Abstract
Anoctamin (ANO) family of proteins, consisting of 10 members in mammals, are transmembrane proteins that have Ca2+-activated Cl- channel activities. Transmembrane channel-like (TMC) family of proteins, consisting of 8 members in mammals, are also transmembrane proteins of which mutations are implicated in various human conditions, such as hearing loss and epidermodysplasia verruciformis. Here we show that ANO and TMC proteins share high sequence similarity and probably the same membrane topology, indicating that these proteins are evolutionarily related. We found many conserved amino acid residues between the two families of proteins, especially in regions spanning the transmembrane domains TM1, TM4-TM5, and TM6-TM7. These findings imply that these proteins form one large family, which we term ANO/TMC superfamily and that TMC proteins may also function as channels for Cl- or possibly other ions. The ANO/TMC superfamily proteins are present in almost all the diverse groups of eukaryotic organisms, suggesting that the proteins function in important biological processes, such as ion homeostasis, in eukaryotic cells.
Keywords: anoctamin, transmembrane channel-like, ion channel, membrane topology
Introduction
The anoctamin (ANO, also known as TMEM16) family of membrane proteins consists of 10 members (ANO1-10) in mammals and are implicated in various cancers and diseases (1). ANO1 (also known as TMEM16A or DOG1) has recently been identified to have a Ca2+-activated Cl- channel activity (2-4), suggesting that other members of this family are Cl- channels (1). ANO1 is highly upregulated in gastrointestinal stromal tumor (5,6) and is amplified in oral (7) and head and neck squamous cell carcinomas (8). Disruption of mouse Ano1 caused abnormal development of the trachea, indicating that the gene is a regulator of epithelial and smooth muscle cell organization in murine development (9). ANO5 (also known as GDD1 or TMEM16E) mutations in humans cause gnathodiaphyseal dysplasia (GDD), a rare skeletal syndrome characterized by bone fragility, sclerosis of tubular bones, and cemento-osseous lesions of the jawbone (10). ANO7 (also known as TMEM16G or NGEP) is selectively expressed in normal and cancerous prostates and mediates cell-cell aggregation (11,12). Axs, an ANO homolog in fruit fly (Drosophila melaogaster), is a membrane protein that colocalizes with components of the endoplasmic reticulum (ER) and is present within a structure ensheathing the meiotic spindle (13). Ist2p, a plasma membrane protein of Baker’s yeast (Saccharomyces cerevisiae) homologous to animal ANO, plays a role in salt balance and transport (14). An extensive analysis of the topology of human ANO7 showed that ANO proteins consist of 8 transmembrane domains with both the N- and C-termini intracellular and with a re-entrant loop between TM5 and 6 (15).
The transmembrane channel-like (TMC) family of proteins contains 8 members (TMC1-8) of which mutations are associated with many human disorders (16,17). Although the gene name implies that these proteins might be ion channels, there is no direct evidence for their channel activities and their molecular function remains unknown. TMC1 protein is present in cochlear hair cells and its mutations are associated with hearing loss (18,19). TMC6 (also known as EVER1) and TMC8 (also known as EVER2) are involved in epidermodysplasia verruciformis (EV) which is a genodermatosis associated with a high risk of skin cancers that results from a selective susceptibility to specific human papillomaviruses (HPVs) (20). TMC6 and 8 have been proposed to regulate Zn2+ balance (21,22). TMC proteins were proposed to have 8 transmembrane domains with both the N- and C-termini intracellular and a re-entrant loop (16).
In this report, we show that ANO and TMC families of proteins share sequence similarities and probably the same membrane topology, suggesting that they are evolutionarily related, possibly having diverged during eukaryotic evolution. We found highly conserved regions encompassing transmembrane domains TM1, TM4-TM5, and TM6-TM7. Some amino acid residues are identical in all members of the two families. The homology implies that TMC proteins also function as Cl- or other ion channels.
Materials and Methods
Sequence analyses
Multiple sequence alignments of human ANO and TMC proteins were generated by using MUSCLE obtained from http://www.drive5.com/muscle/ (23), ClustalX2 downloaded from http://www.clustal.org/ (24), and T-Coffee available at http://www.tcoffee.org/ (25). The consensus-based coloring of the multiple alignments was performed by using the CHROMA program downloaded from http://www.llew.org.uk/chroma/ (26). The Sequence Logo representations (27) of the alignments for the selected conserved regions were produced by using the WebLogo server available at http://weblogo.berkeley.edu/ (28). Transmembrane domain predictions were performed by using the TMHMM 2.0 server at http://www.cbs.dtu.dk/services/TMHMM/ (29).
Identification of ANO and TMC homologs
Conventional BLAST and PSI-BLAST at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/) were used to search non-redundant protein sequence database for ANO or TMC protein homologs in proteomes of various organisms. The HMMER version 2.3.2 software package (http://hmmer.janelia.org/) was used to build profile hidden Markov models (HMMs) (30) for the conserved regions and to search the non-redundant protein sequence database for similar sequence matches.
Results and Discussion
ANO and TMC proteins have conserved sequences and form a superfamily
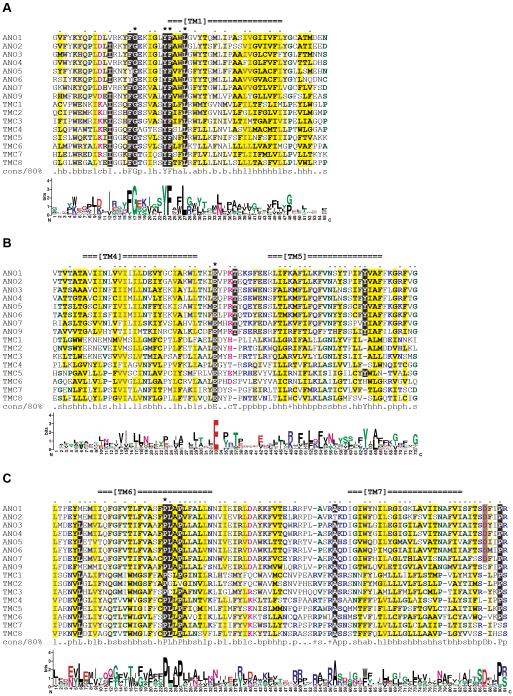
Previously, we identified a human membrane protein ANO7 (reported as NGEP) which was detected only in prostate cancer and normal prostate (11). We noted that TMC proteins (16) possessed the same number of transmembrane domains and a similar proposed membrane topology compared to ANO proteins although conventional BLAST or PSI-BLAST searches using ANO family members as queries failed to detect TMC proteins, and vice versa. To test whether ANO and TMC proteins are actually related, we performed comprehensive pairwise and multiple sequence comparisons of 10 ANO proteins and 8 TMC proteins from human. The proteins from the two families showed overall alignment over the entire length of the sequences. Especially, the regions encompassing transmembrane domains TM1, TM4-TM5, and TM6-TM7 exhibited high sequence similarities among the ANO and TMC family members (Fig. 1). The segments showing sequence similarities were extracted from 8 ANO and TMC proteins of each and were subjected to multiple alignment generation. ANO8 and 10 were excluded from the multiple alignment because they are most divergent compared to other members. A position is considered conserved if a single identical amino acid residue occurs in 80% or more, that is, 13 or more sequences out of the 16 proteins examined.
Figure 1.
Conserved regions of ANO and TMC proteins. ANO and TMC proteins show sequence similarity in regions spanning TM1 (A), TM4-TM5 (B), and TM6-TM7 (C). The multiple alignments were generated from 8 ANO proteins and 8 TMC proteins by using MUSCLE and colored by using CHROMA (see Table I for database accession numbers and coordinates of the aligned regions). The transmembrane domain positions of the human ANO1 predicted by TMHMM are indicated at the top. Asterisks (*) and dots (.) above the alignment mark identical and conserved residues, respectively. The Sequence Logo representation, generated using WebLogo, is presented beneath each alignment block. The 80% consensus (‘cons/80%’) abbreviations are: +, positive (HKR, blue lettering on a white background); a, aromatic (FHWY, blue on dark yellow); b, big (EFHIKLMQRWY, blue on light yellow); c, charged (DEHKR, pink on white); h, hydrophobic (ACFGHILMTVWY, black on dark yellow); l, aliphatic (ILV, grey on dark yellow); p, polar (CDEHKNQRST, light blue on white); and s, small (ACDGNPSTV, dark green on white).
The TM1 region is the most conserved segment and contains several identical residues (Fig. 1A). A highly conserved pattern, [IV]-x(3)-[FY]-G-x(5)-Y-F-x(2)-L, was inferred from the multiple alignment. The pattern is located in the intracellular juxtamembrane region and the intracellular end of TM1. All the human ANO and TMC proteins exhibit this pattern, suggesting that this segment plays a crucial role in the molecular function of both ANO and TMC proteins.
The region around the TM5 and 6 also shows sequence conservation among ANO and TMC proteins (Fig. 1B). The transmembrane segments TM5 and 6 are rich in hydrophobic residues and are linked by a 13- or 14-amino acid-long loop. In the middle of this cytoplasmic loop, all the ANO and TMC proteins have a conserved glutamic acid (E) residue.
The third conserved segment is present around the TM6 and 7 (Fig. 1C). There are two distinctively conserved proline (P) residues spaced by two amino acids, that is, P-x(2)-P, in the middle of the TM6. Most of ANO proteins (8 out of 10) have the sequence PLAP, whereas majority of TMC proteins (5 out of 8) have PLLP at the corresponding region. Leucine (L) residue is present in the extracellular part of the juxtamembrane region of the TM6 in 13 proteins. The TM7 is flanked by conserved alanine (A) residue at the cytoplasmic side and proline (P) residue at the extracellular side.
ANO and TMC proteins may share the same membrane topology
Transmebrane domain analyses predicted that ANO proteins have 9 hydrophobic helices (15). A conserved highly probable N-linked glycosylation site was found within the loop right before the last (9th) hydrophobic helix, indicating that the loop is outside and the C-terminus is inside. Because ANO proteins do not have signal peptides, it is highly likely that the N-terminus is also intracellular. Both the N and C-termini being intracellular, we deduced that ANO proteins should have an even number of transmembrane domains and either the 6th or the 7th helix is a re-entrant loop, a membrane-penetrating region that enters and exits the membrane on the same side (31). Based on this information, we extensively analyzed the topology of human ANO7 by inserting epitope tags into predicted intra- and extracellular loops (15). These studies confirm that it is the 6th hydrophobic helix that forms a re-entrant region between the TM5 (the 5th helix) and the TM6 (the 7th helix) and that human ANO7 has 8 transmembrane domains as predicted. It is highly likely that all of ANO proteins share this topology (2).
TMC proteins are also predicted to have 9 hydrophobic helices. The proposed membrane topology of TMC has 8 transmembrane domains and the 7th helix as a re-entrant loop (16). However, because the ANO and TMC proteins have the same number of transmembrane domains and show high sequence similarity between most of the transmembrane domains, particularly TM6, we consider it probable that TMC proteins actually have the same membrane topology as ANO proteins.
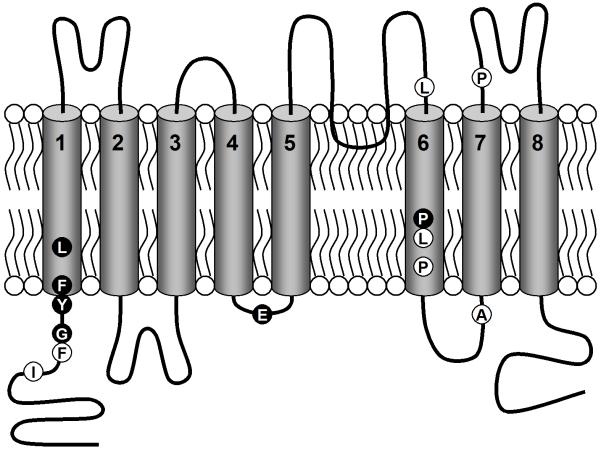
A model of the membrane topology of the ANO/TMC superfamily of proteins is depicted in Fig. 2, in which the approximate locations of the conserved residues are marked. We expect that these conserved amino acids in and around the transmembrane domains play an important role in the molecular function of the ANO/TMC superfamily members.
Figure 2.
Proposed membrane topology of ANO and TMC proteins. A model of the membrane topology probably shared by both the ANO and TMC proteins is depicted. The 8 transmembrane domains are represented as barrels embedded in the membrane. A loop dipping into the membrane between TM5 and 6 is the re-entrant loop. Highly conserved residues identified in the multiple alignments in Fig. 1 are presented as balls, white lettering on a black background, identical in all the sequences; black on white, identical in 13 or more.
TMC proteins may have ion channel activity
The ANO1 was recently identified as Ca2+-activated Cl- channels (2,3,9), suggesting that all the members of the ANO protein family may have Cl- channel activity (1). TMC proteins share the same membrane topology and show sequence conservations in and around the transmembrane domains with ANO proteins. Re-entrant loops are most commonly found in channel proteins forming the pore regions (31). There are some dominant negative mutations found in the ANO/TMC proteins: for examples, human ANO5 (10), Drosophila Axs (13), human TMC1 (20), and mouse Tmc1 (19). Often, mutations in channel proteins exhibit dominant negative effects (32). Taken together, this evidence suggests that the TMC proteins also have channel activity. TMC proteins could be Ca2+-activated Cl- channels as ANO proteins are. Recently, TMC6 and 8 proteins were proposed to be involved in Zn2+ balance (21, 22). Therefore, it is possible that TMC proteins directly participate in Zn2+ transport and hence are Zn2+ channels. Alternatively, TMC proteins could be Zn2+-modulated Cl- or other anion channels as ANO proteins are Cl--channels activated by Ca2+.
ANO/TMC proteins are present in diverse eukaryotic organisms
Sequence similarity and shared membrane topology clearly show that human ANO and TMC proteins are evolutionarily related. We searched the proteomes of human and other organisms for proteins that are related to the human ANO/TMC superfamily. First, we built an HMM profile for each of the 3 regions shown in Fig. 1 using HMMER (30). Next, we searched the non-redundant protein data set downloaded from the NCBI database using the HMMs. All the human ANO and TMC family members and many non-human ANO and TMC homologs were successfully retrieved. However, there were no hits from outside of either the ANO or TMC families from human or other organisms, indicating that the ANO and the TMC families are closest to each other.
When we performed conventional BLAST and PSI-BLAST searches of non-redundant protein database by using the representative human ANO and TMC proteins as queries, we successfully found ANO homologs and TMC homologs in proteomes of various organisms. The ANO homologs are found in very diverse eukaryotic organisms which belong to 5 of 6 eukaryotic super-groups (33). The super-groups with ANO proteins include Amoebozoa (a slime mold, Dictyostelium discoideum), Archaeplastida (an angiosperm, Arabidopsis thaliana, and a moss, Physcomitrella patens), Chromalveolata (ciliates, Paramecium tetraurelia and Tetrahymena thermophila, and a diatom Phaeodactylum tricornutum), Excavata (a Euglenozoan hemoflagellate, Leishmania infantum), and Opisthokonta (animals and fungi). The wide range of ANO homolog distribution indicates that the ancestral ANO protein appeared very early in eukaryotic evolution, before the divergence of major eukaryotic super-groups.
The TMC homologs, however, are only found in animals and a choanoflagellate, Monosiga brevicollis, a unicellular eukaryote closest to animals. This suggests that the TMC protein family had emerged and diverged from the ANO family by duplication during early choanoflagellate/animal evolution, although it is possible that TMC arose earlier but that some members diverged too far to be detected by BLAST searches.
In conclusion, we have shown that ANO and TMC proteins belong to a larger superfamily that plays a role in ion homeostasis in diverse eukaryotic cells. Our findings are useful for the study of molecular functions of ANO proteins and, especially, TMC proteins.
Table I.
Proteins analyzed in this study
| Protein | Accession a | TM1 b | TM4-TM5 b | TM6-TM7 b |
|---|---|---|---|---|
| ANO1 | NP_060513.4 | 309-363 | 536-608 | 699-788 |
| ANO2 | NP_065106.1 | 337-391 | 575-647 | 738-827 |
| ANO3 | NP_113606.2 | 373-427 | 593-665 | 751-840 |
| ANO4 | NP_849148.1 | 293-347 | 513-585 | 674-763 |
| ANO5 | NP_998764.1 | 281-335 | 512-584 | 669-758 |
| ANO6 | NP_001020527.1 | 276-330 | 506-578 | 663-752 |
| ANO7 | NP_001001891.2 | 324-378 | 543-615 | 704-793 |
| ANO9 | NP_001012302.2 | 168-222 | 376-448 | 537-626 |
| TMC1 | NP_619636.2 | 166-220 | 399-470 | 573-661 |
| TMC2 | NP_542789.2 | 230-284 | 462-533 | 632-720 |
| TMC3 | NP_001074001.1 | 119-173 | 352-423 | 557-645 |
| TMC4 | NP_653287.1 | 135-189 | 380-451 | 522-610 |
| TMC5 | NP_001098718.1 | 427-481 | 650-722 | 773-862 |
| TMC6 | NP_009198.3 | 226-280 | 465-537 | 589-678 |
| TMC7 | NP_079123.2 | 141-195 | 400-471 | 540-628 |
| TMC8 | NP_689681.2 | 87-141 | 330-401 | 471-559 |
NCBI accession numbers and version numbers.
Coordinates of the conserved regions shown in Fig. 1.
Acknowledgments
This research was partially supported by the Intramural Research Program of the National Cancer Institute, National institutes of Health.
References
- 1.Hartzell HC, Yu K, Xiao Q, Chien LT, Qu Z. Anoctamin/TMEM16 family members are Ca2+-activated Cl- channels. J Physiol. doi: 10.1113/jphysiol.2008.163709. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang YD, Cho H, Koo JY, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caputo A, Caci E, Ferrera L, et al. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008;322:590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- 5.West RB, Corless CL, Chen X, et al. The novel marker, DOG1, is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am J Pathol. 2004;165:107–113. doi: 10.1016/S0002-9440(10)63279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espinosa I, Lee CH, Kim MK, et al. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surg Pathol. 2008;32:210–218. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- 7.Huang X, Godfrey TE, Gooding WE, McCarty KS, Jr., Gollin SM. Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer. 2006;45:1058–1069. doi: 10.1002/gcc.20371. [DOI] [PubMed] [Google Scholar]
- 8.Carles A, Millon R, Cromer A, et al. Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene. 2006;25:1821–1831. doi: 10.1038/sj.onc.1209203. [DOI] [PubMed] [Google Scholar]
- 9.Rock JR, Futtner CR, Harfe BD. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev Biol. 2008;321:141–149. doi: 10.1016/j.ydbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Tsutsumi S, Kamata N, Vokes TJ, et al. The novel gene encoding a putative transmembrane protein is mutated in gnathodiaphyseal dysplasia (GDD) Am J Hum Genet. 2004;74:1255–1261. doi: 10.1086/421527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bera TK, Das S, Maeda H, et al. NGEP, a gene encoding a membrane protein detected only in prostate cancer and normal prostate. Proc Natl Acad Sci USA. 2004;101:3059–3064. doi: 10.1073/pnas.0308746101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S, Hahn Y, Nagata S, et al. NGEP, a prostate-specific plasma membrane protein that promotes the association of LNCaP cells. Cancer Res. 2007;67:1594–1601. doi: 10.1158/0008-5472.CAN-06-2673. [DOI] [PubMed] [Google Scholar]
- 13.Kramer J, Hawley RS. The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat Cell Biol. 2003;5:261–263. doi: 10.1038/ncb944. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Chattopadhyay S, Locke S, Pearce DA. Interaction among Btn1p, Btn2p, and Ist2p reveals potential interplay among the vacuole, amino acid levels, and ion homeostasis in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:281–288. doi: 10.1128/EC.4.2.281-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S, Hahn Y, Walker DA, et al. Topology of NGEP, a prostate-specific cell:cell junction protein widely expressed in many cancers of different grade level. Cancer Res. 2008;68:6306–6312. doi: 10.1158/0008-5472.CAN-08-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 2003;4:24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82:300–308. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 18.Kurima K, Peters LM, Yang Y, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 19.Vreugde S, Erven A, Kros CJ, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 20.Ramoz N, Rueda LA, Bouadjar B, Montoya LS, Orth G, Favre M. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat Genet. 2002;32:579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- 21.Lazarczyk M, Favre M. Role of Zn2+ ions in host-virus interactions. J Virol. 2008;82:11486–11494. doi: 10.1128/JVI.01314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarczyk M, Pons C, Mendoza JA, Cassonnet P, Jacob Y, Favre M. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J Exp Med. 2008;205:35–42. doi: 10.1084/jem.20071311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 26.Goodstadt L, Ponting CP. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics. 2001;17:845–846. doi: 10.1093/bioinformatics/17.9.845. [DOI] [PubMed] [Google Scholar]
- 27.Schneider TD, Stephens RM. Sequence Logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 30.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 31.Viklund H, Granseth E, Elofsson A. Structural classification and prediction of reentrant regions in alpha-helical transmembrane proteins: application to complete genomes. J Mol Biol. 2006;361:591–603. doi: 10.1016/j.jmb.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 32.Jentsch TJ, Friedrich T, Schriever A, Yamada H. The CLC chloride channel family. Pflügers Arch. 1999;437:783–795. doi: 10.1007/s004240050847. [DOI] [PubMed] [Google Scholar]
- 33.Adl SM, Simpson AG, Farmer MA, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]