Abstract
Selenium is known for its antioxidant properties, making selenoproteins candidate molecules for mitigation of neurological disorders in which oxidative stress has been implicated. The selenium transport protein, selenoprotein P, is essential for neuronal survival and function. We sought to determine whether selenoprotein P expression is associated with Alzheimer’s disease pathology. We examined postmortem tissue from individuals with the hallmark lesions of Alzheimer’s disease and individuals without these lesions. Selenoprotein P immunoreactivity was co-localized with amyloid-β plaques and neurofibrillary tangles. Dense-core and other non-diffuse amyloid-β plaques were nearly always associated with selenoprotein P immunopositive cells. Analysis of spatial distribution showed a significant association between amyloid-β plaques and selenoprotein P. Numerous cells also exhibited immunoreactivity to selenoprotein P and intraneuronal neurofibrillary tangles. Confocal microscopy confirmed co-localization of amyloid-β protein and selenoprotein P. These findings suggest an association of selenoprotein P with Alzheimer’s pathology.
Keywords: Alzheimer’s disease, amyloid-β, antioxidant, human cortex, neurofibrillary tangle, oxidative stress, selenoprotein, selenoprotein P
INTRODUCTION
Neurons are highly vulnerable to damage from oxidative stress due to the high-oxygen usage for energy metabolism and presence of metals such as iron and zinc [20]. Chronic oxidative damage has been strongly implicated in Alzheimer’s disease (AD) and other neurodegenerative diseases. Selenoproteins, which contain the element selenium (Se), decrease oxidative stress [3].
Se is distributed throughout the body via plasma selenoprotein P (SelP) [25]. Although originally identified as a plasma protein, SelP is abundant in neurons and ependymal cells in the human brain [22], and is a survival-promoting factor for cultured neurons [33]. Mice with a deletion of the SelP gene have deficiencies in hippocampal synaptic function and are deficient in memory tasks and longterm potentiation, a cellular model for learning and memory [19]. SelP chelates zinc, mercury and other metals and binds toxic metals in sera [8]. SelP and several important antioxidant selenoproteins are expressed in brain, and may help mitigate neurodegeneration, although specific roles for many of these proteins have not been elucidated [8].
Expression of SelP in human brain increases with aging[16]. A recent comprehensive analysis of gene expression in AD demonstrated that SelP was among a subset of genes with increased expression in AD, even after taking into account the age-dependent increase [17]. To understand the role of SelP in AD, it is important to know if the protein is co-localized with lesions associated with the disorder. Here we investigate the expression of SelP in postmortem brain tissue from decedents in whose neocortices high levels of Alzheimer lesions [amyloid-β (Aβ) plaques and neurofibrillary tangles (NFT)] were observed, and an equal number of individuals without neorcortical Aβ plaques or NFT.
MATERIALS AND METHODS
Formalin-fixed human brain tissue was provided by the Honolulu Asia Aging Study (HAAS), an ongoing project that has monitored the health and lifestyle of Japanese-American men born between 1900 and 1919 and residing on Oahu, Hawaii [32]. Studies were approved by the Internal Review Board of Kuakini Medical Center. Sections (8 μm) of medial temporal gyrus (MT) from four decedents with abundant AD lesions and four individuals without significant lesions were used in this study. For simplicity we refer to those brains with abundant Alzheimer lesions as ‘AD brains’ and those without such lesions as ‘normal brains’.
To test the SelP antibody, recombinant human SelP was expressed in HEK-293 cells and purified from culture media using a nickel-agarose column as previously described [29]. Recombinant SelP, media samples from non-transfected HEK-293 cells, and human plasma samples were analyzed by western blot. Protein was diluted with reduced Laemmeli buffer, boiled at 95°C for 10 min, cooled on ice, and loaded into wells of 10–14.5% polyacrylamide gels (Bio-Rad). Protein was transferred to PVDF membranes, which were blocked for 1 h with 5% yeast extract, and then probed for 90 min with primary anti-SelP antibody diluted 1:2,000 (Lab Frontier) followed by treatment with an anti-mouse HRP-conjugated secondary antibody (GE Healthcare) diluted 1:10,000, and detected using ECL Plus (GE Healthcare).
Cross-linked antigens in deparaffinized sections were “unmasked” by immersing in an alkaline solution containing EDTA (Trilogy, Cell Marque), heating at 95°C and 15 psi in a pressure cooker for 20 min, and then immersing in 90% formic acid for 3 min. Tissue was blocked in PBS with 5% serum species-matched to secondary antibody, and incubated with mouse monoclonal antibodies recognizing human SelP (LabFrontier, diluted 1:100), followed by incubation with biotinylated anti-mouse secondary antibody and ABCTM reagent (Vector Laboratories), and development with 3,4-diaminobenzamidine hydrochloride (DAB, Vector Laboratories).
For double labeling with Aβ, tissue was subsequently blocked in 5% normal horse serum (NHS), followed with separate blocking steps in streptavidin and biotin solutions (from ABC kit) for five minutes each. Tissue was incubated in a mouse monoclonal anti-Aβ antibody (Chemicon, 1:200) followed by secondary antibody and ABC. To distinguish between the two antigens, Aβ immunoreactivity was developed with DAB containing nickel chloride (DAB-Ni, Vector Labs). For double labeling of SelP with NFT, tissue was labeled for SelP as above and subsequently incubated with a rabbit polyclonal anti-NFT antibody (produced using SDS-solubilized protein from AD brain, Chemicon, 1:200). Following secondary antibody application, an alkaline phosphatase-based ABC kit was used to visualize immunoreactivity (Vector Laboratories).
For double labeling of SelP and Aβ for confocal microscopy, tissue was incubated in SelP antibody, then blocked with rabbit anti-mouse IgG FAb fragment (Jackson ImmunoResearch Laboratories) diluted 1:100 in 5% NHS prior to secondary antibody incubation. For double labeling of SelP and NFT, primary antibodies were incubated together. Secondary antibodies labeled with Alexafluor 488 (for green label) or Alexafluor 596 (red label) were used to visualize immunoreactivity. Endogenous fluorescence was reduced by treating with an autofluorescence eliminator reagent (Chemicon). Tissue was mounted in Vectashield containing DAPI to label nuclei (Vector Laboratories). For all double labeling, controls excluding each of the primary antibodies confirmed absence of cross-reactivity.
Sampling of areas for analysis was performed by placing a grid of 1 mm × 1 mm squares randomly over tissue slides, and taking images of every 5th square in every 5th row. Analysis of spatial association was achieved using the software Proxan (proximity analysis), which is available online from Bendor Research at www.bendor.com.au/Proxan. A description of the algorithms used by this software can be found at this site. Proxan uses cluster analysis to determine the degree of spatial association between ‘active’ and ‘reference’ objects with distinct labels in an image [9]. In our analysis, SelP-immunopositive cells were paired with the nearest ‘reference’ Aβ plaque, and Aβ plaques were paired with the nearest SelP-positive cell and any labeled cells not closer to another plaque. Proxan compares the minimum Euclidian distances between the two labels to the minimum distances between the reference objects and randomly redistributed active objects using a χ2 test. If the different labeled objects are associated spatially, then the distribution of minimum distances between these objects will be smaller than those of random objects.
Unbiased stereological estimates of cell numbers were achieved using the dissector method [28]. Briefly, randomly-sampled images from adjacent sections were aligned, and the number of labeled cells in the ‘reference’ section was determined after excluding cells that could also be found in the ‘look-up’ section.
RESULTS
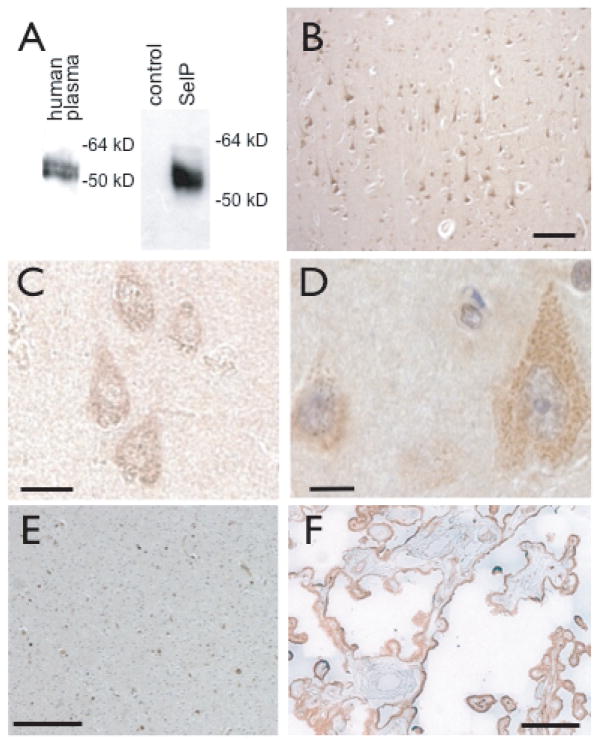
We tested a monoclonal SelP antibody (Labfrontier) for specificity by western blot. The antibody recognized a doublet of 55–60 kD in human plasma (Fig. 1A, left), representing different glycosylated forms previously described. The antibody also recognized a band of about 55 kD in media from HEK-293 cells transfected with a plasmid containing human SelP, but not in media from untransfected HEK-293 cells (Fig. 1A, right).
Fig. 1.
SelP immunoreactivity in human medial temporal gyrus and association with amyloid-β deposition. A: Specificity of SelP antibody. Anti-SelP recognizes a doublet of 55–60 kD in human plasma (left) and a large band of approximately 55 kD in media from HEK-293 cells expressing a recombinant SelP (right, right lane), but not in media from non-transfected cells (right, left lane). B–D: SelP expression in cortex of non-AD brain. B: SelP expression in neurons in upper layers of cortex (brown staining). C, D: SelP expression in cortical neurons. E: SelP in white matter and F: SelP in choroid plexus. Scale bars: B, E, F 100 μm, C, D, 10 μm.
We examined SelP expression in sections of medial temporal gyrus collected postmortem from human brains. SelP expression was present in neurons throughout the cortical layers (Fig. 1B–D). SelP immunoreactivity marked dendritic fibers in upper layers, particularly in apical dendrites of pyramidal neurons. Granular structures immunoreactive for SelP were detected throughout the cytoplasm of pyramidal cell bodies (D). SelP immunoreactivity was also detected in white matter (E) and in choroid plexus (F), confirming findings of a previous study [22].
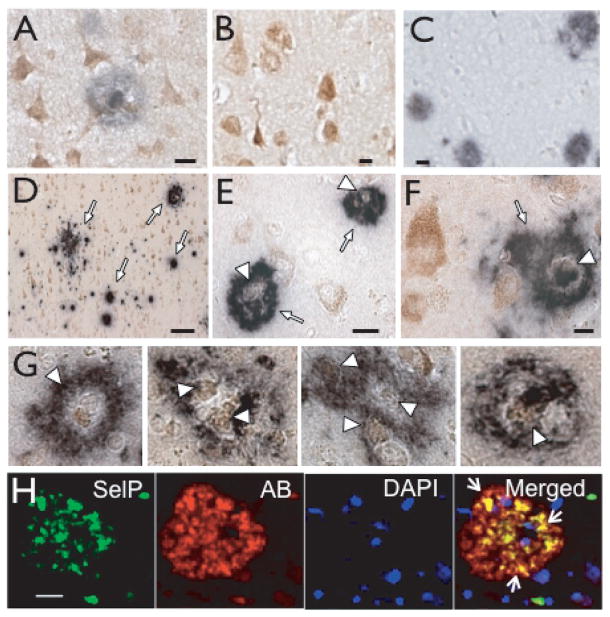
To determine spatial relationship between SelP and deposition of Aβ, tissue was immunostained with antibodies to both SelP and Aβ. As shown in Fig. 2A and 2D–F, the Aβ antibody revealed plaques, a defining pathologic characteristic of AD. We observed marked co-localization of Aβ immunoreactivity with cells expressing SelP. Numerous ring-like plaques throughout the cortex encircled SelP-positive cells (Fig. 2D–F). Controls where either primary antibody was omitted demonstrated that the co localization was not due to cross-reactivity of secondary antibodies (Fig. 2B, C). Non-diffuse Aβ plaques were consistently associated with contiguous SelP-positive cells. Fig. 2G shows several examples from AD cortex demonstrating the association between SelP and Aβ. We also examined the relationship between SelP and Aβ using confocal microscopy. Fig. 2H shows SelP immunoreactivity (green) within or associated with Aβ plaques (red). Nuclei are labeled with DAPI (blue). Yellow indicates the blending of red and green due to the co-localization of SelP with Aβ.
Fig. 2.
SelP immunoreactivity within amyloid plaques. A: Double labeling for SelP and Aβ. B: Negative control for double labeling with Aβ antibody omitted. C: Negative control with SelP antibody omitted. D–F: SelP immunoreactivity (brown) and amyloid plaques (indigo) in AD brain. D: Aβ plaques (white arrows) in upper cortical layers. E–F: Dense-core plaques with SelP-positive cells within and at the border (white arrowheads). G: Enlarged examples of plaques associated with SelP. H: SelP (green, left column) and Aβ (red, second from left). Nuclei are stained with DAPI (second from right). Merged images are shown in right column. Yellow color indicates co-localization of SelP with Aβ (white arrows). Scale bars: D, 50 μm, all others 10 μm.
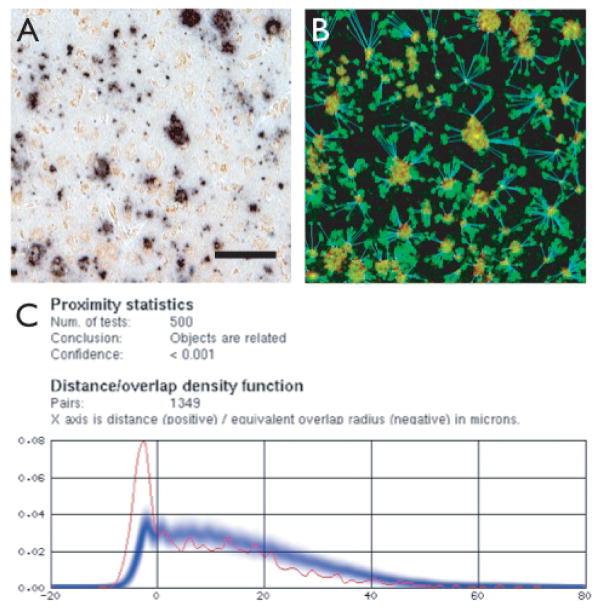
To estimate the probability that the observed association between SelP and Aβ depositions could have occurred by chance, we used a proximity analysis program (Proxan [9]). This software determines the minimum measured distances between each object of a certain class to the nearest object of a reference class. Cluster analyses are then performed on the distribution of these distances by comparison with distributions generated from random assignment of the ‘active’ class of objects with the original locations of the ‘reference’ class of objects. Fig. 3A shows an image before analysis, with minor adjustments to contrast in order to maximize label recognition by the software. Fig. 3B shows the same image after linking each ‘active’ object (SelP-labeled cells) with the nearest ‘reference’ object (Aβ plaques), with the background subtracted and objects identified by color (red for Aβ, green for SelP), and minimum distances calculated and plotted (blue lines). Proxan was able to correctly identify and distinguish the two labels after minor adjustments in saturation and contrast to the original images. No analyses were performed for control subjects as no plaques were apparent in these sections. Similarly, in images from AD subjects where one primary antibody was omitted, the software recognized only the remaining label, and analyses could not be performed. Figure 3C shows the output from analyzing the image in Fig. 3A. The graph indicates the measured minimum distances between the two labels. Since the distance between objects that make contact at their borders is defined as zero, the distance between objects with overlapping borders are defined as the negative value of the radius of overlap. Red shows the measurements between objects in the image, whereas blue indicates the distances between the reference object (in this case Aβ) and randomly distributed SelP-labeled objects. In all tests from AD subjects, we found a significant association between SelP-immunoreactive cells and Aβ plaques.
Fig. 3.
Analysis of spatial distribution of Aβ plaques and SelP-positive cells using Proxan software. A is an image of cortex from an AD subject with contrast enhanced to maximize label identification. B shows the same image after running analysis. Background is filtered, red hatch indicates plaques, green hatch indicates SelP label, blue shows co-localization and minimum distance between SelP cells and closest Aβ plaque. C shows the output from Proxan, showing the number of random object generations, the analysis conclusion and P value, number of Aβ-SelP pairs measured. The graph is a histogram showing the fraction of SelP-Aβ pairs as a function of the minimum distance between the pairs. Red shows the actual measured distances between labeled objects. When objects overlap, the radius of overlap is measured as a negative number. Blue is the average of 500 tests where SelP-labeled objects were randomly redistributed throughout the image. The darkest region indicates the mean, while the width of the blue line indicates ± SEM. All tests of images with both SelP and Aβ labeling indicated a statistically significant association between objects with these two labels. Scale bars: 50 μm.
We measured the SelP-positive numeric cell density in the medial temporal gyrus from AD afflicted individuals and individuals with no evidence of the disease. Using the dissector method to obtain objective cell counts of SelP immunopositive cells in randomly chosen fields from adjacent sections, the numerical density of SelP-positive cells was found to be significantly higher in AD brain compared with non-disease brain (Table 1).
Table 1.
| Control | AD | |
|---|---|---|
| Age range | 79.5 to 90.5 | 76.8 to 95.7 |
| Age at death (YR) | 80.8 ± 3.5 | 87.0 ± 3.9 |
| Post-mortem interval (HR) | 15.3 ± 2.5 | 13.0 ± 5.0 |
| Median (and range) braak stage | 2.5 (2 to 3) | 5 (all) * |
| Neuritic plaques/mm2 | 0 | 6.8 ± 1.2* |
| Diffuse plaques/mm2 | 0.2 ± 0.3 | 6.5 ± 1.2* |
| Tangles/mm2 | 0 | 33.6 ± 3.1* |
| SelP positive cells/mm3 | 11.5 ± 1.3 | 18.5 ± 1.9* |
indicates significantly different from control (p ≤ 0.05)
HAAS subject information obtained at autopsy (methods published previously [32]) and density of SelP-immunopositive cells. All decedents were American-born males of Japanese ancestry living in Hawaii. Data are mean ± SEM unless indicated otherwise.
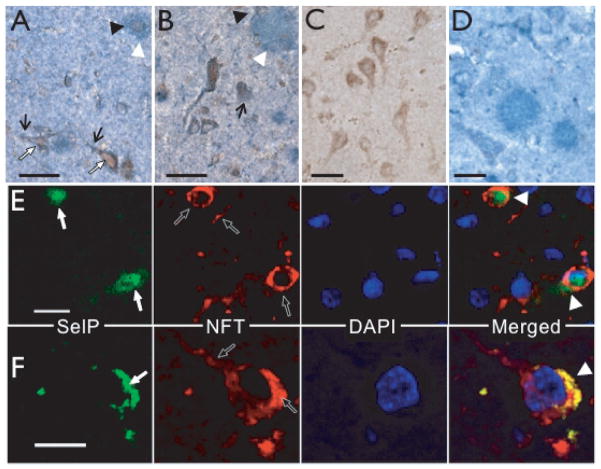
We additionally compared SelP localization with the presence of NFT, another hallmark of AD. Immunoreactivity for NFT clearly correlated with SelP expression (Fig. 4A–B). Controls where either primary antibody was omitted demonstrated specificity of primary antibodies (Fig. 4C, D). Neurons that were immunoreactive with the NFT antibody (white arrows) contained SelP protein in the cell bodies. The NFT antibody also recognized dystrophic neurites (black arrows) that associated with SelP-positive cells (black arrowheads). We examined immunoreactivity with fluorescent-labeled secondary antibodies using confocal microscopy (Fig. 4E, F). SelP (green, white arrows) and NFT (red, black arrows) immunoreactivity are found within the same neuron with some co-localization (yellow, white arrowheads). These data indicate that SelP may directly interact with proteins and other molecules involved in AD pathology.
Fig. 4.
Association of SelP with NFT immunoreactivity. A,B: NFT-immunopositive pyramidal cells with apical dendrites showing abnormal architecture (white arrows) also expressed SelP. The NFT antibody also labeled dystrophic neurites (black arrows) that were associated with SelP-positive cells (black arrowheads). C: Negative control for double labeling with NFT antibody omitted. D: Negative control for double labeling with SelP antibody omitted. E–F: SelP (green, white arrows) and NFT immunoreactivity (red, black arrows), with DAPI-stained nuclei (blue). Merged images on lower right show yellow co-localization of SelP with NFTs (white arrowheads). Scale bars: A–E, 20 μm; C, 10 μm.
DISCUSSION
Our findings have revealed an association of SelP in neuronal cells with markers for AD, indicating a possible role for SelP in the progression or mitigation of the disorder. SelP expression increases with both aging and AD [16,17]. A recent study of gene expression in AD determined that SelP was one of 240 genes upregulated in AD compared to age-matched controls (see supplementary material in[17]). It is thus an important finding that SelP expression is directly associated with the pathology of AD, which suggests a direct involvement in the response to or progression of the disorder.
What could be the function of SelP in AD? SelP is a very unique protein, with 10 selenocysteine residues that require recoding of the UGA stop codon for incorporation [5]. This recoding requires several additional factors and thus synthesis of SelP requires a considerably greater energy investment than other proteins, suggesting it has an important role [12,26]. The strong antioxidant properties and the redox motif of the first selenocysteine suggest a role in relieving oxidative stress [4,8], and SelP has been shown to protect cultured human astrocytes from oxidative stress [27]. SelP has a signal peptide targeting it for secretion, and can bind to heparin residues of glycoproteins on cell membrane surfaces [1,14]. SelP transports Se to the brain and other organs, and is likely to also transport Se within brain [11,18]. Se is required for other selenoproteins such as the glutathione peroxidase family, enzymes that break down hydroperoxydases [8,21]. Thus SelP may have an important role in the global response to oxidative stress, and may be involved in other neurodegenerative disorders as well.
An alternative hypothesis is a potentially harmful effect of SelP being involved in the progression of the disease. Although harmful actions of SelP have not been demonstrated, Se is a toxin at higher doses [31]. Thus increased expression of SelP coupled with proteolysis could result in deposits of organic Se that potentiate the pathology of AD. Although the strong antioxidant and metal chelating properties of SelP suggest a mitigating role for the protein [7], the risk of toxicity and a potential role in pathology also need to be investigated.
Recent studies demonstrate the importance of SelP for normal brain function. Mice with a deletion of the SelP gene have deficiencies in synaptic function in the hippocampus, a region involved in memory, and are deficient in memory tasks and longterm potentiation, a cellular model for learning and memory [19]. These mice develop behavioral abnormalities such as impaired mobility and seizures, and die soon after weaning [10,23]. Normal function can be rescued with a diet supplemented with high levels of Se [24]. Although Se supplemented mice survive and appear normal, they exhibit severe brain stem axonal degeneration [30]. Recent evidence suggests SelP is transported into brain by the apolipoprotein E (ApoE) receptor, ApoER2. Apo-ER2 deficient mice have increased sensitivity to Se deficiency and exhibit many of the same neurological and behavioral deficiencies as SelP knockout mice [6]. The ApoE4 allele is a genetic risk factor for AD [2], and interaction or competition between ApoE4 and SelP could alter selenoprotein transport and expression in individuals with this allele.
Genetic deletion of SelP results in decreased levels of mRNAs encoding selenoproteins in brain, most notably glutathione peroxidase 4, and selenoproteins H, M and W [13]. Selenoprotein M has been implicated in AD through microarray analysis of a mouse model that overexpresses a mutated form of presenilin-2 [15]. Thus, the increased SelP associated with AD lesions may reflect a defensive response to local oxidative stress to provide Se for production of selenoprotein M and other selenoproteins.
Our findings indicate a strong link between AD and SelP expression. SelP may act directly as an antioxidant to prevent oxidative stress, or may provide Se for biosynthesis of other antioxidant selenoproteins. Further investigation of this connection will help us to understand the progression of this disorder, and may lead to improved therapies and treatments.
Acknowledgments
We thank Kristen Ewell for tissue sectioning, Jeffrey Squires for in vitro SelP expression, Andrea Takemoto and Fernando Liquido for assistance, and Dr. Linda Chang for helpful suggestions. Supported by NIH R01 NS40302 to MJB, Hawaii Community Foundation Grant 20061490 to FPB, NIH U01 AG019349 to LRW, and NIH G12 RR003061 for the JABSOM Microscopy and Imaging core facility.
References
- 1.Arteel GE, Franken S, Kappler J, Sies H. Binding of selenoprotein P to heparin: characterization with surface plasmon resonance. Biol Chem. 2000;381:265–268. doi: 10.1515/BC.2000.034. [DOI] [PubMed] [Google Scholar]
- 2.Bennett DA. Part I. Epidemiology and public health impact of Alzheimer’s disease. Dis Mon. 2000;46:657–665. doi: 10.1016/s0011-5029(00)90028-2. [DOI] [PubMed] [Google Scholar]
- 3.Burk RF. Selenium, an antioxidant nutrient. Nutr Clin Care. 2002;5:75–79. doi: 10.1046/j.1523-5408.2002.00006.x. [DOI] [PubMed] [Google Scholar]
- 4.Burk RF, Hill KE, Motley AK. Selenoprotein metabolism and function: evidence for more than one function for selenoprotein P. J Nutr. 2003;133:1517S–1520S. doi: 10.1093/jn/133.5.1517S. [DOI] [PubMed] [Google Scholar]
- 5.Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 6.Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, Austin LM. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J Neurosci. 2007;27:6207–6211. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Yu H, Zhao J, Li B, Qu L, Liu S, Zhang P, Chai Z. The roles of serum selenium and selenoproteins on mercury toxicity in environmental and occupational exposure. Environ Health Perspect. 2006;114:297–301. doi: 10.1289/ehp.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Berry MJ. Selenium and selenoproteins in the brain and brain diseases. J Neurochem. 2003;86:1–12. doi: 10.1046/j.1471-4159.2003.01854.x. [DOI] [PubMed] [Google Scholar]
- 9.Cullen KM, Kocsi Z, Stone J. Pericapillary haem-rich deposits: evidence for microhaemorrhages in aging human cerebral cortex. J Cereb Blood Flow Metab. 2005;25:1656–1667. doi: 10.1038/sj.jcbfm.9600155. [DOI] [PubMed] [Google Scholar]
- 10.Hill KE, Zhou J, McMahan WJ, Motley AK, Burk RF. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J Nutr. 2004;134:157–161. doi: 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- 11.Hill KE, Zhou J, Austin LM, Motley AK, Ham AJ, Olson GE, Atkins JF, Gesteland RF, Burk RF. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J Biol Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann PR, Berry MJ. Selenoprotein synthesis: a unique translational mechanism used by a diverse family of proteins. Thyroid. 2005;15:769–775. doi: 10.1089/thy.2005.15.769. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann PR, Hoge SC, Li PA, Hoffmann FW, Hashimoto AC, Berry MJ. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hondal RJ, Ma S, Caprioli RM, Hill KE, Burk RF. Heparin-binding histidine and lysine residues of rat selenoprotein P. J Biol Chem. 2001;276:15823–15831. doi: 10.1074/jbc.M010405200. [DOI] [PubMed] [Google Scholar]
- 15.Hwang DY, Cho JS, Oh JH, Shim SB, Jee SW, Lee SH, Seo SJ, Lee SK, Lee SH, Kim YK. Differentially expressed genes in transgenic mice carrying human mutant presenilin-2 (N141I): correlation of selenoprotein M with Alzheimer’s disease. Neurochem Res. 2005;30:1009–1019. doi: 10.1007/s11064-005-6787-6. [DOI] [PubMed] [Google Scholar]
- 16.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 17.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama A, Hill KE, Austin LM, Motley AK, Burk RF. All regions of mouse brain are dependent on selenoprotein P for maintenance of selenium. J Nutr. 2007;137:690–693. doi: 10.1093/jn/137.3.690. [DOI] [PubMed] [Google Scholar]
- 19.Peters MM, Hill KE, Burk RF, Weeber EJ. Altered hippocampus synaptic function in selenoprotein P deficient mice. Mol Neurodegener. 2006;1:12. doi: 10.1186/1750-1326-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen RB, Nunomura A, Lee HG, Casadesus G, Perry G, Smith MA, Zhu X. Signal transduction cascades associated with oxidative stress in Alzheimer’s disease. J Alzheimers Dis. 2007;11:143–152. doi: 10.3233/jad-2007-11202. [DOI] [PubMed] [Google Scholar]
- 21.Sahin E, Gumuslu S. Stress-dependent induction of protein oxidation, lipid peroxidation and anti-oxidants in peripheral tissues of rats: comparison of three stress models (immobilization. cold and immobilization-cold) Clin Exp Pharmacol Physiol. 2007;34:425–431. doi: 10.1111/j.1440-1681.2007.04584.x. [DOI] [PubMed] [Google Scholar]
- 22.Scharpf M, Schweizer U, Arzberger T, Roggendorf W, Schomburg L, Kohrle J. Neuronal and ependymal expression of selenoprotein P in the human brain. J Neural Transm. 2007;114:877–884. doi: 10.1007/s00702-006-0617-0. [DOI] [PubMed] [Google Scholar]
- 23.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweizer U, Michaelis M, Kohrle J, Schom-burg L. Efficient selenium transfer from mother to offspring in selenoprotein-P-deficient mice enables dose-dependent rescue of phenotypes associated with selenium deficiency. Biochem J. 2004;378:21–26. doi: 10.1042/BJ20031795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweizer U, Streckfuss F, Pelt P, Carlson BA, Hatfield DL, Kohrle J, Schomburg L. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 27.Steinbrenner H, Alili L, Bilgic E, Sies H, Brenneisen P. Involvement of selenoprotein P in protection of human astrocytes from oxidative damage. Free Radic Biol Med. 2006;40:1513–1523. doi: 10.1016/j.freeradbiomed.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Sterio DC. The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc. 1984;134:127–136. doi: 10.1111/j.1365-2818.1984.tb02501.x. [DOI] [PubMed] [Google Scholar]
- 29.Tujebajeva RM, Harney JW, Berry MJ. Selenoprotein P expression, purification, and immunochemical characterization. J Biol Chem. 2000;275:6288–6294. doi: 10.1074/jbc.275.9.6288. [DOI] [PubMed] [Google Scholar]
- 30.Valentine WM, Hill KE, Austin LM, Valentine HL, Goldowitz D, Burk RF. Brainstem axonal degeneration in mice with deletion of selenoprotein p. Toxicol Pathol. 2005;33:570–576. doi: 10.1080/01926230500243045. [DOI] [PubMed] [Google Scholar]
- 31.Whanger PD. Selenium and the brain: a review. Nutr Neurosci. 2001;4:81–97. doi: 10.1080/1028415x.2001.11747353. [DOI] [PubMed] [Google Scholar]
- 32.White L, Petrovitch H, Ross GW, Masaki KH, Abbott RD, Teng EL, Rodriguez BL, Blanchette PL, Havlik RJ, Wergowske G, Chiu D, Foley DJ, Murdaugh C, Curb JD. Prevalence of dementia in older Japanese-American men in Hawaii: The Honolulu-Asia Aging Study. Jama. 1996;276:955–960. [PubMed] [Google Scholar]
- 33.Yan J, Barrett JN. Purification from bovine serum of a survival-promoting factor for cultured central neurons and its identification as selenoprotein-P. J Neurosci. 1998;18:8682–8691. doi: 10.1523/JNEUROSCI.18-21-08682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]