Abstract
Yeast self-perpetuating amyloids (prions) provide a convenient model for studying the cellular control of highly ordered aggregates involved in mammalian protein assembly disorders. The very ability of an amyloid to propagate a prion state in yeast is determined by its interactions with the stress-inducible chaperone Hsp104. Prion formation and propagation are also influenced by other stress-related chaperones (Hsp70 and Hsp40), and by alterations of the protein trafficking and degradation networks. Some stress conditions induce prion formation or loss. It is proposed that prions arise as byproducts of the reversible assembly of highly ordered complexes, protecting certain proteins during unfavorable conditions.
Keywords: amyloid, protein aggregation, chaperone, heat shock protein, ubiquitin, Saccharomyces cerevisiae
Introduction
Transmissible spongiform encephalopathies (TSEs), also called prion diseases, attracted attention after the “mad cow” disease outbreak in Europe in the 1990s, although other examples of these diseases, such as sheep scrapie, human Creutzfeldt-Jacob disease, etc. were known long ago (for review, see [1]). A most unusual feature of these diseases is that infection is transmitted not by bacteria or viruses, but by protein-based infectious particles (termed prions). The “protein only” model of TSE transmission, accepted by most researchers in the field, postulates that a specific mammalian protein (named PrP) in an abnormal (“prion”) shape becomes an infectious agent as it acquires an ability to convert the normally folded host protein of the same sequence into a prion shape. According to one of the modifications of the prion model, the prion isoform of PrP represents an ordered aggregate that can proliferate by immobilizing the soluble PrP molecules [2]. Indeed, PrP generates fibrous β-rich ordered aggregates (called amyloids) in the brains of infected animals [1].
Amyloids formed by PrP resemble non-infectious amyloids or amyloid-like aggregates associated with some other diseases, including such neurodegenerative disorders as Alzheimer disease (AD), Huntington disease (HD), Parkinson disease (PD), etc. (for review, see [3]). Some of these diseases are caused by mutations, e. g. by expansions of a polyglutamine (poly-Q) stretch in a protein called huntingtin, in case of HD. Other diseases (e. g. most cases of AD) occur sporadically, so that triggering mechanisms remain unknown. Some amyloidoses (including AD) are clearly age-dependent, pointing to a potential connection between amyloid formation and aging. Therefore, the importance of these diseases grows dramatically with eradication of other diseases and increase of life expectancy in the human populations. It is worth mentioning that prion diseases and many other amyloidoses (including AD and HD) remain fatal and incurable thus far. Without any exaggeration, it is easy to forecast that AD and similar disorders have a potential of becoming the major cause of death for the next generations of humans.
Lower eukaryotes, such as yeast and other fungi, also contain self-perpetuating transmissible amyloids that possess prion-like properties (for review, see [4–6]). These amyloids manifest themselves as non-Mendelian elements that control specific traits, heritable in cell generations and infectious via cytoplasmic exchange. Although fungal amyloid-forming proteins are not homologous to mammalian PrP, they are also usually termed “prion proteins”. Fungal prions do not necessarily kill their carriers. However, recent data demonstrate that at least some fungal prions are pathogenic to a certain extent (reviewed in [4, 7]).
In the yeast Saccharomyces cerevisiae, several proteins have been proven to generate self-perpetuating amyloid-based prions. These include: 1) translational termination factor Sup35 (also called eRF3); 2) regulatory protein in the nitrogen metabolism pathway, Ure2; 3) protein of unknown function, Rnq1 (for review, see [4–7]). Prion forms of these proteins are termed [PSI+], [URE3] and [PIN+] (or [RNQ+]), respectively. Yeast prion proteins contain N-terminal or C-terminal regions, termed prion domains or PrDs, that are required and sufficient for prion formation and propagation, and are dispensable for the normal cellular function of a prion protein in cases when this function is known (for review, see [5, 6]). While known yeast prion proteins are not homologous to each other, they contain common sequence elements, some of which also resemble certain regions of mammalian amyloidogenic proteins. For example, all known yeast PrDs contain QN-rich stretches that are similar to the poly-Q stretch of mammalian huntingtin, and some yeast PrDs contain oligopeptide repeats (ORs) similar to those found in mammalian PrP (reviewed in [4–6]). Mechanisms of amyloid formation in yeast and other fungi appear to be very similar to those described in mammalian systems. Therefore, yeast prions provide easy and efficient experimental assays for studying the factors and conditions influencing amyloid formation and propagation.
As amyloids are protein aggregates, the question arises whether cellular defense systems aimed at protecting the cells from aggregation of stress-damaged proteins can also recognize amyloids. This review summarizes data, obtained in the yeast models, that implicate stress-related proteins as major modulators of amyloid formation and propagation in a eukaryotic cell.
Role of heat shock proteins in formation and propagation of the yeast prions
Role of Hsp104 in prion propagation
The first evidence connecting a component of the cellular stress defense system to a prion was obtained in the yeast model. A prion isoform of the yeast protein Sup35 was shown to be maintained only at certain levels of the chaperone protein Hsp104 [8]. If Hsp104 was inactivated or overproduced, the initially prion-containing culture ([PSI+]) produced cells containing only a non-prion isoform of Sup35 ([psi−]). Later, it was shown that intact Hsp104 is also required for the maintenance of all other known yeast prions, even though prion forms of the proteins other than Sup35 are not “cured” by Hsp104 overproduction (for review, see [9]).
Hsp104, a member of the evolutionarily conserved Clp/Hsp100 family and a yeast counterpart of bacterial ClpB, is a stress-inducible chaperone that functions as a homohexamer and promotes solubilization of the aggregated misfolded proteins and in this way, protects yeast cells from heat shock and some other environmental stresses (for review, see [9]). One possibility is that Hsp104 is required for “shearing” prion aggregates into smaller oligomeric “seeds”, initiating new rounds of prion propagation [10] (Fig. 1). Indeed, the immediate consequence of Hsp104 inactivation in the prion-containing cells is an increase in the size of Sup35 aggregates [11, 12]. Moreover, the prion isoforms of Sup35 that are maintained only at high levels of Hsp104 were either generated by genetic alterations of the Sup35 protein [13] or found among various prion “variants” (or “strains”) generated by the intact Sup35 protein [14], and shown in both cases to form abnormally large aggregates.
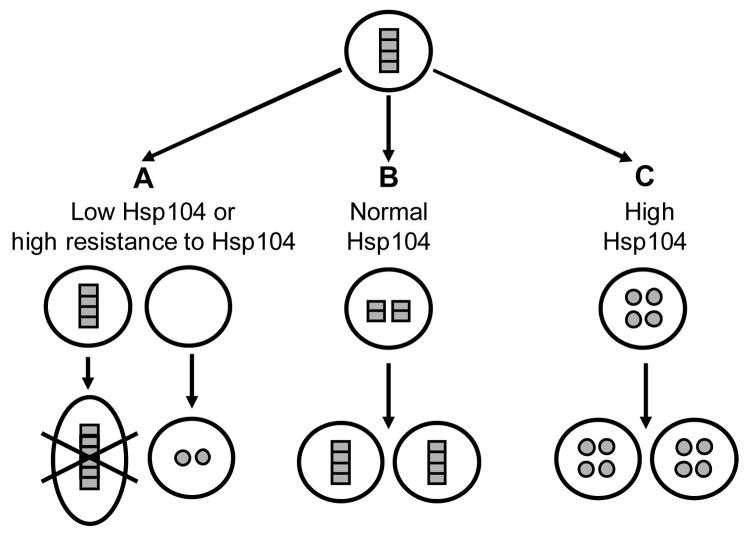
Figure 1. Role of Hsp104 in propagation of the prion form of Sup35.
Non-prion isoform (soluble monomer) is designated as a circle, prion isoform (unit of an amyloid polymer) - as a rectangle. A – At low levels (or activity) of Hsp104, or in case of the amyloid aggregate which is insensitive to Hsp104, a polymer stays in the mother cell and is growing in an uncontrollable fashion, possibly resulting in the eventual death of the polymer-containing cell. A daughter cell which does not get an amyloid “seed” after the cell division produces de novo synthesized protein in a non-prion form. B – At normal levels of Hsp104, initial polymer is being broken into oligomeric “seeds”, that are transmitted to both cells after cell division and initiate new rounds of prion propagation. C – At high levels of Hsp104, amyloid polymers are solubilized into monomers, that are converted into a non-prion form. Note that for the sake of simplicity, fate of only one initial polymer is followed on each part of the figure (A, B or C). In a reality, each prion-bearing cell usually contains a number of propagating polymers. Therefore, both prion loss and mother cell death depicted in part A should normally occur only after several cell divisions in the conditions where disaggregation is impaired. See text for details and references.
On the other hand, results of the direct biochemical experiments aimed at studying interactions between Hsp104 and prion proteins such as Sup35 remain somewhat contradictory. While some data indicate that Hsp104 can indeed break Sup35-based aggregates in vitro producing oligomeric “seeds” [15], other results disagree [16] or suggest that Hsp104 only does this with the help of the other chaperones [17]. There is evidence that Hsp104 somewhat promotes initial aggregation of the yeast prion proteins in vitro [15], an observation that has not so far been confirmed in vivo.
A number of the mutant derivatives of Hsp104 have been checked for their effects on thermotolerance and prion maintenance (reviewed in [9]). In most cases, mutations affecting thermotolerance also affected prion maintenance, and vice versa. However, several exceptions of this rule were described (e. g., [18]). Interestingly, the majority of Hsp104 mutations affecting maintenance of the Sup35 prion but not thermotolerance arise from substitutions of amino acids located along the lateral channel of the Hsp104 hexamer [19]. This suggests that a lateral channel may play a specific role in the interaction of the Hsp104 hexamer with prion aggregates.
Regions of Sup35 involved in interactions with Hsp104
Despite clear and indisputable evidence confirming the crucial role of Hsp104 in prion propagation, numerous attempts to prove physical interactions between Hsp104 and prion proteins such as Sup35 failed (for example, see [20]), while in vitro results were contradictory (for examples, see [15–17]). It is likely that Hsp104 specifically recognizes only the aggregated form of a prion protein, and its interaction with such an aggregate is of a transient nature, so that it breaks the aggregate and dissociates from it. The question arises whether Hsp104 recognizes any specific region of a prion protein within the aggregate. Based on the considerable experimental evidence, it has been proposed that PrDs of the yeast prion proteins can be subdivided into “aggregation” and “propagation” modules (Fig. 2; for review, see [6]). In case of Sup35, the “aggregation” module appears to correspond to the N-terminal QN-rich stretch, while the adjacent region of oligopeptide repeats is required for the efficient propagation of the prion state in vivo [21]. As prion propagation is achieved via breakage of aggregates by Hsp104 (see above), the simplest possibility appeared to be that the repetitive structure is recognized by Hsp104. However, some yeast prions (e. g. Ure2) do not have repeats. Moreover, reshuffling of the Sup35 PrD sequence that eliminated the repetitive pattern of its organization did not eliminate the prion-propagating ability [22]. Apparently, even if the region of repeats is indeed involved in interaction with Hsp104, its repetitive organization is dispensable for such an interaction. An alternative explanation could be that the “propagation” module influences some patterns of aggregates that control accessibility by Hsp104, but does not physically interact with a chaperone on its own. Another region of Sup35 suspected to either directly or indirectly interact with Hsp104 is the middle (M) region. This region, containing a high concentration of charged residues, is required for neither normal cellular function of the Sup35 protein (that is, translational termination) nor prion formation and maintenance (reviewed in [5, 6]). However, recent studies indicate that at least some residues of Sup35M are involved in interactions within the amyloid fiber [23]. This agrees with the observation that certain alterations of the M region change patterns of the Hsp104 dependence [24]. Moreover, prions formed in S. cerevisiae by the Sup35 protein of the closely related species S. paradoxus that contains a divergent (87% of amino acid identity) M region possess a propagation defect, which is eliminated in case if the M region of S. paradoxus is substituted by the M region of S. cerevisiae [25]. As of now, the exact target region of Sup35 recognized by Hsp104 remains unknown.
Figure 2. Structural and functional organization of Sup35 protein and Sup35 prion aggregates.
A – Structural and functional organization of the Saccharomyces cerevisiae Sup35 protein. Sup35N, Sup35M and Sup35C refer to the N-proximal, middle and C-proximal regions, respectively. Sup35N is a prion domain, while Sup35C is a release factor domain responsible for the major cellular function of Sup35 (termination of translation). QN – gluatamine/asparagine-rich stretch, OR – oligopeptide repeats. Numbers correspond to amino acid positions. N/M boundary is arbitrarily chosen, as different papers located it at different positions between the amino acid residues 113 and 137 (for review, see [6]). B – Structural organization of the Sup35 amyloid fiber. The axis of a fiber is kept together by intermolecular cross-β interactions between the prion domains, while Sup35C regions are exposed on the side of a fiber. An Hsp104 hexamer invades the axis and initiates breakage of a fiber, presumably via interactions that involve a lateral channel of the hexamer. It is not yet clear which exact parts of Sup35N (and possibly Sup35M) are involved in axis formation and/or binding of Hsp104 (see text for more details).
Effects of the Hsp70 and Hsp40 proteins on yeast prions
The Hsp70 and Hsp40 chaperones assist Hsp104 in protecting yeast cells from environmental stresses: while Hsp104 is thought to promote disaggregation and solubilization of the stress-damaged proteins, Hsp70 with the help of Hsp40 is thought to refold these proteins back into their native state [26]. Hsp70 is also shown to play a crucial role in the propagation of yeast prions, specifically of the prion forms of Sup35 and Ure2 (for detailed review, see [9]). Interestingly, different members of the yeast Hsp70 family have different effects on the prion form of Sup35. The proteins of the stress-inducible Ssa subfamily generally aid in propagation of the Sup35 prion, while proteins of the constitutively expressed Ssb subfamily manifest themselves as prion antagonists. These differences are determined, in a significant part, by the peptide-binding domains of these chaperones [20]. Both Ssa and Ssb are shown to interact with Sup35 directly.
Despite significant amount of mutational analysis data, accumulated especially in D. Masison lab, molecular mechanism of the Hsp70 effect on yeast prions remains unclear (for recent models, see [9]). Even less is known about the role of Hsp40 proteins, usually acting as co-chaperones of Hsp70s. Two Hsp40 co-chaperones of Ssa, Ydj1 and Sis1, exhibit differential effects on prion forms of some yeast proteins, such as Rnq1 [27] and Ure2 [28], as well as on polyglutamine aggregates produced in yeast [29].
Overall, existing evidence indicates that proteins of the same complex that is involved in protection of the yeast cells against aggregates of the stress-damaged proteins (Hsp104, Hsp70 and Hsp40) also play a crucial role in propagation of prion aggregates. It appears that the balance between the components of this complex is a key regulator of prion propagation, as increased production of Hsp70-Ssa can ameliorate the ability of overproduced Hsp104 to eliminate the Sup35 prion [20, 30]. Thus, prions employ chaperone machinery of a yeast cell for the purpose of their own propagation, and alteration of the chaperone balance can modulate acquisition or loss of the prion state. As Hsp levels and balance are regulated by environmental and physiological stresses, these data provide a molecular mechanism connecting stress to amyloid-like aggregation.
Yeast prions and proteolytic pathways
As an alternative to refolding, cells can eliminate stress-damaged proteins via proteolytic degradation. Two major proteolytic systems are responsible for protein degradation in a eukaryotic cell (for review, see [31]): 1) ubiquitin-proteasome system (UPS); 2) lysosome, or (in yeast) vacuole. Cytoplasmic proteins are usually degraded by UPS, but can also be directed to the lysosome/vacuole via starvation-induced or stress-induced process called autophagy [32]. Misfolded proteins are targeted for proteasomal degradation via covalent attachment of “tails” composed of a polymerized small protein called ubiquitin. Mono- or polyubiquitin attachment is also involved in targeting some proteins for lysosomal/vacuolar degradation, as well as in regulation of protein activity and directing proteins to some non-degradative fates. Efficiency and specificity of ubiquitination is controlled by a variety of ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3). In yeast, the vast majority of protein ubiquitination during stress is mediated by the related ubiquitin-conjugating enzymes Ubc4 and Ubc5 [33].
Little is known about degradation of prions and other amyloid aggregates in eukaryotic cells. It is unlikely that the proteasome is capable of degrading huge protein aggregates. Moreover, some amyloidogenic proteins such as polyglutamines are not efficiently degraded by proteasomes even in a monomeric state [34]. On the other hand, accumulation of amyloidogenic proteins overwhelms UPS, that induces formation of so-called aggresomes, huge complexes assembled with the help of cytoskeletal structure and apparently sequestering ubiquitin and other UPS components [35]. Autophagy appears to be involved in aggresome degradation when UPS function is inhibited [36].
In yeast, alterations of some UPS components, specifically deletion of the gene coding for the major ubiquitin-conjugating enzyme Ubc4, decrease the loss of a prion state of the Sup35 protein in the presence of excess Hsp104 and increase spontaneous de novo formation of the Sup35 prion [37]. Once again, the same UPS component that plays a major role during environmental stress also modulates prion behavior. Interestingly, deletion of the gene coding for the enzyme Ubc5, that is functionally similar to Ubc4, does not appear to increase spontaneous prion formation. It is not clear whether this difference reflects different functional specificities of Ubc4 and Ubc5 or different modes of their expression: it is possible that effects on the prion are most pronounced at the stage when Ubc4 rather than Ubc5 is predominantly present.
Mechanism of the UPS effect on a yeast prion remains unclear. The simplest explanation would be that impairment of ubiquitin-mediated degradation increases accumulation of misfolded protein which is directed to prion aggregates. However, evidence for the direct ubiquitination and UPS-directed degradation of Sup35 is lacking thus far. It is therefore possible that Ubc4 and other UPS components modulate prion formation and maintenance via ubiquitination of some auxillary components that influence Sup35 protein. Indeed, some of the chaperones involved in prion propagation (for example, some members of the Hsp70 family) as well as some other prion proteins (for example, Rnq1) are found among ubiquitination targets [38], and effect of the ubc4 deletion on de novo formation of the Sup35 prion depends on Rnq1 [37]. Another possibility is that UPS defect induces “aggresome” formation that may in fact promote prion aggregation. Aggresome-like complexes of Sup35 co-localized with some components of the cytoskeletal networks are indeed identified in the yeast cells [39], although they appear to interact with cortical actin networks rather than with microtubules implicated in the assembly of mammalian aggresomes. Moreover, formation of some types of these cytologically detectable Sup35 inclusions is apparently elevated in the ubc4 deletion strains [37]. It is worth noting that two explanations of the Ubc4 role in prion formation and maintenance that were mentioned above are not mutually exclusive. Further experiments are needed to decipher the molecular pathway connecting UPS to yeast prions, as well as to determine whether and how prion aggregates are degraded in the yeast cell.
Prions and environmental stresses
As components of the cellular stress-defense systems modulate prion formation and propagation in yeast, the question arises whether environmental stress can lead to prion appearance or elimination. Detailed study performed in B. Cox’s lab (reviewed in [40]) indicates that at least the prion form of Sup35 can be eliminated (“cured”) by some environmental stresses, such as severe heat shock, ethanol or methanol stress, and some osmotic stressors. Moreover, mutational alteration of the Hsp70 chaperone Ssa1 increases susceptibility of the Sup35 prion to heat shock [41]. Our recent data1 demonstrate that efficient elimination of the Sup35 prion is observed only during short-term heat shock, while longer exposure to the high temperature treatment results in prion recovery. Moreover, both prion elimination and prion recovery coincide, respectively, with alteration and restoration of the balance between the major chaperones shown to be involved in prion propagation. These data indicate that chaperone balance regulates maintenance of the Sup35 prion not only during normal growth but also in the stress conditions. Whether this is applicable to other yeast prions, remains to be seen.
Less is known about environmental conditions that can promote de novo prion formation. One treatment clearly demonstrated thus far to do so is long-term storage of the starving yeast culture in the refrigerator, that induces generation of the prion forms of Ure2, Rnq1 and Sup35 (reviewed in [5]). Molecular mechanisms of this phenomenon have not been investigated. Starving non-dividing cells are likely to accumulate increased amounts of aggregated proteins, while modification of the chaperone expression patterns at low temperature may provide intracellular environment that favors aggregate conversion into self-perpetuating prions. However, it is still to be determined which chaperones are involved in the process in this specific case. It is known that depletion of the Ssb chaperone of Hsp70 family [42] or increased levels of the Ssa proteins of the same family [20] facilitate formation of the Sup35 prion. Also, alterations of heat shock factor, that change patterns of Hsp regulation, exhibit distinctive effects on the de novo formation of the Sup35 prion [43]. However, it is still unknown whether (and which) chaperones contribute to the prion-inducing effect of low temperature.
There was also a report of increased formation of the Ure2 prion in the presence of the antibiotic G418 that induces translational errors [44]. Once again, it remains to be seen whether this is a direct consequence of the accumulation of a misfolded protein in result of mistranslation, or this effect is mediated by stress-related proteins induced in response to it.
Parallels and differences between yeast and animal amyloids and inclusions
As yeast prion propagation is controlled by prion proteins and influenced by environmental stresses, the question arises whether the same is true for prions and other amyloid-like aggregates found in mammals and other animals. Yeast chaperones involved in propagation of the endogenous yeast prions, specifically members of the Hsp40 family (Ydj1 and Sis1) and some mutant derivatives of Hsp104, are also shown to influence aggregation and toxicity of mammalian polyglutamines in yeast, as demonstrated for the N-terminal fragment of human huntingtin, containing the expansion of the polyglutamine stretch and expressed in the yeast strain containing an endogenous QN-rich prion, apparently providing the initial nucleus for huntingtin aggregation [29]. Moreover, some effects of Sis1 and Ydj1 observed in yeast parallel the effects of their mammalian orthologs, Hdj1 and Hdj2 respectively, detected in the animal models for polyglutamine disorders [45, 46]. Relationship between stress-related proteins and polyglutamine aggregation is also established in the nematode Caenorhabditis elegans (for review, see [47]). Remarkably, introduction of yeast Hsp104 can counteract polyglutamine aggregation in nematode [48]. These data confirm that at least some patterns of interactions between chaperones and amyloids or amyloid-like inclusions are conserved between yeast and animals, and further signify importance of the yeast-based model for studying the chaperone effects on amyloidogenic and prion-forming proteins.
On the other hand, animal genomes sequenced to date lack the ortholog of yeast Hsp104, even though such orthologs are found in plants and even bacteria. It is not yet clear which protein plays the role of Hsp104 in protection of mammalian cells against aggregates of stress-damaged proteins. Although alterations in expression and distribution of some cytoplasmic heat shock proteins, e. g. mammalian members of the Hsp70 family, are associated with the infection by PrP prion in the mammalian systems (for review, see [49]), it remains to be determined whether these chaperones influence prion formation and/or propagation of the prion form of PrP. The matter is further complicated by the fact that PrP is, at least in its prion form, an extracellular protein. It is therefore possible that other chaperone proteins, expressed in compartments that are associated with the secretory or endocytic pathways, take Hsp104’s place in “breaking” the mammalian prion aggregates and converting them to infectious prion units. However, recent data also indicate that alterations in Hsp levels are associated with toxicity of certain intracellular amyloids, for example in the case of Parkinson disease (for review, see [49]). Moreover, one should note that mammalian genomes contain a significant number of proteins with QN-rich regions similar to yeast PrDs. It can not be ruled out that some of these proteins can form intracellular prions, controlled by the cytoplasmic chaperones in a yeast-like fashion.
Stress and possible biological roles of prions
Intimate involvement of some stress-related proteins in prion formation and/or propagation, as well as effects of environmental stresses on yeast prions suggest a possible explanation for the conservation of prion domains in evolution. Indeed, the N-proximal domain of Sup35 (which behaves as PrD in yeast) is present in the majority of organisms studied, despite its apparent dispensability for the major cellular function of Sup35 in termination of translation (reviewed in [7]). Moreover, although PrD region evolves faster than the functional part of Sup35 protein, PrD amino acid sequence still remains under the purifying selection pressure, indicating that this portion of Sup35 plays a certain adaptive role. Although it has been suggested that the prion form of Sup35, which is defective in termination, could play an adaptive role by allowing readthrough of open reading frames interrupted by nonsense-mutations, such a model was challenged by the data indicating that Sup35 prion can not be found in the natural populations of yeast (for review, see [4, 7]). Moreover, such a mechanism could not apply to other yeast prions that have nothing to do with translation. On the other hand, it is possible that adaptive functions are performed by metastable prion variants that persist only in certain conditions and are converted back to a non-prion form after conditions are changed. Recent data provide a proof of existence of the metastable variant of the Sup35 prion that can be maintained only at high levels of the Hsp104 chaperone [14]. One could suggest that metastable or reversible aggregates of prion-like type are used to preserve certain proteins through unfavorable periods, so that such proteins could be quickly activated after conditions change back to normal and regular cellular metabolism is resumed. Indeed, such a mechanism is demonstrated in mammalian cells for the so-called stress granules, huge complexes containing mRNAs associated with the small ribosomal subunits and some translation initiation factors that are transiently assembled during stresses (such as heat shock) in order to protect these components from stress-induced damage. Remarkably, protein TIA-1 involved in assembly of stress granules contains a QN-rich domain which is needed for the granule formation, resembles PrD of Sup35 and can be functionally substituted by attachment of Sup35 PrD to the TIA-1 protein lacking this domain [50]. As Sup35 is an important protein that also has to be present in the active form immediately after the stress is over, it seems logical to propose that reversible formation of the ordered aggregates of Sup35, mediated by its PrD, could protect this protein from damage and destruction during stress conditions. Both aggregation and subsequent disaggregation of Sup35 would have to be mediated by stress-regulated chaperones, that could explain emergence of the “prion propagation machinery”, composed of Hsp104, Hsp70 and Hsp40, in the yeast cell, and of equivalents of this machinery (if found) in other eukaryotic cells. Increased prion formation in the starving cells kept at low temperature could then be due to increased “protective” aggregation of Sup35 in these conditions. In such a model, stably inherited (irreversible) prions would represent “diseases” originated as rare by-products of generally reversible protective aggregation (Fig. 3). Such a mechanism could also apply to other prion-forming proteins, assuming they are playing certain roles (not necessarily essential roles) in the cells during (or immediately following) recovery from stress.
Figure 3. Model for the biological role of prion-like aggregation.
At normal conditions, a prion-forming protein exists as a soluble monomer and perform its normal cellular function. In certain unfavorable conditions, ordered protein complexes are formed via interactions between prion domains (designated as filled ovals). Formation of these complexes, which is possibly assisted by some Hsps and/or cytoskeletal assembly proteins (CSK), downregulates the enzymatic function of a prion-forming protein and protects this protein from damage and destruction. Upon return to normal conditions, complexes are solubilized by Hsp104, so that a prion-forming protein is converted back into the active state. However, prion polymers can be occasionally generated as by-products of such protective aggregation. These prion polymers are no longer solubilized to monomers at normal levels of Hsp104. Instead, they undergo repetitive breakage and propagation cycles as shown in more detail on Fig. 1.
Acknowledgments
I thank G. Newnam for help in preparing the manuscript for publication, as well as T. Chernova and N. Romanova for the critical reading of manuscript and helpful suggestions. This work was supported by grants from National Institutes of Health (R01GM5763), National Science Foundation (MCB-0614772) and Hereditary Disease Foundation.
Footnotes
G. Newnam and Y. Chernoff, in preparation
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lansbury PT, Caughey B. The chemistry of scrapie infection: implications of the ‘ice 9’ metaphor. Chem Biol. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 3.Thompson AJ, Barrow CJ. Protein conformational misfolding and amyloid formation: characteristics of a new class of disorders that include Alzheimer’s and Prion diseases. Curr Med Chem. 2002;9:1751–1762. doi: 10.2174/0929867023369123. [DOI] [PubMed] [Google Scholar]
- 4.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T, Engel A, McCann L, Kryndushkin D. Yeast prions: evolution of the prion concept. In: Chernoff YO, editor. Protein-Based Inheritance. Austin: Landes Bioscience; 2007. [Epub http://eurekah.com/chapter/3051] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernoff YO. Mutation processes at the protein level: is Lamarck back? Mutation Res. 2001;488:39–64. doi: 10.1016/s1383-5742(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 6.Chernoff YO. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Current Opinion in Chemical Biology. 2004;8:665–671. doi: 10.1016/j.cbpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Inge-Vechtomov SG, Zhouravleva GA, Chernoff YO. Biological roles of prion domains. In: Chernoff YO, editor. Protein-Based Inheritance. Austin: Landes Bioscience; 2007. [Epub http://eurekah.com/chapter/3104] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 9.Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and non-prion aggregates. In: Chernoff YO, editor. Protein-Based Inheritance. Austin: Landes Bioscience; 2007. [Epub http://eurekah.com/chapter/3242] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kushnirov VV, Ter-Avanesyan MD. Structure and replication of yeast prions. Cell. 1998;10:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 11.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 13.Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new seeds. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borchsenius AS, Müller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–29. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 15.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;4:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J Biol Chem. 2004;279:52319–52323. doi: 10.1074/jbc.M408159200. [DOI] [PubMed] [Google Scholar]
- 17.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi A, Hara H, Kurahashi H, Nakamura Y. A systematic evaluation of the function of the protein-remodeling factor Hsp104 in [PSI+] prion propagation in S. cerevisiae by comprehensive chromosomal mutations. Prion. 2007;1:69–77. doi: 10.4161/pri.1.1.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:e86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci USA. 2002;99:16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci USA. 2007;104:2791–2796. doi: 10.1073/pnas.0611158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:1–20. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 27.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;23:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 30.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf DH. From lysosome to proteasome: the power of yeast in the dissection of proteinase function in cellular regulation and waste disposal. Cell Mol Life Sci. 2004;61:1601–1614. doi: 10.1007/s00018-004-4134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalfan WA, Klionsky DJ. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr Opin Cell Biol. 2002;14:468–475. doi: 10.1016/s0955-0674(02)00343-5. [DOI] [PubMed] [Google Scholar]
- 33.Sommer T, Seufert W. Genetic analysis of ubiquitin-dependent protein degradation. Experientia. 1992;48:172–178. doi: 10.1007/BF01923510. [DOI] [PubMed] [Google Scholar]
- 34.Venkatraman P, Wetzel R, Tanaka Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 35.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 36.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. Effects of the ubiquitin system alterations on the de novo formation and loss of a yeast prion. J Biol Chem. 2007;282:3004–3013. doi: 10.1074/jbc.M609597200. [DOI] [PubMed] [Google Scholar]
- 38.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 39.Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 41.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a “protein mutator” in yeast: Role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park KW, Hahn JS, Fan Q, Thiele DJ, Li L. De novo appearance and “strain” formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics. 2006;173:35–47. doi: 10.1534/genetics.105.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatin I, Bidou L, Cullin C, Rousset JP. Translational errors as an early event in prion conversion. Cell Mol Biol (Noisy-le-grand) 2001;47:OL23–8. Online Pub. [PubMed] [Google Scholar]
- 45.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 46.Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, Rubinsztein DC. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington’s disease. Proc Natl Acad Sci USA. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brignull HR, Morley JF, Morimoto RI. The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv Exp Med Biol. 2007;594:167–189. doi: 10.1007/978-0-387-39975-1_15. [DOI] [PubMed] [Google Scholar]
- 48.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winklhofer KF, Tatzelt J. The role of chaperones in Parkinson’s disease and prion diseases. Handb Exp Pharmacol. 2006;172:221–258. doi: 10.1007/3-540-29717-0_10. [DOI] [PubMed] [Google Scholar]
- 50.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]