Abstract
When correlation is measured in the presence of noise, its value is decreased. In single-neuron recording experiments, for example, the correlation of selectivity indices in a pair of tasks may be assessed across neurons, but, because the number of trials is limited, the measured index values for each neuron will be noisy. This attenuates the correlation. A correction for such attenuation was proposed by Spearman more than 100 yr ago, and more recent work has shown how confidence intervals may be constructed to supplement the correction. In this paper, we propose an alternative Bayesian correction. A simulation study shows that this approach can be far superior to Spearman's, both in accuracy of the correction and in coverage of the resulting confidence intervals. We demonstrate the usefulness of this technology by applying it to a set of data obtained from the frontal cortex of a macaque monkey while performing serial order and variable reward saccade tasks. There the correction results in a substantial increase in the correlation across neurons in the two tasks.
INTRODUCTION
A central theme in the statistical analysis of neuronal data is the appropriate accounting for uncertainty. This often involves the inclusion of sources of variability that might otherwise be omitted. Although in some cases taking into account additional sources of variability may decrease the magnitude of an effect (Behseta et al. 2005), in other cases, the effect of interest may actually increase. An important example of this second situation involves estimation of correlation in the presence of noise. Suppose θ and ξ are random variables having a positive correlation ρθξ and ɛ and δ are independent “noise” random variables that corrupt the measurement of θ and ξ producing X = θ + ɛ and Y = ξ + δ. A simple mathematical argument (see appendix) shows that
 |
where ρXY is the correlation between X and Y. In words, the presence of noise decreases the correlation. Thus in many circumstances, a measured correlation will underestimate the strength of the actual correlation between two variables. However, if the likely magnitude of the noise is known, it becomes possible to correct the estimate. The purpose of this note is to provide a Bayesian correction and to show that it can produce good results when examining correlations derived from multi-trial spike counts.
We apply the method in the context of single-neuronal recording experiments. Broadly speaking, it is sometimes necessary to compare the selectivity of a neuron for a particular variable across two task contexts. For example, one might wish to compare shape selectivity across blocks of trials in which the shape has different colors (Edwards et al. 2003) or compare selectivity for saccade direction across blocks of trials in which the saccade is selected according to different rules (Olson et al. 2000). It is also sometimes necessary to compare selectivity for two different variables as measured in separate task contexts. For example, we might wish to compare selectivity for the direction of motion of a visual stimulus viewed during passive fixation with selectivity for saccade direction in a task requiring eye movements (Horwitz and Newsome 2001). The standard approach to making such comparisons is to compute, for multiple neurons, index 1 in context 1 and index 2 in context 2 and then to compute the correlation between the two indices across neurons. The correlation may be statistically significant but smaller than one might expect, which raises the question: is the small correlation due to a genuine discordance between the two forms of selectivity, or is it due to noise arising from random trial-to-trial variability in the neuronal firing rates? This is the kind of question the methods of this article are designed to answer.
The idea of introducing a “correction for attenuation” of the correlation goes back at least to Spearman (1904). He did not at that time, however, have the technology to provide confidence intervals associated with his proposed technique. Frost and Thompson (2000) reviewed some solutions to the problem of constructing confidence intervals for the slope of a noise-corrupted regression line, and Charles (2005) gave procedures for obtaining confidence intervals for the correlation based on Spearman's formula. We performed a computer simulation study to compare the Bayesian correction with Spearman's correction and the Bayesian confidence intervals with those based on Spearman's correction. We found the Bayesian method to be far superior. We then applied the method to data from the frontal cortex of a macaque monkey recorded while the monkey was performing serial order and variable reward saccade tasks.
METHODS
Notation
Let Xi = θi + ɛi, and Yi = ξi + δi, where θi and ξi represent the underlying true values, and ɛi and δi are the associated error values (representing noise) for the ith observation, for i = 1, 2,…, n. Let 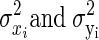 represent the variance for Xi and Yi respectively. To construct the Bayesian model (see following text), we assume that ɛi ∼ N(0,σɛi2), and δi ∼ N(0,σδi2). In neurophysiological applications,
represent the variance for Xi and Yi respectively. To construct the Bayesian model (see following text), we assume that ɛi ∼ N(0,σɛi2), and δi ∼ N(0,σδi2). In neurophysiological applications, 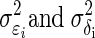 may be estimated from repeated trials and then treated as known, separately for each neuron, as we discuss in data analysis and also in the appendix. This is referred to as inhomogeneous variances. Otherwise, in the case of homogeneous variances, since
may be estimated from repeated trials and then treated as known, separately for each neuron, as we discuss in data analysis and also in the appendix. This is referred to as inhomogeneous variances. Otherwise, in the case of homogeneous variances, since 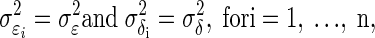 we will use the notation
we will use the notation 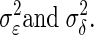 Finally, we let
Finally, we let 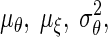 and σξ2 be the means and the variances of θi and ξi, respectively.
and σξ2 be the means and the variances of θi and ξi, respectively.
Spearman's method
Spearman (1904) tackled the problem of correcting the attenuation of the correlation coefficient through a series of intuitive steps (see Charles 2005). Spearman's formula for attenuation correction is given by
 |
(1) |
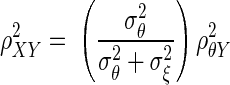
where ρθξ is the corrected correlation, ρXY is the correlation between X and Y, and ρXX, and ρYY are known as reliabilities, defined as 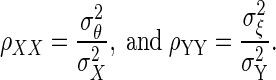 The derivation of Eq. 1 is given in the appendix.
The derivation of Eq. 1 is given in the appendix.
In practice, we estimate (1) with the sample corrected correlation as follows
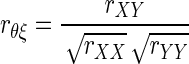 |
(2) |
where rXY is the usual (Pearson) correlation given by 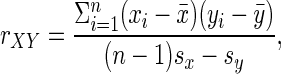 in which x̄ and ȳ are the sample means, and sx and sy are the sample SDs of X and Y. Also
in which x̄ and ȳ are the sample means, and sx and sy are the sample SDs of X and Y. Also 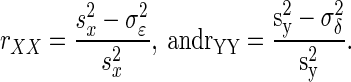 According to Spearman (1904), to calculate
According to Spearman (1904), to calculate 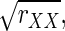
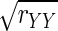 in Eq. 2, one would need repeated measurements of Xi and Yi for each i = 1,…, n. This is important because in some applications, such as in data analysis in the following text, the quantities Xi and Yi are indices derived from complete sets of trials; repeated measurements are not available.
in Eq. 2, one would need repeated measurements of Xi and Yi for each i = 1,…, n. This is important because in some applications, such as in data analysis in the following text, the quantities Xi and Yi are indices derived from complete sets of trials; repeated measurements are not available.
It is possible for 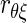 to be >1. Spearman was aware of this, but the method was received quite unfavorably by Karl Pearson (1904), who firmly believed that correlation formulas that yield values larger than one are essentially faulty. The Bayesian method we introduce in the following text does not suffer from this defect.
to be >1. Spearman was aware of this, but the method was received quite unfavorably by Karl Pearson (1904), who firmly believed that correlation formulas that yield values larger than one are essentially faulty. The Bayesian method we introduce in the following text does not suffer from this defect.
Confidence intervals for Spearman's method
The most commonly applied confidence interval for ρθξ is as follows (Charles 2005; Hunter and Schmidt 1990; Winne and Belfrey 1982)
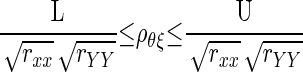 |
(3) |
in which by L and U, we refer to the lower and upper bounds of the confidence interval for ρxy. To calculate L and U in Eq. 3, one can use the asymptotic confidence interval of the so-called Fisher's z transformation (Fisher 1924). In general, if ρ and r are the theoretical and the sample correlations, respectively, then it can be shown that 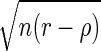 converges in distribution to a normally distributed random variable with mean 0 and variance (1 − ρ2)2, and that the asymptotic variance of
converges in distribution to a normally distributed random variable with mean 0 and variance (1 − ρ2)2, and that the asymptotic variance of 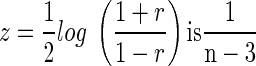 which does not depend on r (see page 52 in DasGupta 2008). Consequently, the lower and the upper bounds of a confidence interval for
which does not depend on r (see page 52 in DasGupta 2008). Consequently, the lower and the upper bounds of a confidence interval for 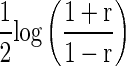 are
are
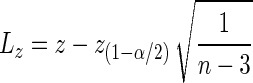 |
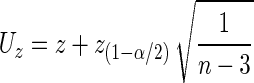 |
where z(1−α/2) is the 100(1 − α/2)-th percentile of the standard normal distribution. Using the inverse transformation to restate the confidence bounds in terms of r we obtain
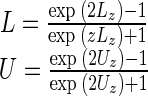 |
Specifically, for data (x1, y1),…, (xn, xn), putting L and U defined in the preceding text in Eq. 3 provides the usual confidence interval based on Spearman's method. The values of Lz and Uz based on Fisher's z-transformation can be obtained using standard software. For example, to calculate L and U in Matlab, one can use the command corrcoef. The command cor.test in the statistical software R will also produce L and U.
Measurement error methods
The problem of correlation attenuation due to noise can be viewed from a simple linear regression perspective. For a thorough review, see the books by Carroll et al. (2006), and Fuller (1987), and the article by Frost and Thompson (2000). Consider the linear regression model
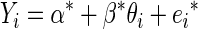 |
for i = 1,…, n, with the familiar assumption 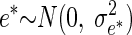 Suppose that instead of observing values for the predictor θ, we record noisy values
Suppose that instead of observing values for the predictor θ, we record noisy values 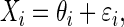 where ɛi is the measurement error for the ith observation. Consequently, we fit
where ɛi is the measurement error for the ith observation. Consequently, we fit
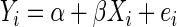 |
Note that unlike previous sections, measurement error is only assumed for the predictor X.
The problem may be formulated further with two assumptions: 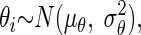 and
and 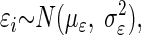 where σθ2 and σɛ2 represent the so-called between objects and within objects uncertainty, respectively. As shown in the appendix, one can estimate the true slope of the regression line β*, via
where σθ2 and σɛ2 represent the so-called between objects and within objects uncertainty, respectively. As shown in the appendix, one can estimate the true slope of the regression line β*, via
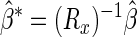 |
where β̂ is the least-squares estimator of β, and 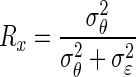 is the reliability or
is the reliability or 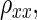 as introduced in the previous text. Note that similar to Spearman's technique, the regression-based model assumes homogeneous variances. As shown in the appendix, when the measurement error is also considered for the response variable, the regression-based method and Spearman's approach will produce the same correction for attenuation. As with Spearman's technology, regression-based techniques are constructed under the assumption that there are repeated measurements in each (Yi,Xi), for i = 1,…, n.
as introduced in the previous text. Note that similar to Spearman's technique, the regression-based model assumes homogeneous variances. As shown in the appendix, when the measurement error is also considered for the response variable, the regression-based method and Spearman's approach will produce the same correction for attenuation. As with Spearman's technology, regression-based techniques are constructed under the assumption that there are repeated measurements in each (Yi,Xi), for i = 1,…, n.
Bayesian method
To address the problem of correlation attenuation, we form a random-effects model, also called a hierarchical model. Following the notation in the previous text, let Xi = θi + ɛi and Yi = ξi + δi, where θi and ξi represent the underlying true values, and ɛi and δi are the associated error values for the ith observation, i = 1, 2,…, n. In the first stage of the model, we have: 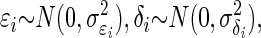 where σɛi, and σδi are estimated for repeated trials and then treated as known for each i = 1,…, n.
where σɛi, and σδi are estimated for repeated trials and then treated as known for each i = 1,…, n.
In the second stage of the model, we join to the distributional assumption for the pairs (Xi, Yi), the additional specification
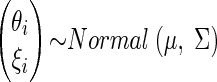 |
in which
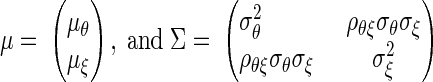 |
where 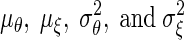 be the means and the variances of θi and ξi respectively.
be the means and the variances of θi and ξi respectively.
The quantity we seek is 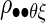 Because we have formulated the model in this way, and because the uncertainty of measurements of interest is explicitly incorporated into
Because we have formulated the model in this way, and because the uncertainty of measurements of interest is explicitly incorporated into 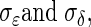 the estimate of ρθξ will be adjusted for those sources of variation. We employ Bayesian methods (Casella and George 1992; Gelman et al. 1995; Wilks et al. 1995) to estimate ρθξ via its posterior distribution. We use conjugate priors for μ and Σ. This will facilitate the application of conventional software (such as WinBugs).
the estimate of ρθξ will be adjusted for those sources of variation. We employ Bayesian methods (Casella and George 1992; Gelman et al. 1995; Wilks et al. 1995) to estimate ρθξ via its posterior distribution. We use conjugate priors for μ and Σ. This will facilitate the application of conventional software (such as WinBugs).
We put independent univariate normal priors on 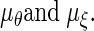 In both cases, we use diffuse Gaussian priors with mean 0 and a large variance, σ2. An Inverted-Wishart prior with the scale matrix R, and degrees of freedom d with the density
In both cases, we use diffuse Gaussian priors with mean 0 and a large variance, σ2. An Inverted-Wishart prior with the scale matrix R, and degrees of freedom d with the density 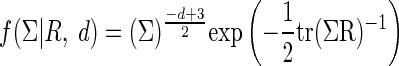 is used for the covariance matrix Σ. To avoid injecting substantial information, we use a very diffuse prior on Σ. Following Kass and Natarajan (2006), we set d = 2 df, and we take the scale matrix R to be the harmonic mean of the observed covariance matrices
is used for the covariance matrix Σ. To avoid injecting substantial information, we use a very diffuse prior on Σ. Following Kass and Natarajan (2006), we set d = 2 df, and we take the scale matrix R to be the harmonic mean of the observed covariance matrices 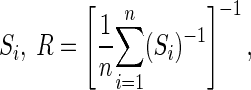 where Si is a 2 × 2 observed covariance matrix associated with neuron i, obtained from trial-to-trial variation in the spike counts.
where Si is a 2 × 2 observed covariance matrix associated with neuron i, obtained from trial-to-trial variation in the spike counts.
By sampling from the posterior distribution of Σ, one can form a confidence interval (often called a Bayesian “credible interval”) for the correlation coefficient ρ. Thus a confidence interval for ρ having coverage probability approximately 1 − α is
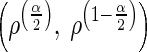 |
where ρ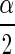 , and ρ1−
, and ρ1−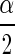 are the
are the 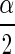 th and the1−
th and the1−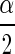 th quantiles of the sample from the posterior distribution of ρ.
th quantiles of the sample from the posterior distribution of ρ.
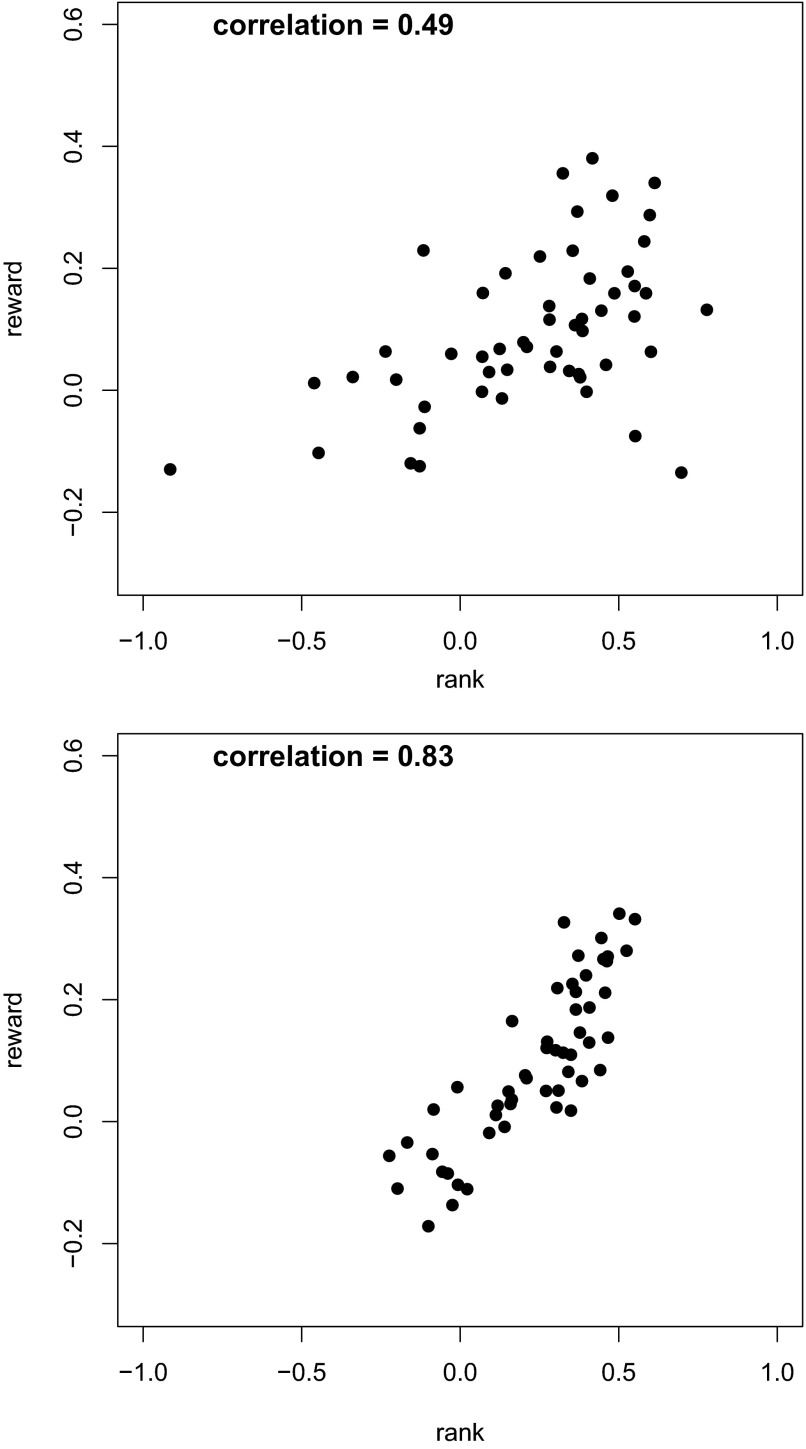
The hierarchical model used here has the property that the resulting posterior means for (θi,ξi) are weighted combinations of the observed values (Xi,Yi) and their weighted means (with weights defined in terms of the relative values of the variances, so that more precisely measured observations have greater weight). This is usually referred to as “shrinkage” because the spread of the values of posterior means for (θi,ξi) will be smaller than the spread of the data (Xi,Yi). This phenomenon is discussed in Bayesian texts, such as Gelman et al. (1995) and will be illustrated in Fig. 1.
FIG. 1.
Plots of reward- vs. rank-selective indices, before and after Bayesian correction. Top: uncorrected indices. The x axis represents the index value for the the serial order saccade task. This is obtained through Irank = (f3 − f1)/(f3 + f1), where f1 and f3 were the mean firing rates measured at the times of the 1st and 3rd saccades, respectively. The y axis indicates the index of selectivity for the size of the anticipated reward: Ireward = (fb − fs)/(fb + fz) where fb and fs were the firing rates during the postcue delay period on big-reward and small-reward trials, respectively. Bottom: plot of posterior means representing values after correction for noise.
RESULTS
Data analysis
We demonstrate the effect of the Bayesian correction of the attenuation using data from an experiment we carried out recently. The aim of the experiment was to characterize the neural mechanisms that underlie serial order performance. We trained monkeys to perform eye movements in a given order signaled by a cue. For example, one cue carried the instruction: look up, then right, then left. Monitoring neuronal activity in frontal cortex during task performance, we found that many neurons fire at different rates during different stages of the task, with some firing at the highest rate during the first, some during the second, and some during the third stage. We refer to this property as rank selectivity. Rank-selective neurons might genuinely be sensitive to the monkey's stage in the sequence. Alternatively, they might be sensitive to some correlated factor. One such factor is expectation of reward. Reward (a drop of juice) was delivered only after all three movements had been completed. Thus as the stage of the trial progressed from one to three, the expectation of reward might have increased.
To test whether rank-selective neurons were sensitive to the size of the anticipated reward, we trained the same monkeys to perform a task in which a visual cue presented at the beginning of the trial signaled to the monkey whether he would receive one drop or three drops of juice after a fixed interval. We reasoned that neuronal activity related to expectation of reward would be greater after the promise of three drops than after the promise of one. We monitored the activity of 54 neurons during the performance of both the serial order task and the variable reward task. We then computed an index of rank selectivity in the serial-order task and of selectivity for the size of the anticipated reward in the variable reward task for each neuron. The rank index was 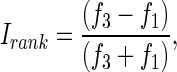 where f1 and f3 were the mean firing rates measured at the times of the first and third saccades, respectively. To obtain the rank index for a given neuron, we collected data from ∼25 trials for each of the nine possible rank-direction combinations (3 directions of eye movements: rightward, upward, or leftward; and 3 ranks: 1st, 2nd, or 3rd). The reward index was
where f1 and f3 were the mean firing rates measured at the times of the first and third saccades, respectively. To obtain the rank index for a given neuron, we collected data from ∼25 trials for each of the nine possible rank-direction combinations (3 directions of eye movements: rightward, upward, or leftward; and 3 ranks: 1st, 2nd, or 3rd). The reward index was 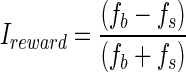 where fb and fs were the firing rates during the post cue delay period on big- and small-reward trials, respectively. To obtain reward index for a given neuron, we collected data from ∼20 trials for each of the six possible reward-direction combinations (3 directions of eye movements: rightward, upward or leftward; and 2 sizes of anticipated reward: 1 or 3 drops of juice).
where fb and fs were the firing rates during the post cue delay period on big- and small-reward trials, respectively. To obtain reward index for a given neuron, we collected data from ∼20 trials for each of the six possible reward-direction combinations (3 directions of eye movements: rightward, upward or leftward; and 2 sizes of anticipated reward: 1 or 3 drops of juice).
On carrying out a conventional correlation analysis, we found that the two measures were positively correlated but that the effect was not strong (r = 0.49). We speculated that the low correlation was in part due to trial-to-trial uncertainty of neuronal firing rate that would affect the correlation even when all controllable factors (such as rank and the size of the anticipated reward) are held constant. We were surprised at the low degree of the measured correlation between the rank and reward indices because we knew that the expectation of reward increases over the course of a serial-order trial and that neuronal activity in the SEF is affected by the expectation of reward (Roesch and Olson 2003). We wondered if our estimate of the correlation between the two indices had been attenuated by noise arising from random trial-to-trial variations in neuronal activity.
To implement the proposed Bayesian technology, we let Ireward and Irank play the role of 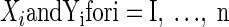 neurons as in the preceding text. Consequently, we would need to estimate
neurons as in the preceding text. Consequently, we would need to estimate 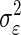 and
and 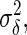 the variances of
the variances of  respectively. The following relationship may be used to calculate
respectively. The following relationship may be used to calculate 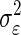 per neuron
per neuron
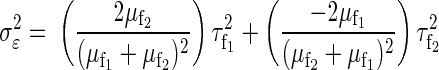 |
(4) |
where μf1, τf12, μf2, and τf22 are the means and the variances of f1 and f2 in I = (f1 − f2)/(f1 + f2), respectively. A similar formulation can be used to estimate σδ2. The derivation of Eq. 4 is given in the appendix. Also considering the magnitude of X, and Y values, we let σ2 = 80 for the variance of normal priors on μθ and μξ.
In Fig. 1, we demonstrate the dramatic effect of the Bayesian attenuation correction for 54 recorded neurons. On the top, we give the scatterplot for the pairs of indices prior to the application of the Bayesian method. The x and y axes are represented by Irank and Ireward, respectively. The bottom part contains the scatterplot for the posterior estimation of the same set of indices. The correlation for the attenuated data are 0.49. The corrected correlation using the Bayesian method increased dramatically to 0.83 with the confidence interval (0.77,0.88). For this data, Spearman's method gave an estimate of 0.85 but the 95% confidence interval was (0.65,0.99). Not only is the Bayesian interval much smaller, but from our results of the simulation study, we expect it to have a probability coverage much closer to 0.95.
Simulation study
METHODS.
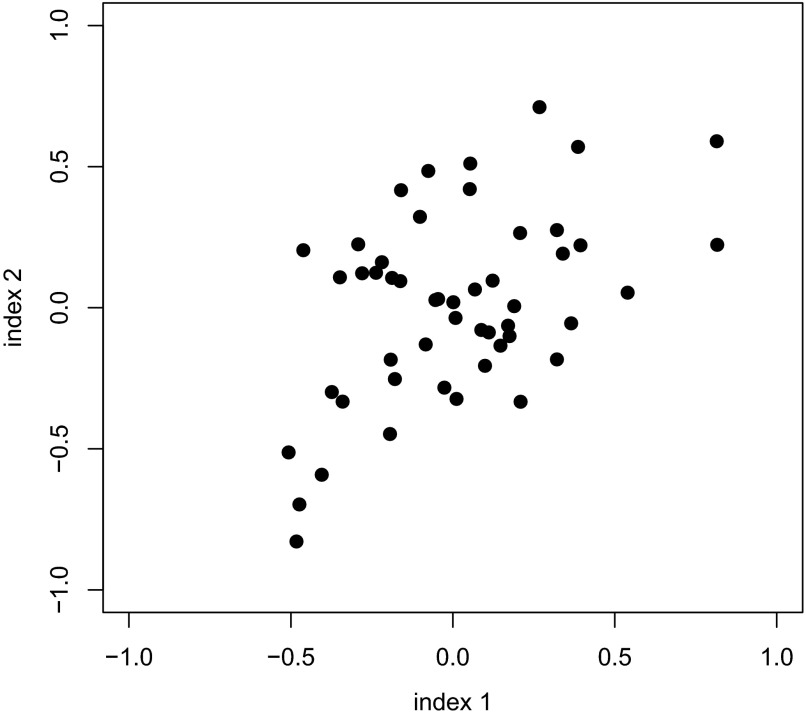
To investigate the accuracy of the proposed Bayesian method, we performed simulation studies repeating the data structure of the previous section under three scenarios. In scenario 1 (Fig. 2), we considered the truth to be a realization of a bivariate normal distribution with
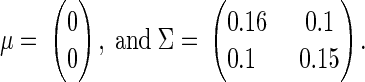 |
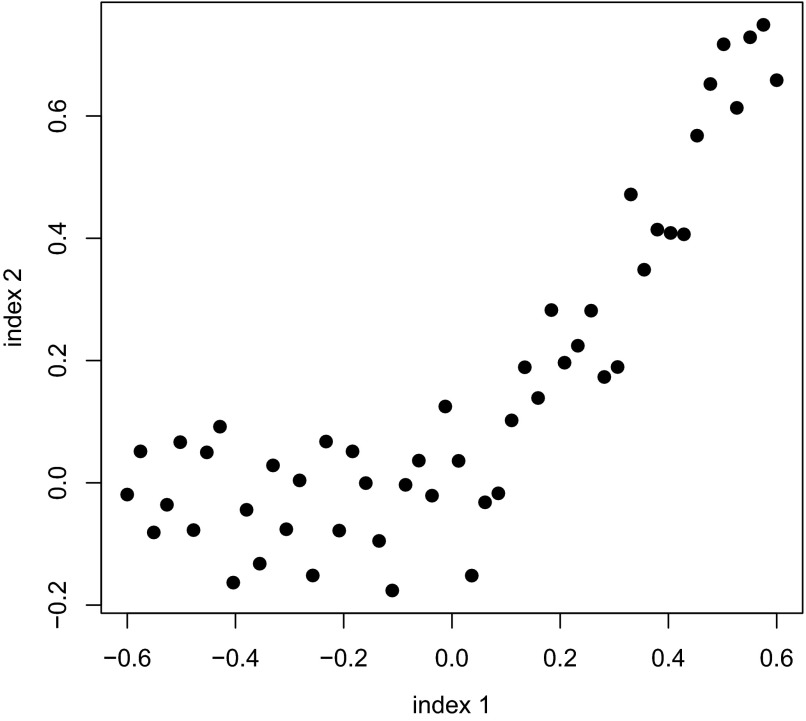
As a result of bivariate normality, the Bayesian method assumes a linear relationship between θ and ξ. However, one might not expect much distortion from this assumption. Correlation is itself a measure of linear association, so a method based on linearity might be expected to behave reasonably well even when the underlying relationship is nonlinear. We used scenario 2 as an initial check on this intuition. Scenario 2 is shown in Fig. 3. In scenario 2, we assumed that the true data points were built around the quadratic equation y = 1.1x2 + 0.6x − 0.004, evaluated at x ∈ (−0.6, 0.6), after adding random noise N(0,1) to each point.
FIG. 2.
Simulation scenario 1. True indices are obtained by simulating from a bivariate normal distribution.
FIG. 3.
Simulation scenario 2. Data points are constructed around the quadratic equation y = 1.1x2 + 0.gx − 0.004, evaluated at x ∈ (−0.6,0.6), after adding random noise N(0,1) to each point.
As may be seen in Fig. 3, scenario 2 provides a strikingly nonlinear relationship between θ and ξ, clearly violating the linearity assumption imbedded in the Bayesian method. Finally, in scenario 3, we took the true values to be the data shown in Fig. 1, bottom, that is, the data obtained from implementing the Bayesian procedure.
The general algorithm for simulating neuronal data in all scenarios can be explained through the following steps: 1) let the bivariate data in Figs. 2 and 3 and Fig. 1, bottom, represent true values for Irank and Ireward in each scenario. 2) Considering that 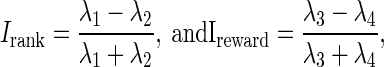 manufacture true values for λ1, λ2, λ3, λ4, associated with each pair of true (Irank, Ireward). 3) Knowing λs, randomly simulate 1,000 sets of 200 Poisson counts: 200 = 50 (neurons) × 4 (λs). 4) For each randomly generated dataset, calculate the associated simulation values of Irank and Ireward . 5) Implement both the Bayesian technology and Spearman's approach on the simulated data. Assess the goodness of each technology.
manufacture true values for λ1, λ2, λ3, λ4, associated with each pair of true (Irank, Ireward). 3) Knowing λs, randomly simulate 1,000 sets of 200 Poisson counts: 200 = 50 (neurons) × 4 (λs). 4) For each randomly generated dataset, calculate the associated simulation values of Irank and Ireward . 5) Implement both the Bayesian technology and Spearman's approach on the simulated data. Assess the goodness of each technology.
To implement step 2, we generated a pair of Poisson intensities, one for each condition. Because we began the simulation process by setting true values for I, we would need to calculate true λ values by applying a reverse procedure. Due to the fact that 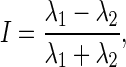 we manufactured a pair of
we manufactured a pair of 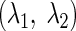 such that they match the equation for I. For example, when I = 0.08, we may take
such that they match the equation for I. For example, when I = 0.08, we may take 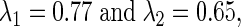 and when −0.036, we let
and when −0.036, we let 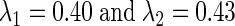 (see the supplementary material for an extended example.)
(see the supplementary material for an extended example.)
In each scenario, to imitate the multi-trial nature of neuronal recordings, we simulated Poisson data with n = 15,30,60,120, where n is the number of trials. For step 3, we generated Poisson counts using the following general idea. Let λ be the true firing rate for a given experimental condition and suppose that interest lies in creating multiple trials of spike occurrences over the span of 1 s. Let n be the number of trials. Then nλ is the expected value of the Poisson counts for that condition.
After simulating data for each scenario, we kept track of the mean square error as well as the SE of the point estimator using the usual correlation (rXY), the corrected rXY or rθξ as shown in formula 1, and the proposed Bayesian method. Additionally, we calculated the coverage of the true correlation in the simulated confidence intervals, using: the z-transformed uncorrected correlation coefficient, the corrected z method as presented in the previous text, and Our proposed Bayesian method.
To calculate the coverage we use
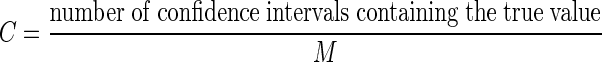 |
where M is the number of simulated data sets. To calculate the mean squared error, we set MSE = E(ρ̂ − ρ)2, where ρ̂ is the point estimator for ρ. For example, in the Bayesian case, since the posterior draws of ρ were fairly symmetric, the posterior mode and the posterior mean were extremely close. Consequently we used the posterior mode as the point estimator. Note that MSE is generally approximately proportional to 1/n. Therefore the practical implication is that if one method reduces MSE by a factor of say 2, this is roughly equivalent to doubling the sample size.
Simulation results are summarized in Tables 1–6. Each table contains the results for the z-transformed method, the corrected z method, and the Bayesian method. The method that produces smaller mean square error is more desirable. Confidence intervals were constructed to have ∼0.95 probability of coverage. Therefore good performance of each method would be indicated by coverage probability close to 0.95.
TABLE 1.
MSE for simulations under scenario 1 (normal data)
| rXY | Spearman | Bayesian | |
|---|---|---|---|
| n = 15 | 0.036 | 0.025 | 0.0037 |
| n = 30 | 0.015 | 0.01 | 0.0016 |
| n = 60 | 0.0061 | 0.0051 | 0.00091 |
| n = 120 | 0.0026 | 0.0025 | 0.00049 |
For example, the MSE of rXY when n = 15 was 0.036. In every case, the Bayesian estimator has much smaller MSE than Spearman's. MSE, mean squared error.
TABLE 6.
Simulations under scenario 3 (based on the SEF data)
| z | Corrected z | Bayesian | |
|---|---|---|---|
| n = 15 | 0.012 | 0.91 | 0.93 |
| n = 30 | 0.07 | 0.92 | 0.94 |
| n = 60 | 0.39 | 0.96 | 0.95 |
| n = 120 | 0.68 | 0.96 | 0.95 |
Entries as in Table 2. In all cases, the Bayesian technique provides accurate coverage probability with much less varibility across datasets.
In each scenario, 25,000 sets of Poisson data were simulated. The results of the mean squared error calculations are given in Table 1. They indicate that the hierarchical Bayesian method yields a huge reduction in mean squared error compared with both of the rXY and rθξ, the corrected rXY method. An explanation for the improvement of the hierarchical method over the other two techniques is that the Bayesian approach, because of its multi-level structure, allows for proper accounting for uncertainty due to error. To give an example, the estimated mean squared error achieved by the Bayesian method at n = 30 is about half of the mean squared error of the other two methods at n = 120. In other words, use of the Bayesian method is roughly equivalent to eightfold increase in sample size with Spearman's method. Further calculations (not included in Tables 1–6) revealed results: mean square error of the Bayesian estimator at n = 15 is nearly equal to the mean squared error at n = 90 when the corrected rXY method is performed; both in terms of the mean square error and the SE, the Bayesian method at n = 45, outperformed Spearman's technique even at a considerably large sample size of n = 120; and, for smaller n (n = 15,30), the mean squared error of the Bayesian technique was 10 times smaller than the mean squared error of the Spearman's method.
As shown in Table 2, both transformed-z, and the corrected technique tend to either underestimate (for small n), or overestimate (for large n), the true coverage of 0.95. The overestimation phenomenon reveals itself sharply especially in the case of larger trial numbers (n ≥ 30). In contrast, the coverage of the Bayesian method is consistently close to 0.95 regardless of the sample size. Additionally, simulation standard errors of the Bayesian method are universally smaller than the ones obtained from the other two methods. For instance, the SE of the coverage at n = 120 is 0.008 for the z-transformed method, 0.009 for the corrected z method, and 0.0008 for the Bayesian method, a reduction of >10 times. This is a suggestion the SE of the coverage in Spearman's method is highly affected when a few of the simulated data sets behave erratically.
TABLE 2.
Simulations under scenario 1 (normal data)
| z | Corrected z | Bayesian | |
|---|---|---|---|
| n = 15 | 0.82 | 0.93 | 0.94 |
| n = 30 | 0.97 | 0.95 | 0.95 |
| n = 60 | 0.99 | 0.99 | 0.95 |
| n = 120 | 1.00 | 1.00 | 0.95 |
Coverage probability of the Fisher's z, corrected z, and Bayesian, 95% confidence intervals. For different sample size values, the Bayesian estimator has a coverage probability closer to 95% than the corrected z. The coverage standard errors are all small. For example, the SE of the Bayesian method is 0.007.
Tables 3 and 4 summarize the results for scenario 2. Again, the proposed Bayesian method outperforms the other methods in terms of the mean squared error, as well as the coverage values. As shown in Table 3, the mean squared error of the Bayesian method at n = 30, which is 0.001, cannot be achieved by either of the other two methods even at a significantly larger trial size of n = 120. According to Tables 3 and 4, the Bayesian coverage is far more accurate than the others. Similarly to scenario 1, both the z-transformed and its corrected version have the undesirable property of underestimating the coverage for data sets with smaller to moderate trial numbers (n ≤ 30), and overestimating the coverage for the ones with even moderate trial sizes (n ≥ 60). The results of the scenario 3 (Tables 5 and 6) are fully consistent with the findings of the other two scenarios.
TABLE 3.
MSE for simulations under scenario 2 (banana-shaped data)
| rxy | Spearman | Bayesian | |
|---|---|---|---|
| n = 15 | 0.091 | 0.034 | 0.0072 |
| n = 30 | 0.035 | 0.012 | 0.0013 |
| n = 60 | 0.014 | 0.0052 | 0.00086 |
| n = 120 | 0.0036 | 0.0026 | 0.00035 |
In all cases, the Bayesian method has much smaller MSE than Spearman's entries as in Table 1.
TABLE 4.
Simulations under Scenario 2 (banana-shaped data)
| z | Corrected z | Bayesian | |
|---|---|---|---|
| n = 15 | 0.092 | 0.85 | 0.94 |
| n = 30 | 0.15 | 0.92 | 0.95 |
| n = 60 | 0.61 | 0.96 | 0.95 |
| n = 120 | 0.96 | 0.97 | 0.95 |
Entries as in Table 2. In all cases, the Bayesian technique provides accurate coverage probability with much less varibility across datasets.
TABLE 5.
MSE for simulations under scenario 3 (based on the SEF data)
| rxy | Spearman | Bayesian | |
|---|---|---|---|
| n = 15 | 0.011 | 0.042 | 0.0098 |
| n = 30 | 0.055 | 0.019 | 0.0049 |
| n = 60 | 0.017 | 0.0061 | 0.00099 |
| n = 120 | 0.009 | 0.0011 | 0.00047 |
The results of simulations 1 and 2 are repeated. For different sample size values, the Bayesian method produces a much smaller MSE.
In addition to the above mentioned simulation studies, Poisson data were generated from a true scenario with extremely low correlation, r ≈ 0. We performed simulations for n = 15 and n = 60. We did this additional simulation to study the performance of the Bayesian correction method when there is no correlation. As with the previous simulations, for both sample sizes, the posterior coverage of 0 was extremely close to 0.95 for the Bayesian test at the 0.05 level. Meanwhile, the simulation's mean squared error was close to 0.01 for n = 15, and 0.001 for n = 60. This provides an example when the Bayesian methodology does not cause an artificial increase in the correlation.
DISCUSSION
Spearman's idea of correcting for attenuation of the correlation coefficient has been around for >100 yr. It deserves wider recognition in neurophysiological practice, but Spearman's method is flawed. In the first place, it can produce correlations larger than 1. Second, Spearman's reliability rxx is only well-defined when the variability within items (here, within neurons) is the same for every item; but the variance of such things as a firing-rate index is likely to change across neurons. Third, as our simulation results showed, it is much less accurate than the Bayesian method we introduced: for normally distributed variables Spearman's method may require 10 times as many trials for comparable accuracy, while in the non-normal case Spearman's method is even worse. Finally, while confidence intervals based on Spearman's method have been developed, they are also much less accurate than the Bayesian intervals we recommend.
The Bayesian method we applied is easily implemented with existing software (see the appendix). In some situations, practitioners worry that the prior information injected via Bayes' Theorem may introduce substantial biases. The priors we used were chosen to be diffuse (not highly informative) and have been shown to have good properties in previously-published work (Kass and Natarajan 2006). The simulation study reported here indicates that the method performs well. From our analysis of SEF data we conclude that the method can have a substantial impact on neurophysiological findings.
GRANTS
This work was supported by National Institutes of Health Grants RO1 MH-064537-04 to R. E. Kass, 1P50MH-084053-01 and 1RO1EY-018620-01 to C. R. Olson.
APPENDIX
Correlation decreases when noise is added
Let θ and ξ be two independent random variables with ɛ and δ as their associated noise. Let X = θ + ɛ, and Y = ξ + δ Because of independence, one can write
 |
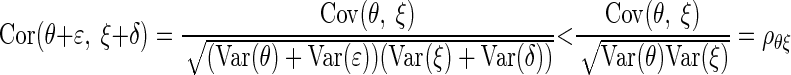 |
Derivation of Spearman's formula
Suppose that we observe pairs of X and Y s, with X = θ + ɛ, and Y = ξ + δ. Note from the definition of correlation that
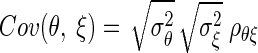 |
Also due to independence between θ, and ξ (previous appendix), we have
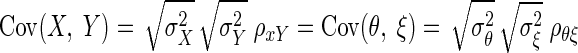 |
therefore
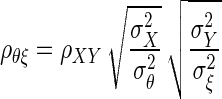 |
let
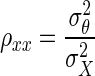 |
Spearman referred to ρxx, the proportion of the observed variance of X's that is due to variance among true values θs, as the reliability of X's. Similarly the reliability of Y's is defined as
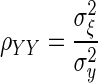 |
Then Spearman's formula for correcting attenuation in correlation is
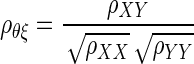 |
(A1) |
Relationship between regression-based approach and Spearman's correction
First, assume that 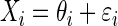 and consider the regression model
and consider the regression model 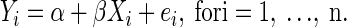 The covariance matrix of (X, Y) is
The covariance matrix of (X, Y) is
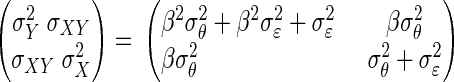 |
Let 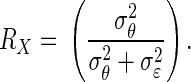 Clearly,
Clearly, 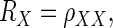 the reliability in Spearman's notation.
the reliability in Spearman's notation.
Suppose now that we are interested in finding the relationship between ρYθ, the true correlation, and ρXY, the attenuated one through the regression model. We have
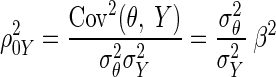 |
(A2) |
on the other hand
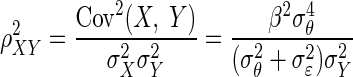 |
(A3) |
Substituting Eq. A2 in A3 gives
 |
One can easily expand this result to the case in which Yi = ξi + δi, where ξi = α + βXi. Note that
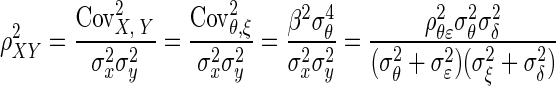 |
therefore
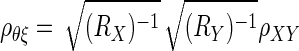 |
which is Spearman's formula.
Derivation of Formula 4
The derivation of σξ2 and to σδ2 is a simple application of the delta method (for details, see section 9.9 in Wasserman 2004.) Let X and Y be two random variables. Consider the function 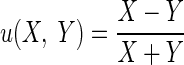 . We have
. We have 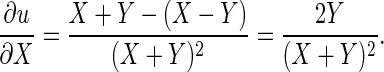
Similarly, 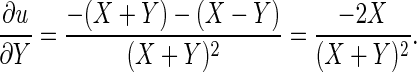 Let
Let 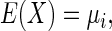 and E(Y) = μ2, Var(X) = σ12, and Var(Y) = σ22. Then assuming independence between X and Y, one can write
and E(Y) = μ2, Var(X) = σ12, and Var(Y) = σ22. Then assuming independence between X and Y, one can write 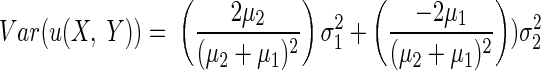
Normal hierarchical modeling in WinBugs
WinBugs is a freeware that can be downloaded from the BUGS Project website at http://www.mrc-bsu.cam.ac.uk/bugs/. The following provides a general way of coding hierarchical normal models as explained in the previous text (Bayesian Method).
model
{ for (i in 1 : N) (
theta {i, 1 : 2} dmnorm (mu []. signal [.])
for (j in 1 :T) (
Y [i. j]dnorm (theta [i, j]. se[i, j])
}
}
mu [1 : 2] dnorm (mean [], prec[,])
sigma [1 : 2, 1 : 2]dwish (r[,], 2)
R. inv [1:2,1:2]< -inverse (sigma [,])
}
In the preceding, N represents the number of objects (neurons), T = 2 is the dimensionality, Y[i, j] represents the experimental data. In our data analysis data analysis), Y[i, j] would be the ith index for the variable reward task (j = 1), and serial-order task (j = 2), theta[i,j] refers to the vector (θi,ξi), se[i,j] represents σɛ2 (j = 1), and σδ2 (j = 2) for the ith observation, mu[], and Sigma[,] are the mean vector μ, and the covariance matrix Σ, as shown in Bayesian model, respectively. Finally, mu[1:2], and R.inv[1:2,1:2] are the priors for the mean vector μ, and the covariance matrix Σ. To run this model, the data need to be formatted appropriately. As an example, to generate the posterior values in the lower panel of Fig. 1, the data need to be formatted as a 54 × 2 array. Additionally, an array of size 54 × 2 of se[i,j] values is needed to run the model.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- Behseta et al. 2005.Behseta S, Kass RE, Wallstrom G. Hierarchical models for assessing variability among functions. Biometrika 92: 419–434, 2005. [Google Scholar]
- Carroll et al. 2006.Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement Error in Nonlinear Models, A Modern Perspective. New York: Chapman and Hall, 2006.
- Carroll and Stefanski 1994.Carroll RJ Stefanski LA. Measurement error, instrumental variables and corrections for attenuation with application to meta-analysis. Stat Med 13: 1265–1282, 1994. [DOI] [PubMed] [Google Scholar]
- Casella and George 1992.Casella G, George EI. Explaining the Gibbs sampler. J Am Stat Assoc 46: 167–174, 1992. [Google Scholar]
- Charles 2005.Charles EP The correction for attenuation due to measurement error: clarifying concepts and creating confidence sets. Psychol Methods 10.2: 206–226, 2005. [DOI] [PubMed] [Google Scholar]
- DasGupta 2008.DasGupta A Asymptotic Theory of Statistics and Probability. New York: Springer-Verlag, 2008.
- Edwards et al. 2003.Edwards R, Xiao D, Keysers C, Foldiak P, Perrett D. Color sensitivity of cells responsive to complex stimuli in the temporal cortex. J Neurophysiol 90: 1245–1256, 2003. [DOI] [PubMed] [Google Scholar]
- Fisher 1924.Fisher RA On a distribution yielding the error functions of several well-known statistics. Proc Int Congr Math 2: 805–813, 1924. [Google Scholar]
- Frost and Thompson 2000.Frost C, Thompson SG. Correcting for regression dilution bias: comparison of methods for a single predictor variable. J R Statist Soc A 163: 173–189, 2000. [Google Scholar]
- Fuller 1987.Fuller WA Measurement Error Models. New York: Wiley, 1987.
- Gelman et al. 1995.Gelman A, Carlin JB, Stern H, Rubin DR. Bayesian Data Analysis. London: Chapman and Hall, 1995.
- Horwitz and Newsome 2001.Horwitz GD, Newsome WT. Target selection for saccadic eye movements: direction-selective visual responses in the superior colliculus. J Neurophysiol 86: 2527–2542, 2001. [DOI] [PubMed] [Google Scholar]
- Hunter and Schmidt 1990.Hunter JE Schmidt FL. Methods of Meta-Analysis. Newbury Park, CA: Sage, 1990.
- Kass and Natarajan 2006.Kass RE, Natarajan R. A default conjugate prior for variance components in generalized linear mixed models (comment on article by Browne and Draper). Bayesian Anal 1: 535–542, 2006. [Google Scholar]
- Olson et al. 2000.Olson CR, Gettner SN, Ventura V, Carta R, Kass RE. Neuronal activity in macaque supplementary eye field during planning of saccades in response to pattern and spatial cues. J Neurophysiol 84: 1369–1384, 2000. [DOI] [PubMed] [Google Scholar]
- Patterson and Thompson 1971.Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika 58: 545–554, 1971. [Google Scholar]
- Pearson 1904.Pearson K On the laws of inheritance in man. II. On the inheritance of mental and moral characters in man, and its comparison with the inheritance of physical characters. Biometrika 2: 131–159, 1904. [Google Scholar]
- Roesch and Olson 2003.Roesch MR, Olson CR. Impact of expected reward on neuronal activity in prefrontal cortex, frontal and supplementary eye fields and premotor cortex. J Neurophysiol 90: 1766–1789, 2003. [DOI] [PubMed] [Google Scholar]
- Spearman 1904.Spearman C The proof and measurement of association between two things. Am J Psychol 15: 72–101, 1904. [PubMed] [Google Scholar]
- Wasserman 2004.Wasserman LA All of Statistics. New York: Springer-Verlag, 2004.
- Wilks et al. 1995.Wilks GR, Richardson S, Spiegelhalter D. Markov Monte Carlo in Practice. London: Chapman and Hall, 1995.
- Winnie and Belfry 1982.Winnie PH, Belfry MJ. Interpretive problems when correcting for attenuation. J Educat Measure 19: 125–134, 1982. [Google Scholar]