Summary of recent advances
Organisms from all kingdoms of life produce a plethora of natural products that display a range of biological activities. One key limitation of developing these natural products into pharmaceuticals is the inability to perform effective, fast, and inexpensive structure-activity relationship studies (SAR). Recently, enzyme engineering strategies have allowed the exploration of metabolic engineering of biosynthetic pathways to create new “natural” products that can be used for SAR. The enzymes that enable the biosynthesis of natural products represent a largely untapped resource of potential biocatalysts. A challenge for the field is how to harness the wealth of reaction types used for natural product metabolism to obtain useful biocatalysts for industrial biotransformations.
Introduction
Humans have long exploited the medicinal value of natural products.[1] For example, the use of morphine isolated from opium poppy dates back to the Neolithic era. Natural products continue to play a key role in modern pharmaceuticals, either as the actual drug substance or as a lead structure for the development of synthetic derivatives.[2–4]
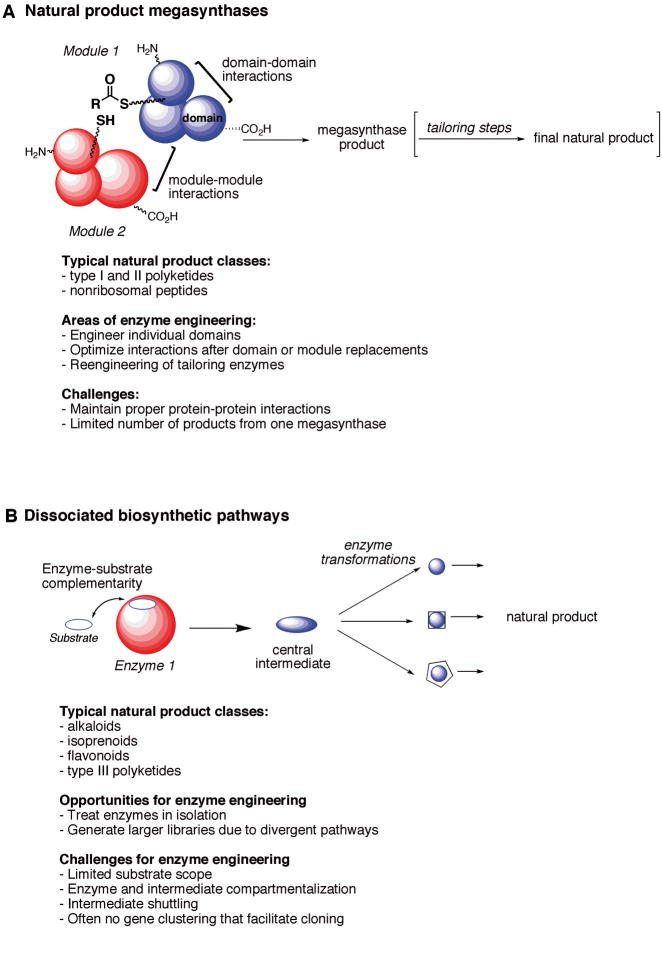
Reprogramming of biosynthetic pathways is an attractive approach to modifying natural product structures.[3] Specifically, the individual pathway enzymes can be engineered to accept altered substrates or to produce new types of products. When thinking about enzyme engineering of natural product machineries, biosynthesis of a natural product can be described to occur in two main ways. (1) A natural product can be made by large protein complexes, or megasynthases, in which the biosynthetic intermediates are shuttled between the enzyme active sites without being released into solution. These pathways are highly dependent on covalent and noncovalent protein-protein interactions for optimal activity. Alternatively, (2) natural product biosynthetic pathways can act in a dissociated fashion, in which individual enzymes do not interact with one another. In this case, tightly controlled enzyme-substrate complementarity ensures the correct sequence of biochemical transformations (Figure 1). The fundamental distinction between megasynthases and dissociated pathways raises key implications for enzyme engineering and application of reengineered proteins in biosynthetic pathways and for biocatalysis. With megasynthases, protein-protein interactions must be considered when implementing the engineering strategy. For stand-alone, dissociated pathways, engineering can focus on modulating substrate specificity of the individual enzymes. However, one must consider if any new chemical intermediates produced by the engineered enzymes are biosynthetically competent with the native downstream biosynthetic enzymes (Figure 1).
Figure 1.
Biosynthetic pathways fall into two categories depending on how the core structure of a natural product is assembled. A. Megasynthases are modular structures where the growing substrate is shuttled between different domains and protein-protein interactions are key for activity. B. Dissociated pathways are arrays of individual enzymes that turn over a set of given substrates. Substrate specificity is often tightly regulated, since enzyme-substrate complementarity determines the order of biosynthetic events.
For over twenty years, advances in genetic and enzyme engineering have allowed manipulation of biosynthetic pathways (metabolic engineering) to produce natural product analogs. This review covers research on megasynthases and dissociated pathway enzymes published between 2006 and 2008, with emphasis on enzyme engineering efforts. We also highlight key mechanistic and structural advances that point the way to future enzyme engineering opportunities. We also refer the interested reader to recent reviews that cover related topics.[3,5,6]
Megasynthase enzyme engineering
Prototypical examples of megasynthases, often found in microbes, are the type I polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs). Colinear gene clusters usually encode megasynthases; this genetic organization can often be used to predict a biosynthetic mechanism simply by protein sequence homology. Libraries of natural products can thus be obtained by rational genetic alterations of the individual catalytic domains of the megasynthase. However, one must consider that productive protein-protein interactions among the domains are key for robust enzyme activity. Small perturbations of contact interfaces (often distant from the active sites) can have detrimental effects on natural product yield.[7,8]
Multimodular polyketide biosynthesis
Multimodular PKSs consist of one or more polypeptide chains (typically >100 kDa) that are composed of several modules; each module contains discrete protein domains that work together to catalyze one cycle of carbon-carbon bond formation (Claisen condensation) and optional reducing steps catalyzed by ketoreducatses (KR) and enoyl reductases (ER). The earliest efforts to reengineer natural product biosynthesis were focused on rationally modifying domains of this type of megasynthase - in particular, 6-deoxy-erythronolide B synthase (DEBS), which makes a precursor of erythromycin.[9,10] More recent efforts have also focused on modifying tailoring enzymes that often act upon natural products after they are released from the megasynthase (see the section on tailoring steps).
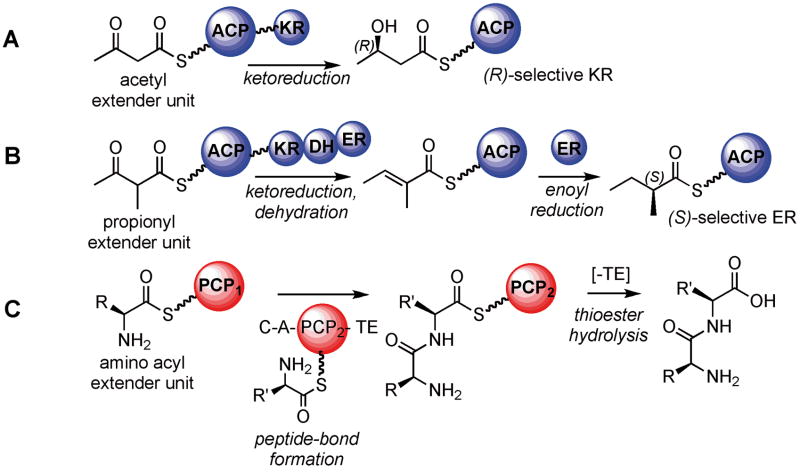
Ketoreductases (KRs) embedded within the PKS megasynthase use NADPH to reduce carbonyl groups in all PKS systems (Figure 2A). Different KR domains have different stereoselectivities and the inactivation or replacement of KR domains to alter product outcome has previously been used to diversify PKs.[11] Recently, KR stereoselectivity could be modulated by site-directed and site-saturation mutagenesis.[12,13] In a separate study, a rule to predict the KR-enforced substituent chirality was proposed based on structural information and sequence pattern analysis.[14] Future experiments will determine if this protocol is generally useful for changing KR specificity and exploring how predictably the reengineered KR domains function in the context of an entire metabolic pathway.
Figure 2.
Simplified schematics illustrating a few of the reactions catalyzed by megasynthase domains. A. KR domains use NADPH to reduce carbonyl groups to alcohols; exemplified here by an acetyl extender unit (malonyl group). Altering the stereoselectivity of KR domains could alter the product outcome. B. After KR and DH domain action on a propionyl extender unit (methyl-malonyl group), the resulting olefin can be reduced with NADPH by the action of stereoselective ER domains. Reengineering the ER stereoselectivity is also a potential means to alter product outcome. C. Illustration of NRPS domain actions on an amino acyl extender unit. A typical module is minimally composed of A (loading an amino acid onto the PCP domain), C (catalyzing the peptide bond formation), and PCP (carrying the amino acyl chain). The TE domain hydrolyzes the thioester linkage to release the natural product. KR: ketoreductase; DH: dehydratase; ER: enoyl-reductase; A: adenylation domain; C: condensation domain; PCP: peptidyl carrier protein domain; TE: thioesterase domain.
Enoyl reductases (ERs) are responsible for the generation of a fully reduced polyketide backbone from an initial β-ketoacyl group (Figure 2B). In the case where a propionate extender unit is used, a methyl substituent will be added to the polyketide backbone (Figure 2B), which can have either (S) or (R) configuration. The stereochemical configuration of the methyl group can now be predicted and rationally altered. Leadlay and co-workers identified a critical tyrosine residue in the ER active site that when present leads to an (S) configuration in the reduced product.[15] They showed that it is possible to switch the stereochemical outcome by mutating Tyr to Val in an erythromycin PKS-derived ER. However, other residues may be responsible for this stereocontrol, since the Val to Tyr mutation in an (R) selective ER was not sufficient to interconvert its stereoselectivity.[15]
Iterative polyketide biosynthesis
Eukaryotic iterative type I PKSs are monomodular megasynthases that produce aromatic polyketides.[16] Crawford et al. recently reported on the deconstruction and reassembly of selected domains in Aspergillus parasiticus (fungal) PksA, which makes a precursor of the environmental carcinogen aflatoxin. By using high-resolution mass spectrometry, the authors revealed how individual domains control PK chain-length, cyclization, and product release. A product template (PT) domain, which had previously been identified in iterative fungal PKSs, was now found to play a central role in driving the reaction to product.[17]
Bacterial iterative type II PKSs, compared to their fungal (type I) counterparts, are not modular. Instead, they consist of individual proteins that form intimate non-covalent associations. Ames et al. obtained a 1.9 Å resolution X-ray structure of an aromatase in Streptomyces lividans tetracenomycin synthase, Tcm.[18] Substrate docking led to hypotheses of which residues possibly control cyclization outcome. Eleven mutations were made at five positions and then tested for different product formation; two residues were deemed essential for anchoring the substrate (Tyr35 and Arg69) and several other residues had a marked effect on the product outcome. Future studies need to show how this enhanced understanding of iterative type I and type II PKS systems can be used to rationally alter cyclization outcome.
Nonribosomal peptide biosynthesis
A prototypical NRPS contains a peptidyl carrier protein (PCP), a condensation (C) domain and an adenylation (A) domain (Figure 2C). The termination module usually contains a thioesterase (TE) domain that releases the natural product from the megasynthase by hydrolysis of a thioester linkage. A major stumbling block in NRPS biosynthesis reengineering is how to introduce new catalytic activity while maintaining the optimal protein-protein interactions necessary for high activity and product yield.
The crystal structure of the intact 144-kDa NRPS termination module (C-A-PCP) of surfactin synthetase, which adds the last amino acid to a hexapeptide precursor, was solved to 2.6 Å resolution (PDB ID: 2VSQ).[19] The crystal structure shows the extensive contact interfaces between the different domains and suggests that there are large structural reorganizations taking place in the catalytic cycle.[19] The authors suggest that a promising approach to generate nonnatural NRPS products by domain swapping is to exchange a C-A domain pairs and then optimize the PCP/C-A interactions using directed evolution.
Two recently solved NMR structures also help to explain dynamic domain interactions. (1) The apo-form of a PCP-thioesterase (PCP-TE) bidomain of EntF in enterobactin synthetase revealed that domain interface structures are modulated by proteins that participate in the biosynthesis.[20] (2) The structure of a type II TE from Bacillus subtilis surfactin synthetase, which is used to repair misprimed PCP-domains, showed that this domain exists in multiple conformations while free in solution. However, only one conformation is selected in the presence of its native substrate, a modified holo-PCP domain.[21] This is consistent with an induced fit model and experimentally validates the effect that appropriate protein-protein interactions can have in megasynthases.
With structural information in hand, one limiting factor is the lack of an appropriate screening or selection strategy. Siderophores provide a unique handle for selection, since they enable host organisms to survive on iron-limiting media. A model system that utilizes this concept is the two-module siderophore-producing NRPS, enterobactin synthetase (EntBDEF) from E. coli.[22] E. coli cells with a gene knockout that results in lack of enterobactin biosynthesis grows slowly on iron-limiting media. EntB is an aryl carrier protein (ArCP) that carries the 2,3-dihydroxybenzoyl starter group. The contact interfaces of EntB with the other domains and the second module were mapped using combinatorial mutagenesis and in vivo selection using an entB− E. coli knockout, where only the cells harboring active enzyme mutants survived the selection process.[23,24] A similar strategy also enabled replacement of the native ArCP with two mutated noncognate ArCPs.[25] Since the noncognate ArCPs do not have optimal protein-protein interactions, slower growth on iron limiting media was observed. However, a non-cognate ArCP with 500-fold improved activity compared to the wild-type ArCP could be evolved by subjecting each of the two orthogonal genes to three rounds of error-prone PCR. The mutations responsible for the increased activity mapped the primary contact sites of EntB.[25]
Dissociated enzyme engineering
The order of biosynthetic events in dissociated biosynthesis is primarily determined by the complementarity between an enzyme and its substrate(s). There are no (or fewer) protein-protein interactions to consider, which may likely make dissociated pathway enzymes better suited as leads for useful biocatalysts. Also, while megasynthases typically give one or a few product(s), many dissociated pathways found in eukaryotes give rise to a central biosynthetic intermediate that is in turn converted into a diverse set of products. If the formation of the central biosynthetic intermediate can be controlled enzymatically, it may be possible to generate a vast array of natural product analogs, provided that the downstream enzymes can turn over the altered biosynthetic intermediate. Because dissociated pathways are primarily found in eukaryotes, one challenge is enzyme compartmentalization, in which enzymes are spatially localized to particular cellular compartments or cell types. Successful engineering therefore also depends on the efficient transport of biosynthetic intermediates across cell membranes. In addition, when the reengineered enzyme is introduced into the host organism, new bottlenecks or unexpected “off-pathway” products may emerge.
Monoterpene indole alkaloid biosynthesis
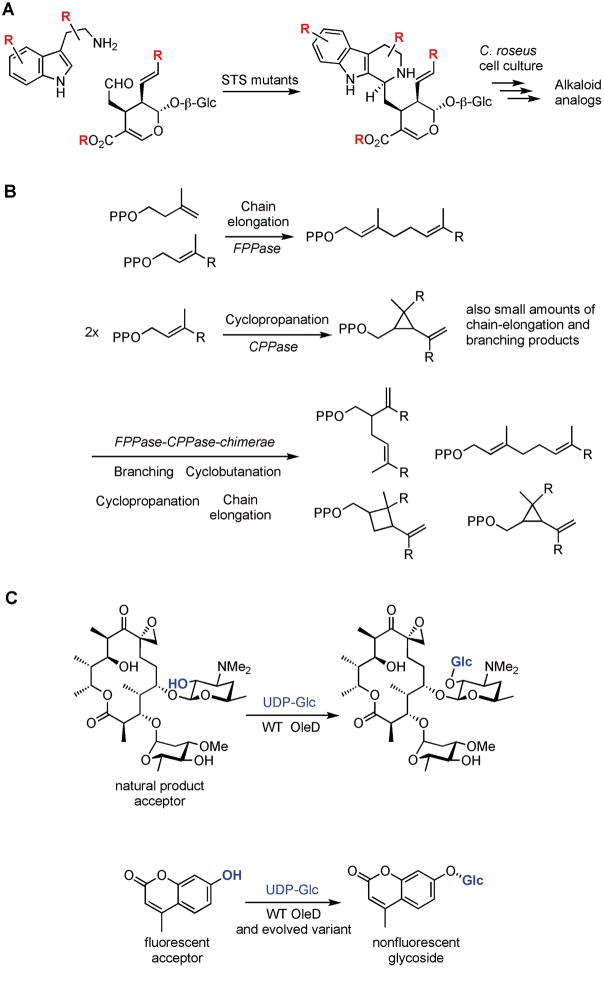
Monoterpene indole alkaloids (MIAs) constitute a large class of natural products, where several members possess important biological activities.[26] For example, Catharanthus roseus produces vinblastine and vincristine, two anticancer agents currently in use.[27] All MIAs are produced from one precursor, strictosidine, which is formed via a stereoselective Pictet-Spengler reaction between tryptamine and the monoterpene secologanin (Figure 3A).[28] This reaction is catalyzed by a highly substrate specific enzyme, strictosidine synthase (STS), which limits precursor directed biosynthesis in plant cell culture to produce natural product analogs.[29] It is possible to expand both the tryptamine and secologanin substrate scope using site-directed or site-saturation mutagenesis in the substrate-binding site (Figure 3A).[30–32] One of these STS mutants (Val214Met) was transformed into C. roseus cell culture and function in the context of the biosynthetic pathway, where it produced modified higher alkaloids not produced by the wild-type plant (manuscript submitted).
Figure 3.
Examples of enzyme engineering work on dissociated pathways. A. Strictosidine synthase (STS) catalyzes the Pictet-Spengler reaction between tryptamine and secologanin. STS variants, found by screening site-directed and saturation mutagenesis libraries, accept substrates not accepted by wild-type STS. B. Isoprenoids are synthesized by four coupling reactions: chain elongation, branching, cyclopropanation, and cyclobutanation. By replacing the sequences of a chain-elongating enzyme with the corresponding homologous sequences of a cyclopropanation enzyme, all four coupling reactions were observed. C. OleD catalyzes the C-O bond formation between the C-2 hydroxyl group on oleandomycin (acceptor) and the anomeric carbon of UDP-glucose (donor). OleD is 300-fold less efficient in transferring UDP-glucose onto 4-methylumbelliferone, a fluorescent surrogate substrate. Variants of OleD with enhanced activity were found by mutagenesis and screening the libraries in spectrophotometric assays for the loss of fluorescence of the acceptor starting material.
Isoprenoid biosynthesis
Isoprenoids constitute the largest fraction of known natural products and include sterols, carotenoids, and terpenes. The carbon skeletons of isoprenoids are constructed from 3-methyl-1-butyl units and four coupling reactions are responsible for all isoprenoid biosynthesis: chain elongation, branching, cyclopropanation and cyclobutanation (Figure 3B). Thulasiram et al. constructed eleven chimerae of chrysanthemyl diphosphate synthase (CPPase), an enzyme with cyclopropanation activity, and farnesyl diphosphate synthase (FPPase) with chain-elongation activity.[33] Using hybrids between chain-elongation and cyclopropanation enzymes, all four coupling reactions were catalyzed (Figure 3B).[33] Therefore, these four coupling reactions are likely evolutionarily related.
A relatively small number of targeted mutations to isoprenoid synthases can lead to significantly modulated activity or specificity. γ-humulene synthase is a promiscuous sesquiterpene cyclase from Abies grandis that makes numerous cyclized products from the farnesyl pyrophosphate precursor.[34] To show that it is possible to alter the product spectrum rationally, Yoshikuni et al. mutated 19 residues near the active site that were hypothesized to determine substrate selectivity. Four residues were found to affect selectivity >100-fold and these mutations were systematically recombined to give desired product profiles. The same method was used to alter the substrate selectivity of seven terpene cyclases, four of which had nonnatural selectivities.[34] The plasticity of the terpene cyclase active site, in its ability to provide different water-free environments for several types of rearrangements and cyclizations, shows the power in using enzyme engineering for constructing new terpenoid scaffolds.
Tailoring steps - natural product glycosyltransferases
Megasynthases are often combined with dissociated enzymes that provide starting materials (such as non-canonical amino acids) or carry out tailoring steps after the basic scaffold of the natural product has been assembled. For example, natural product glycosylation by means of glycosyltransferases (GTs) is a common tailoring step that occurs after megasynthase product release. Since glycosylation is often key for bioactivity, custom glycosylation of natural products could therefore offer a means to diversify bioactivity. However, the GTs that transfer a sugar from a donor to a natural product acceptor are often restricted in substrate scope. Williams et al. showed that the substrate scope of a natural product GT, OleD, from Streptomyces antibioticus can be expanded. They used three rounds of error-prone PCR with an average mutation frequency of one to two amino acids per gene product,[35] and screened for activity using a fluorescent surrogate acceptor substrate, which OleD is 300-fold less active towards than its natural acceptor oleandomycin (Figure 3C).[36] Using this strategy, a triple mutant of OleD (Pro67Thr/Ser132Phe/Ala242Val) increased the activity ~60-fold using the unnatural acceptor. Interestingly, this also translated into an enhanced conversion of a range of donor and some alternate acceptor substrates.[35] Notably, the triple mutant accepted twelve out of 22 nucleotide sugar donors that were not substrates for wild-type OleD. These experiments show that directed evolution can be used to highlight key residues for activity that can then be further optimized towards alternate substrates. To this end, Williams et al. generated site-saturation mutagenesis to optimize the specificity towards a nonnatural acceptor.[37]
Conclusions
While domain or module swapping can replace mutational protein engineering work on megasynthases, a challenge of practical importance is the significantly reduced yields associated with a lack of proper protein-protein interactions. Recent structural work, particularly on NRPS modules and domains, forms a basis for understanding contact interface optimization. Protein engineering, and in particular directed evolution, can be used to optimize interfaces provided that a suitable selection or screening assay is available; there is a need to develop novel high-throughput screening or selection systems for different megasynthases in order to obtain a broader range of natural product modifications.
Standalone dissociated pathway enzymes do not usually require intimate protein-protein interactions. As such, they are excellent targets for replacement with engineered or homologous enzymes with expanded substrate scope without contact interface optimization. However, eukaryotic compartmentalization often influences the biosynthesis of natural products and more work remains to understand how this spatial organization impacts engineering of biosynthetic pathways. Progress on customizing tailoring enzymes, such as glycosyl transferases, offers a possibility to further process megasynthase natural products. However, these biosynthetic tailoring enzymes could also be used as biocatalysts for elaborating dissociated pathway products, which may further expand bioactivity space.
Acknowledgments
We gratefully acknowledge support from NIH GM074820 and the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dewick P. Medicinal Natural Products: A Biosynthetic Approach. 2. Wiley; 2002. [Google Scholar]
- 2.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson B, Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nat Chem Biol. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 4.Baker DD, Chu M, Oza U, Rajgarhia V. The value of natural products to future pharmaceutical discovery. Nat Prod Rep. 2007;24:1225–1244. doi: 10.1039/b602241n. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard S, Thorson JS. Enzymatic tools for engineering natural product glycosylation. Curr Opin Chem Biol. 2006;10:263–271. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Thibodeaux CJ, Melancon CE, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 7*.Weissman KJ. The structural basis for docking in modular polyketide biosynthesis. Chembiochem. 2006;7:485–494. doi: 10.1002/cbic.200500435. Correct protein-protein interactions among the PKS domains and modules are critical to the fidelity of biosynthesis. It has been proposed that “docking domains” via the formation of a four-α-helix bundle mediate PKS docking. This article describes the replacement of a specific helical segment, and shows support for the docking model. Furthermore, the paper identified key residues that can be targeted to alter the efficiency or specificity of docking interactions. [DOI] [PubMed] [Google Scholar]
- 8.Weissman KJ. Single amino acid substitutions alter the efficiency of docking in modular polyketide biosynthesis. Chembiochem. 2006;7:1334–1342. doi: 10.1002/cbic.200600185. [DOI] [PubMed] [Google Scholar]
- 9.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 10.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 11.Marsden AFA, Wilkinson B, Cortes J, Dunster NJ, Staunton J, Leadlay PF. Engineering broader specificity into an antibiotic-producing polyketide synthase. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 12.Baerga-Ortiz A, Popovic B, Siskos AP, O’Hare HM, Spiteller D, Williams MG, Campillo N, Spencer JB, Leadlay PF. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:277–285. doi: 10.1016/j.chembiol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.O’Hare HM, Baerga-Ortiz A, Popovic B, Spencer JB, Leadlay PF. High-throughput mutagenesis to evaluate models of stereochemical control in ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:287–296. doi: 10.1016/j.chembiol.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 15*.Kwan DH, Sun Y, Schulz F, Hong H, Popovic B, Sim-Stark JC, Haydock SF, Leadlay PF. Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem Biol. 2008;15:1231–1240. doi: 10.1016/j.chembiol.2008.09.012. Enoyl reductases that act on a propionate extender unit generate a methyl substituent with either (S) or (R) configuration. The authors found a correlation between chirality of the methyl substituent and the presence or absence of a tyrosine residue in the active site. They also used this observation to convert the stereoselectivity of a (S)-methyl generating reductase into a reductase that gives the (R) configuration. [DOI] [PubMed] [Google Scholar]
- 16.Schneider G. Enzymes in the biosynthesis of aromatic polyketide antibiotics. Curr Opin Struct Biol. 2005;15:629–636. doi: 10.1016/j.sbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 17**.Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. An iterative polyketide synthase was expressed as individual domains, which were then reconstituted in vitro in various combinations. These experiments revealed the roles of the individual catalytic domains and in particular highlighted the importance of a product template domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC. Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. Proc Natl Acad Sci U S A. 2008;105:5349–5354. doi: 10.1073/pnas.0709223105. The cyclase domains of iterative polyketide biosynthesis help diversify aromatic polyketides. In this paper, a 1.9 Å crystal structure of a cyclase/aromatase was solved and analyzed. By using site-directed mutagenesis, the authors identified which residues are key for cyclization specificity and which are important for product distribution. This information leads to insights into the molecular basis of regioselective cyclization and is a first step towards achieving rational control of cyclization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. This first structure of a complete megasynthase (termination) module offers unique insights into the architecture of the individual domains and the nature of domain-domain interactions. This information, in combination with NMR structures isolated domains or domain-pairs give information that could be applied to alter or replace these protein domains to obtain new functions. [DOI] [PubMed] [Google Scholar]
- 20.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koglin A, Lohr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Guntert P, Wagner G, Walsh CT, et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roche ED, Walsh CT. Dissection of the EntF condensation domain boundary and active site residues in nonribosomal peptide synthesis. Biochemistry. 2003;42:1334–1344. doi: 10.1021/bi026867m. [DOI] [PubMed] [Google Scholar]
- 23.Lai JR, Fischbach MA, Liu DR, Walsh CT. Localized protein interaction surfaces on the EntB carrier protein revealed by combinatorial mutagenesis and selection. J Am Chem Soc. 2006;128:11002–11003. doi: 10.1021/ja063238h. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, Lai JR, Walsh CT. Interdomain communication between the thiolation and thioesterase domains of EntF explored by combinatorial mutagenesis and selection. Chem Biol. 2006;13:869–879. doi: 10.1016/j.chembiol.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 25*.Zhou Z, Lai JR, Walsh CT. Directed evolution of aryl carrier proteins in the enterobactin synthetase. Proc Natl Acad Sci U S A. 2007;104:11621–11626. doi: 10.1073/pnas.0705122104. The recognition of carrier proteins by multiple catalytic partners is key in the biosynthesis of fatty acids, polyketides, and nonribosomal peptides. By developing a clever selection system based on the characteristics of the natural product enterobactin, the authors set the stage for reengineering a nonribosomal peptide synthetase. They evolved two non-natural aryl carrier proteins to be recognized by the native partner enzymes and identified likely contact interfaces. The up to 500-fold improvement in the production of enterobactin indirectly showed that it is possible to increase activity by optimizing contact interfaces. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 27.van Der Heijden R, Jacobs DI, Snoeijer W, Hallard D, Verpoorte R. The Catharanthus alkaloids: pharmacognosy and biotechnology. Curr Med Chem. 2004;11:607–628. doi: 10.2174/0929867043455846. [DOI] [PubMed] [Google Scholar]
- 28*.Maresh JJ, Giddings LA, Friedrich A, Loris EA, Panjikar S, Trout BL, Stockigt J, Peters B, O’Connor SE. Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme. J Am Chem Soc. 2008;130:710–723. doi: 10.1021/ja077190z. Strictosidine synthase catalyzes a Pictet-Spengler reaction at the first committed step in indole alkaloid biosynthesis. By using a combination of theoretical analysis and experimental results, the authors demonstrated that the C-2 proton abstraction to restore aromaticity was actually the rate-limiting step in both the chemical and enzymatic reaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy E, O’Connor SE. Directed biosynthesis of alkaloid analogs in the medicinal plant Catharanthus roseus. J Am Chem Soc. 2006;128:14276–14277. doi: 10.1021/ja066787w. [DOI] [PubMed] [Google Scholar]
- 30.Chen S, Galan MC, Coltharp C, O’Connor SE. Redesign of a central enzyme in alkaloid biosynthesis. Chem Biol. 2006;13:1137–1141. doi: 10.1016/j.chembiol.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 31*.Bernhardt P, McCoy E, O’Connor SE. Rapid identification of enzyme variants for reengineered alkaloid biosynthesis in periwinkle. Chem Biol. 2007;14:888–897. doi: 10.1016/j.chembiol.2007.07.008. This article describes the first structure-based engineering of an enzyme in indole alkaloid biosynthesis that allowed tryptamine analogs to be accepted that are not accepted by the wild-type enzyme. The products produced by these mutant enzymes were fed to plant cell culture and were converted into higher alkaloids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loris EA, Panjikar S, Ruppert M, Barleben L, Unger M, Schubel H, Stockigt J. Structure-based engineering of strictosidine synthase: Auxiliary for alkaloid libraries. Chem Biol. 2007;14:979–985. doi: 10.1016/j.chembiol.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 33*.Thulasiram HV, Erickson HK, Poulter CD. Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science. 2007;316:73–76. doi: 10.1126/science.1137786. Isoprenoid skeletons are constructed by four fundamental coupling reactions. Enzymes that catalyze chain elongation and cyclopropanation are well studied, while enzymes that catalyze branching and cyclobutanation are not identified. By constructing chimerae of two homologous enzymes specialized in either chain elongation or cyclopropanation, the authors catalyzed all four coupling reactions suggesting an evolutionary relationship between all four coupling reactions by relatively minor changes in the catalytic site. [DOI] [PubMed] [Google Scholar]
- 34.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 35**.Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat Chem Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. By increasing the activity of a natural product glycosyl transferase towards a non-natural fluorescent sugar acceptor Williams et al generated a triple mutant enzyme that could process a range of sugar donors and some sugar acceptors that the wild-type enzyme cannot turn over These results can be used to optimize the activity around selected sugar acceptors or donors. [DOI] [PubMed] [Google Scholar]
- 36.Williams GJ, Thorson JS. A high-throughput fluorescence-based glycosyltransferase screen and its application in directed evolution. Nat Protoc. 2008;3:357–362. doi: 10.1038/nprot.2007.538. [DOI] [PubMed] [Google Scholar]
- 37.Williams GJ, Goff RD, Zhang C, Thorson JS. Optimizing glycosyltransferase specificity via “hot spot” saturation mutagenesis presents a catalyst for novobiocin glycorandomization. Chem Biol. 2008;15:393–401. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]