Biosci. 2000;5:D813–820. doi: 10.2741/albers. [DOI] [PubMed] [Google Scholar]
- 116.Sydow U, Wohland P, Wolke I, Cypionka H. Bioenergetics of the alkaliphilic sulfate-reducing bacterium Desulfonatronovibrio hydrogenovorans. Microbiology. 2002;148:853–860. doi: 10.1099/00221287-148-3-853. [DOI] [PubMed] [Google Scholar]
- 117.De Vrij W, Bulthuis RA, Konings WN. Comparative study of energy-transducing properties of cytoplasmic membranes from mesophilic and thermophilic Bacillus species. J Bacteriol. 1988;170:2359–2366. doi: 10.1128/jb.170.5.2359-2366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guffanti AA, Krulwich TA. Features of apparent nonchemiosmotic energization of oxidative phosphorylation by alkaliphilic Bacillus firmus OF4. J Biol Chem. 1992;267:9580–9588. [PubMed] [Google Scholar]
- 119.Häse CC. Virulence and sodium bioenergetics. Trends Microbiol. 2000;8:490–491. doi: 10.1016/s0966-842x(00)01870-9. [DOI] [PubMed] [Google Scholar]
- 120.Huq A, West PA, Small EB, Huq MI, Colwell RR. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar 01 associated with live copepods in laboratory microcosms. Appl Environ Microbiol. 1984;48:420–424. doi: 10.1128/aem.48.2.420-424.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bakeeva LE, Chumakov KM, Drachev AL, Metlina AL, Skulachev VP. The sodium cycle. III. Vibrio alginolyticus resembles Vibrio cholerae and some other vibriones by flagellar motor and ribosomal 5S RNA structures. Biochim Biophys Acta. 1986;850:466–472. doi: 10.1016/0005-2728(86)90115-5. [DOI] [PubMed] [Google Scholar]
- 122.Dibrov P, Dibrov E, Pierce GN, Galperin MY. Salt in the wound: a possible role of Na+ gradient in chlamydial infection. J Mol Microbiol Biotechnol. 2004;8:1–6. doi: 10.1159/000082075. [DOI] [PubMed] [Google Scholar]
- 123.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]



