Abstract
Common variants at only two loci, FTO and MC4R, have been reproducibly associated with body mass index (BMI) in humans. To identify additional loci, we conducted meta-analysis of 15 genome-wide association studies for BMI (n > 32,000) and followed up top signals in 14 additional cohorts (n > 59,000). We strongly confirm FTO and MC4R and identify six additional loci (P < 5 × 10−8): TMEM18, KCTD15, GNPDA2, SH2B1, MTCH2 and NEGR1 (where a 45-kb deletion polymorphism is a candidate causal variant). Several of the likely causal genes are highly expressed or known to act in the central nervous system (CNS), emphasizing, as in rare monogenic forms of obesity, the role of the CNS in predisposition to obesity.
Obesity is a major public health problem, resulting in increased morbidity and mortality and severe economic burdens on health-care systems1,2. Excessive energy intake and diminished physical activity contribute to the increasing prevalence of obesity, but genetic factors strongly modulate the impact of the modern environment on each individual. Indeed, family and twin studies have shown that genetic factors account for 40–70% of the population variation in BMI3,4. BMI is the most commonly used quantitative measure of adiposity, and adults with high values of BMI (>30 kg/m2) are termed obese.
Until recently, genetic variants known to influence BMI were largely restricted to mutations in several genes that cause rare, often severe monogenic syndromes with obesity as the main feature5. Mutations in these genes are thought to act through the CNS, and in particular the hypothalamus, to influence energy balance and appetite, thereby leading to obesity. However, it is not known whether genetic variation in similar pathways is also relevant to the common form of obesity and population variation in BMI.
In the past year, large-scale searches for genetic determinants of BMI revealed previously unreported associations with common variants at two loci, FTO and MC4R6-10. Common variants at these loci are associated with modest effects on BMI (0.2–0.4 kg/m2 per allele) that translate into odds ratios of 1.1–1.3 for obesity (defined as BMI ≥ 30 kg/m2)6-10. Common variation in PCSK1 has been strongly associated with the risk of extreme obesity11, but this association has not yet been independently replicated.
Together, common variants at FTO and MC4R and rare variants known to cause obesity explain only a small fraction of the inherited contribution to population variation in BMI. To expedite the identification of alleles associated with variation in BMI, obesity and other anthropometric traits, we formed the GIANT (Genetic Investigation of ANthropometric Traits) consortium to facilitate large-scale meta-analysis of data from multiple genome-wide association studies (GWAS). Here, we report a meta-analysis of 15 GWAS totaling 32,387 individuals and test for association between BMI and ~2.4 million genotyped or imputed SNPs. We then follow up 35 SNPs drawn from the most significantly associated loci by a combination of de novo genotyping in up to 45,018 additional individuals and analysis of these SNPs in another 14,064 individuals already genotyped as part of other GWAS. These studies show that variants at six previously unreported loci in or near TMEM18, KCTD15, SH2B1, MTCH2, GNPDA2 and NEGR1 are reproducibly associated with BMI.
RESULTS
Initial meta-analysis of GWAS studies of BMI (stage 1)
We carried out a GWA meta-analysis of a total of 32,387 individuals of European ancestry from 15 cohorts of 1,094 to 5,433 individuals using two parallel analytic strategies (Supplementary Fig. 1 and Supplementary Tables 1–3 online). First, we carried out a weighted z-score–based meta-analysis combining P values from cohort-specific analysis strategies. Second, we also performed an inverse-variance meta-analysis using regression coefficients and their standard errors obtained by applying a uniform analysis strategy across all studies. The results for these two strategies were highly congruent (Supplementary Fig. 2 online). Here we report results of the weighted P value analysis, as it was completed first and used to select SNPs for follow-up genotyping.
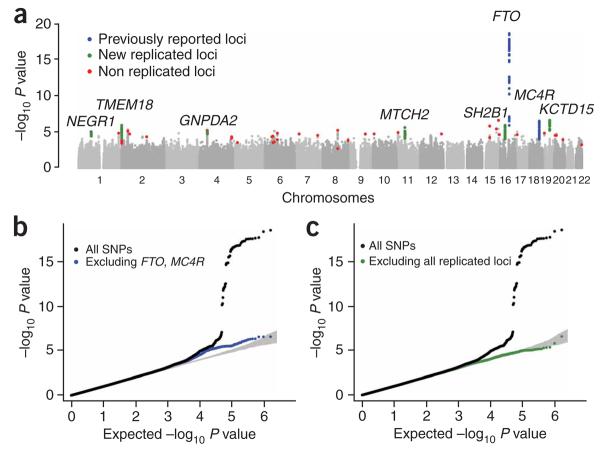
SNPs that reached P < 5 × 10−8 (a threshold that corresponds to P < 0.05 after adjusting for ~1 million independent tests) in this stage 1 analysis all mapped within the FTO gene (association peak at rs1421085, P = 2.6 × 10−19), were in linkage disequilibrium (LD) with each other (r2 > 0.51), and strongly confirm previous reports of association at this locus6-8. A locus located near MC4R (rs17782313, P = 3.9 × 10−7) and recently associated with BMI9,10 was the fourth most significant region in the stage 1 data (Fig. 1). Even after excluding SNPs in these two established BMI loci, we observed an excess of SNPs with small P values compared to chance expectations, suggesting that some of the remaining loci with strong but not definitive evidence of association in stage 1 are truly associated with BMI (Fig. 1b).
Figure 1.
Genome-wide association results for GIANT (stage 1). (a) Manhattan plot showing the significance of association of all SNPs in the stage 1 GIANT meta-analysis with BMI. SNPs are plotted on the x axis according to their position on each chromosome; association with BMI is indicated on the y axis (as −log10 P value). SNPs previously reported to show association with BMI are shown in blue, signals examined but not confirmed in stage 2 samples are shown in red and the six new regions described here are highlighted in green. (b) Quantile-quantile plot of SNPs after stage 1 GIANT meta-analysis (black) and after removal of any SNPs within 500 kb of FTO- or MC4R-associated SNPs (blue). (c) Quantile-quantile plot of SNPs in the stage 1 GIANT meta-analysis for association with BMI (black) and after removal any SNPs surrounding FTO, MC4R and the six new loci reported here (green).
Additional analysis of the strongest associations (stage 2)
To validate potential associations with BMI, we designed a pool of 35 variants for further genotyping, drawn from among the most strongly associated independent loci (for technical reasons, these SNPs do not correspond perfectly to the top 35 loci; see Methods). We genotyped these SNPs in up to 45,018 additional individuals of European ancestry from nine stage 2 samples (Supplementary Fig. 1, Supplementary Tables 1 and 4 and Supplementary Note online). We also obtained in silico association results for these SNPs from five BMI GWAS on 14,064 additional individuals of European ancestry (Supplementary Fig. 1, Supplementary Tables 1 and 4 and Supplementary Note). Meta-analysis of these stage 2 results combined with stage 1 data revealed SNPs from five previously unreported loci near TMEM18, KCTD15, SH2B1, MTCH2 and GNPDA2 that are strongly associated with BMI (P < 5 × 10−8; Table 1, Fig. 2 and Supplementary Table 5 online). Two additional loci, represented by rs2815752 (near NEGR1) and rs10769908 (near STK33) had supporting evidence in stage 2 samples but did not reach the P < 5 × 10−8 threshold (P = 6.0 × 10−8 and P = 1.3 × 10−6, respectively). Among these two, rs2815752 also showed a highly significant independent association with severe obesity in a pediatric cohort (P = 2.2 × 10−7; Supplementary Table 6 online), strongly suggesting that this variant represents a sixth newly discovered locus influencing BMI. For each of the six loci, multiple SNPs showed highly significant association in the stage 1 data (Fig. 2), and the associations were observed across multiple cohorts genotyped on different platforms (Supplementary Table 7 online), suggesting that idiosyncratic genotyping artifacts are unlikely to explain our results. Furthermore, the consistent association signals across different European-ancestry samples, each with low genomic control inflation factors (Supplementary Table 3), also suggest that population structure is unlikely to account for these associations. Finally, five of the six associated variants (near TMEM18, KCTD15, SH2B1, MTCH2 and NEGR1, but not GNPDA2) had Illumina proxies in high LD (r2 > 0.66) with our best SNPs that were included in an independent GWAS by Thorleifsson et al.12; for all five, they observed confirmatory evidence of association with BMI (Table 1), providing strong validation of these newly discovered associations.
Table 1.
SNPs with genome-wide significant evidence for association with BMI
| SNP | Nearby gene |
Chr. | Position (bp) |
Alleles |
Frequency of effect allele (%) |
Per-allele change in BMI (kg/m2) beta (95% CI) |
Explained variance (%) |
Stage 1 |
Stage 2 |
Stage 1 + 2 |
With deCODE |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect Other | n | P | n | P | n | P | n | P | ||||||||
| rs9939609a | FTO | 16 | 52378028 | A | T | 41 | 0.33 (0.27, 0.39) |
0.34 | 32,329 | 6.3E–17 | 51,055 | 3.3E–36 | 83,384 | 4.3E–51 | 113,204 | 4.9E–74 |
| rs6548238 | TMEM18 | 2 | 624905 | C | T | 84 | 0.26 (0.19, 0.34) |
0.13 | 32,256 | 1.2E–06 | 52,567 | 1.6E–13 | 84,823 | 1.4E–18 | 114,643 | 3.2E–26 |
| rs17782313 | MC4R | 18 | 56002077 | C | T | 21 | 0.20 (0.12, 0.28) |
0.10 | 32,385 | 3.9E–07 | 48,362 | 1.9E–12 | 80,747 | 4.6E–18 | 110,567 | 1.1E–20 |
| rs10938397 | GNPDA2 | 4 | 45023455 | G | A | 45 | 0.19 (0.13, 0.25) |
0.13 | 32,387 | 1.0E–05 | 49,371 | 4.5E–12 | 81,758 | 3.4E–16 | n.d. | n.d. |
| rs7498665a | SH2B1 | 16 | 28790742 | G | A | 41 | 0.15 (0.08, 0.21) |
0.08 | 32,361 | 5.4E–06 | 54,316 | 1.7E–06 | 86,677 | 5.1E–11 | 116,497 | 2.2E–14 |
| rs10838738 | MTCH2 | 11 | 47619625 | G | A | 34 | 0.07 (0.01, 0.13) |
0.02 | 32,387 | 7.1E–06 | 48,530 | 9.6E–05 | 80,917 | 4.6E–09 | 110,737 | 1.9E–11 |
| rs11084753 | KCTD15 | 19 | 39013977 | G | A | 67 | 0.06 (−0.01, 0.13) |
0.01 | 32,335 | 2.6E–07 | 39,371 | 4.1E–03 | 71,706 | 2.3E–08 | 101,526 | 4.5E–12 |
| rs2815752a | NEGR1 | 1 | 72524461 | A | G | 62 | 0.10 (0.04, 0.16) |
0.03 | 32,387 | 9.3E–06 | 51,112 | 6.8E–04 | 83,499 | 6.0E–08 | 113,319 | 1.0E–12 |
Nearby gene is the gene closest to the SNP in column 1. Chromosome and position (in Build 35) of SNPs used in analyses are listed. ‘Effect’ allele, BMI-increasing allele; ‘Other’ allele, BMI-decreasing allele. Frequency of effect allele, weighted frequency of the effect allele (and the proxy SNP for those loci where more than one SNP was typed) in genotyped population-based cohorts (EPIC, FINRISK97, BPPP, METSIM). Per-allele change in BMI, the change in BMI per increase in the effect allele (with 95% confidence interval) from stage 2 genotyped population-based cohorts. Explained variance, the average of the variance explained for the SNP used for analysis in stage 2 genotyped population-based cohorts. Stage 1, data from stage 1 GIANT meta-analysis; stage 2; data from meta-analysis of genotyped and in silico studies; stage 1 + 2, data from stage 1 GIANT analyzed with genotyped and in silico studies from stage 2. With deCODE, data from meta-analysis of stages 1 and 2 and data from ref. 12. n.d., not determined because of unavailability of good Illumina proxy for rs10938397.
SNPs rs1121980 (FTO r2 = 0.96 to rs9939609), rs9931989 (SH2B1 r2 = 0.68 to rs7498665) and rs2568958 (NEGR1 r2 = 1 to rs2815752) were used in some stage 2 samples.
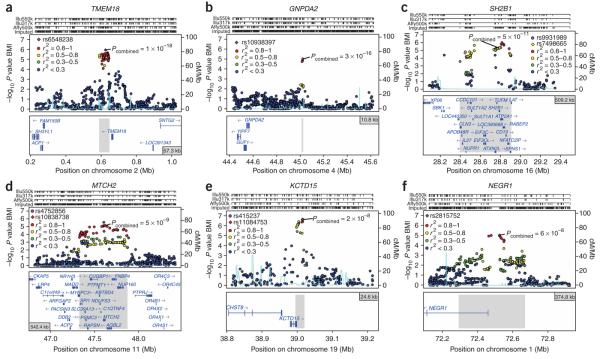
Figure 2.
Regional association plots showing signals in stage 1 samples for replicating loci around TMEM18, GNPDA2, SH2B1, MTCH2, KCTD15 and NEGR1. (a–f) SNPs are plotted by position on chromosome against association with BMI (−log10 P value). The figures highlight the most significant SNP after stage 1 meta-analysis (in purple) and the SNP selected for follow-up (diamond) in stage 2 analyses, labeled with its combined P value (stage 1 + stage 2). In most cases, the SNP followed up is the most significant SNP in the region (therefore, a purple diamond). Otherwise, the LD between the followed-up SNP and the most significant SNP in the region is indicated by the color of the diamond. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant SNP (purple diamond) are color-coded to reflect their LD with this SNP as in the inset (taken from pairwise r2 values from the HapMap CEU database). Genes and the position of exons, as well as the direction of transcription, are noted below the plots (data from UCSC genome browser) with a gray area marking the extent of the region that includes any SNP with r2 ≥ 0.3 relative to the most significant SNP (purple diamond). Hashmarks represent SNP positions on each genotyping array used by any individual study and also show SNP positions after imputation. In e, rs11084753 was selected as the reference SNP for the KCTD15 region and shows essentially identical results to rs415237. The two SNPs are virtually superimposed on the association plot.
Of the variants showing strong association with BMI, only rs9939609 (in FTO) showed nominally significant evidence of heterogeneity across cohorts (P = 0.02, Supplementary Table 5), and none of the associations showed significantly different effects by sex (P > 0.16, Supplementary Table 5). We did not observe any significant evidence supporting the recently reported BMI associations with SNPs near INSIG2 (rs7566605, P = 0.98) and CTNNBL1 (rs6013029, P = 0.34)13,14. We did observe modest evidence for association between BMI and variation in PCSK1 (rs6232, P = 0.03 in the appropriate direction), which has previously been associated with severe obesity11.
Impact on BMI, obesity, related traits and complications
The effects of the associated variants on BMI were estimated using data solely from genotyped stage 2 samples, to lessen the impact of the ‘winner's curse’; they ranged from 0.06 kg/m2 to 0.33 kg/m2 per allele, corresponding to a change of 173–954 g in weight per allele in adults who are 160–180 cm tall (Table 1). In our stage 2 samples, the six newly discovered loci together account for 0.40% of the variance of BMI, and in conjunction with the known associations at FTO and MC4R account for 0.84% of the variance (Table 1). We also estimated the allelic odds ratios for these six newly discovered variants on the risk of being overweight (BMI ≥ 25 kg/m2) or obese (BMI ≥ 30 kg/m2) compared to non-overweight controls (BMI < 25 kg/m2). According to data from the newly genotyped stage 2 samples, the allelic odds ratios for being overweight for each of the six variants ranged from 1.03 to 1.14, and for being obese from 1.03 to 1.25 (Supplementary Table 8 online).
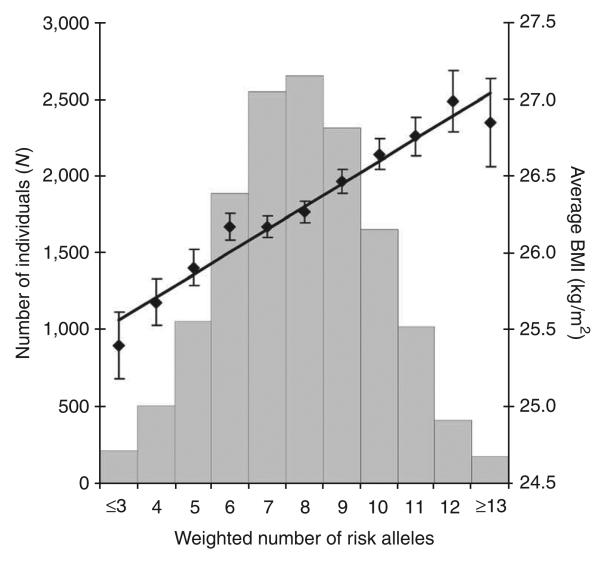
To estimate the combined impact of these variants on BMI, we examined our largest population-based stage 2 sample (the EPIC–Norfolk cohort), analyzing the 14,409 individuals who had no missing genotypes for associated SNPs at any of the eight validated loci (TMEM18, KCTD15, SH2B1, MTCH2, NEGR1 and GNPDA2, plus FTO and MC4R). We calculated a genotype score for each individual, weighting the number of BMI-increasing alleles by their relative effect sizes (so that FTO alleles had the largest weight and MTCH2 alleles the smallest). In this cohort, the 1.2% (n = 178) of the sample with 13 or more ‘standardized’ BMI-increasing alleles across these eight loci is on average 1.46 kg/m2 (equivalent to 3.7–4.7 kg for an adult 160–180 cm in height) heavier than the 1.4% (n = 205) of the sample with ≤3 standardized BMI-increasing alleles, and 0.59 kg/m2 (1.5–1.9 kg for an adult 160–180 cm in height) heavier than the average individual in our study (Fig. 3).
Figure 3.
Combined impact of risk alleles on average BMI in the population-based EPIC-Norfolk cohort. All eight confirmed SNPs were successfully genotyped in the EPIC-Norfolk cohort (14,409 individuals with complete genotype data). For each individual, the number of risk alleles (0,1,2) per SNP was weighted for their relative effect sizes estimated from the stage 2 cohort data only. Subsequently, the weighted risk alleles were summed for each individual, and the overall individual sum was rounded to the nearest integer to represent the individual's risk allele score. Along the x axis, individuals in each risk allele category are shown (grouped ≤3 and ≥13 at the extremes), and the mean BMI (± s.e.m.) is plotted (y axis on right), with the line representing the regression of the mean BMI values across the risk allele scores. The histogram (y axis on left) represents the number of individuals for each risk-score category.
Further follow-up of the confirmed SNPs in a large geographically based cohort of children (ALSPAC Study, n = 4,951 children with BMI information at age 11) showed significant and directionally consistent associations between BMI and the variants near TMEM18 (P = 3.4 × 10−5), KCTD15 (P = 0.0010) and GNPDA2 (P = 0.018) (Supplementary Table 6). Comparison of extreme childhood obesity cases (n = 1,308, SCOOP-UK) to all children in the ALSPAC cohort (n = 8,369 in the full cohort) revealed an increased risk of extreme childhood obesity for the BMI-increasing alleles near TMEM18 (OR = 1.41, P = 7.9 × 10−7), GNPDA2 (OR = 1.20, P = 1.5 × 10−4) and NEGR1 (OR = 1.29, P = 2.2 × 10−7). The absense of significant associations with childhood BMI or extreme childhood obesity for the variants near MTCH2 and SH2B1 could reflect the relatively smaller sample sizes and lower statistical power of our childhood cohorts, or perhaps a differential effect of these variants on the risk of childhood and adult-onset obesity.
Although BMI is a well accepted and commonly used measure of obesity, it is an indirect and approximate measure of adiposity. BMI has two components, weight and height, and can also be influenced by lean and/or fat mass. To determine which aspect(s) of BMI are influenced by the variants we identified, we analyzed their association with the different anthropometric components of BMI, and also with a more direct measure of adiposity, percentage fat mass. All of the variants had much stronger associations with weight than with height (Supplementary Tables 6 and 8), with the exception that for KCTD15 and MTCH2 the small effects on BMI in stage 2 samples limited our ability to dissect the effect on BMI into its constituent components. Variation at MC4R was significantly associated with adult height, as previously reported9. To measure more directly the effects on adiposity, we tested these variants for association with percentage fat mass in a meta-analysis of three cohorts of adults in which percent fat mass was assessed (EPIC-Norfolk, Botnia PPP and METSIM; total n = 18,279), and also in the children from ALSPAC in whom percent body fat mass was measured at age 11 (n = 4,876). As was seen previously for FTO and MC4R7,9, the BMI-increasing alleles at all new loci were also associated with or trended with increased fat mass in both the combined samples of adults and the childhood cohort (Supplementary Tables 6 and 8; each variant had a P value <0.1 in the appropriate direction in either adults, children or both). Thus, the associations with BMI are largely driven by effects on weight rather than height, and seem to act at least in part through an effect on adiposity.
We used publicly available results of GWAS for known obesity complications, including type 2 diabetes15, lipid levels16 and coronary artery disease (CAD)17,18, to assess the impact of the newly discovered obesity loci on these traits. Two of the loci were associated with diabetes15: GNPDA2 (P = 6.6 × 10−5) and TMEM18 (P = 7.5 × 10−4) (Supplementary Table 8). Most of the BMI-associated variants were not significantly associated with these BMI-related traits, most likely because of low power to detect very small effects in the public datasets and the incomplete correlation between BMI and these traits19.
Impact of copy number polymorphism (CNP) on BMI
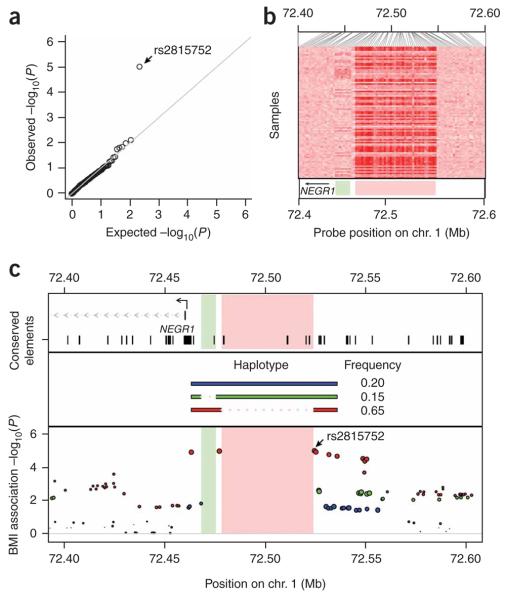
A large fraction of human copy number variation arises from common, diallellic polymorphisms20. Most of these CNPs are in LD with adjacent SNPs, so their contribution to phenotypes can be assessed via these SNPs20. We used these SNP–CNP LD relationships to assess the extent to which this subset of human copy number variation might influence BMI (see Methods). The distribution of BMI association P values in stage 1 samples for CNP-tagging SNPs conforms closely to the distribution expected under the null hypothesis, except for a single SNP (rs2815752, P = 9.3 × 10−6) (Fig. 4a).
Figure 4.
Contribution of copy number polymorphism to BMI. (a) Quantile-quantile plot of the −log10 P values for SNPs (n = 261) that tag copy number polymorphisms (r2 > 0.8) in the stage 1 genome-wide meta-analysis data. The data generally conform to the uniform distribution expected under the null hypothesis of no association, with the exception of a strong association to the CNP-tagging SNP rs2815752 (near NEGR1). (b) Copy number variation in the NEGR1 association region near rs2815752. Heat map representation of the hybridization intensity of copy number probes (SNP 6.0 array) across the NEGR1 association region in 90 HapMap CEU samples. Darker shades of red indicate reduced hybridization intensity. The data indicate two regions of copy number variation (pale green and pink rectangles in lower panel) upstream of NEGR1. (c) Structural haplotypes and BMI association signal in the NEGR1 region. Two deletion polymorphisms (a 10-kb and a 45-kb deletion affecting nonoverlapping sequences upstream of NEGR1) segregate on distinct haplotypes. Both deletions remove conserved elements upstream of NEGR1 (top panel). In the bottom panel, the color of each SNP indicates the structural haplotype with which it is in strongest LD; the size of each circle indicates the strength of this LD. The 45-kb deletion is immediately flanked and perfectly tagged (r2 = 1.0) by the two most strongly BMI-associated SNPs; these SNPs are separated by 47.3 kb on the reference genome sequence but by only 1.7 kb on the BMI-associated deletion haplotype (red). These SNPs flank but are not contained within the copy number variable region.
We noticed that this SNP is the most strongly associated variant at one of our six validated loci, NEGR1. To understand better common patterns of structural variation at NEGR1, we analyzed hybridization data from 270 HapMap samples, finding that two distinct genomic segments upstream of NEGR1 are copy number variable (Fig. 4b). Haplotype analysis indicated that two deletion polymorphisms—a 10-kb deletion and a 45-kb deletion—are segregating at the locus on distinct haplotypes (Fig. 4c). The two most significantly BMI-associated SNPs immediately flank the 45-kb deletion and are in perfect LD with it (r2 = 1.0) across all HapMap analysis panels. Indeed, what initially seemed to be a long associated haplotype (the 47.3 kb spanned by these SNPs on the reference genome sequence) is in fact a short haplotype whose major feature is the absence of 45.6 kb of the reference sequence (Fig. 4c). The 45-kb deletion is therefore a strong candidate to explain the association signal at NEGR1. Although the deletion region consists entirely of noncoding sequence, the deletion allele lacks several conserved elements upstream of NEGR1 that are present on the other structural haplotypes at the locus (Fig. 4c).
Possible role in the CNS of genes near associated variants
The newly discovered variants showing strong associations with BMI lie in or downstream of KCTD15, SH2B1, TMEM18, MTCH2 and GNPDA2, and upstream of NEGR1 (Fig. 2). SH2B1 is a strong prior candidate for regulating body weight. SH2B1 is implicated in leptin signaling21, and Sh2b1-null mice are obese21. Notably, the obesity in Sh2b1-null mice can be reversed by targeted Sh2b1 expression in neurons21, suggesting that the effects of this gene on obesity are mediated through the CNS. KCTD15, TMEM18 and GNPDA2 have unknown functions, whereas MTCH2 encodes a putative mitochondrial carrier protein that may function in cellular apoptosis22,23, and NEGR1 has a role in neuronal outgrowth24,25. Although fine mapping and other experimental approaches will be required to identify and confirm the causal variant(s) and gene(s) for each locus, we note that, with the exception of SH2B1, our newly associated loci do not include obvious or previously studied candidate genes26. Thus, a large sample size and an unbiased genome-wide approach has not only increased the number of known obesity loci, but also highlighted new aspects of the biology of body weight regulation.
To provide additional data on where these genes may function, we measured the expression of the genes nearest to our best SNP association signals in a panel of different human tissues. We found that, in our data, all genes except MTCH2 were highly expressed in the brain and/or hypothalamus (Supplementary Fig. 3 online). Additionally, MTCH2 mRNA expression is observed in the brain in publicly available expression data27, and in these data, variant rs17788930 (r2 = 1 with lead SNP rs10838738) was associated (P = 1.3 × 10−6) with MTCH2 mRNA levels (Supplementary Table 8). These expression data suggest that, as is seen in monogenic forms of obesity, inherited variation influences common human obesity through effects in the CNS, although effects in other tissues for at least some of these genes remain possible.
DISCUSSION
Through meta-analysis of GWA data from >32,000 samples, followed by additional large-scale follow-up, we have identified six new loci that show compelling associations with adult BMI. Four of these loci (TMEM18, GNPDA2, SH2B1 and NEGR1) also show compelling evidence of association with obesity in adults or children. In general, definitive identification of the specific mechanisms through which these loci influence BMI and obesity will require detailed fine mapping and subsequent functional characterization. With the exception of SH2B1, the genes most strongly implicated on the basis of colocalization with the association signal have limited prior candidacy.
We compared our results with those obtained in another large GWAS of BMI, described in an accompanying manuscript by Thorleifsson et al.12. For the five of our six newly identified loci where a comparison was possible (those that had strongly correlated proxies on the Illumina 317K genotyping platform at TMEM18, KCTD15, SH2B1, MTCH2 and NEGR1), the data of Thorleifsson et al. also showed strong evidence of association (Table 1); for GNPDA2, no adequate proxy was available. None of the other top SNPs for which we attempted replication and which had adequate proxies showed evidence of associations in the study by Thorleifsson et al. (Supplementary Table 5; results provided by U. Thorsteinsdottir, G. Thorleifsson and K. Stefansson on behalf of Thorleifsson et al.). After the six validated loci (and SNPs in LD with them) were removed from our analysis, we no longer observed a clear excess of P values smaller than expected by chance (Fig. 1c). One might conclude from this that few detectable BMI loci remain to be found. However, we are encouraged in further pursuit because among the remaining data are two additional loci reported by Thorleifsson et al. (BDNF and ETV5); both of these loci show strong confirming evidence for association in our stage 1 meta-analysis (P values of 0.00035 and 0.00043).
Many of our associated loci highlight genes that are highly expressed in the brain (and several particularly so in the hypothalamus), consistent with an important role for CNS processes in weight regulation. We found that TMEM18, KCTD15, SH2B1, GNPDA2 and NEGR1 are expressed at high levels in brain and hypothalamus (as are FTO and MC4R; Supplementary Fig. 3). The remaining gene, MTCH2, has evidence of expression in the brain in published data27, as does BDNF28, a locus identified by Thorleifsson et al.12. These results extend and confirm previous observations with respect to FTO and MC4R, and are consistent with insights derived from monogenic forms of obesity and functional studies. Disruption in mice of Mc4r, Sh2b1 and Bdnf (all genes that seem to be involved in signaling in the brain) results in hyperphagia and/or obesity, and both Fto and Sh2b1 show diet- or obesity-related changes in expression in hypothalamus21,29-34. Further general support for a neuronal basis for obesity comes from the observation that NEGR1 is thought to affect neuronal outgrowth24,25. Finally, the effect of variants that map to a gene desert between GNPDA2 (Supplementary Fig. 3) and GABRA2 (ref. 35) might be mediated by GABRA2, which affects addiction behavior36-38. Abundant evidence supports multiple possible roles of the CNS on body weight regulation, including on appetite, energy expenditure and other behavioral aspects39. Determining the precise mechanism of action of these loci will require further experimentation.
Our analyses explicitly interrogate only a minority of common sequence variants in a given region; we expect therefore that the causal variant is, for some loci at least, yet to be examined. Although many variants are strongly correlated at each locus, precluding definitive identification of a causal variant, several loci have intriguing candidates. These include a large polymorphic deletion in the association interval upstream of NEGR1 (Fig. 4), and missense variants rs7498665 (A484T) at SH2B1 (r2 = 0.71 to best SNP) and rs1064608 (A290P) at MTCH2 (r2 = 1.0 to best SNP), which also disrupts a predicted SC35 exonic splicing enhancer site40,41.
We cannot be sure which of the nearby genes are causally involved in influencing BMI. As a source of additional clues to likely causal mechanisms, we exploited publicly available eQTL data for lymphocytes42 and brain27, and tested for association between the eight replicated variants and mRNA levels of the nearby genes (Supplementary Fig. 3 and Supplementary Table 9 online). Other than variants in the MTCH2 locus (associated with MTCH2 mRNA levels in brain and NDUFS3 levels in lymphocytes) and in the SH2B1 locus (associated with EIF3C levels in lymphocytes and brain and with TUFM levels in lymphocytes only), these studies did not yield indications of the likely causal gene(s). The SH2B1 result also illustrates some of the difficulties in interpreting associations with gene expression levels, as the presence of a missense SNP in SH2B1 and the strong prior candidacy of this gene would seem to implicate strongly alteration in SH2B1 function as the causal mechanism for influencing obesity. One possibility is that the SH2B1 variant has a causal role but happens to be in LD with a different variant that influences EIF3C and TUFM mRNA levels; alternatively, regulation of EIF3C or TUFM mRNA levels could have a causal role, instead of or in addition to variation in SH2B1.
Logistically, one important challenge in executing our study was coordinating analysis strategies and phenotype modeling across 15 different cohorts, each with specific genotyping, phenotyping, trait modeling and analytical strategies. Given this challenge, we decided to start by carrying out a meta-analysis of results from study-specific analyses, relying only on knowledge of the BMI-increasing allele and P value for each study, before completing a uniform analysis across all studies. Notably, we found very similar results between the study-specific analysis, in which different adjustments for covariates and analytical procedures were performed in different studies, and the uniform analysis, in which these procedures were harmonized across all studies (Supplementary Fig. 2). Thus, at least for this phenotype, the association analysis is robust enough to differences in phenotypic modeling so that differences in study design or analytic strategies do not preclude discovery of new loci using meta-analysis.
The effect sizes attributable to the associated variants range from 0.06 to 0.33 BMI units per allele, and each explains only a small proportion of the variance in adult BMI. As might be expected, given these modest effects and the smaller size of the relevant available datasets, we did not consistently observe measurable effects on the risk of diseases in which obesity is one of several contributing factors (such as type 2 diabetes). It is also possible that some of these variants influence BMI but have negligible effects on the downstream risk of obesity-related disease. Despite these small effects on BMI, when we combined information from the eight validated loci, we were able to identify small groups of individuals who differ appreciably with respect to mean BMI. However, at the population level, the value of these signals in predicting obesity remains quite limited (Supplementary Fig. 4 online).
These results raise the question as to why the variants detected in this large study only explain a small fraction of the inherited variability in BMI. There are several possible explanations, which require further experimentation to explore. First, there may be many more loci with common variants that influence BMI. We can predict that additional loci will be discovered by similarly sized studies in new samples: because we had only 5–10% power to detect variants such as those at KCTD15, MTCH2 and NEGR1, dozens of additional variants with comparable effect sizes likely remain unidentified. The number of common variants with smaller effects, and which might be detected with larger samples, is harder to predict, as this depends on the allelic architecture; if the number of causal variants increases as effect sizes decrease, then increasing sample size will be especially productive. Modifying effects such as interactions with environmental factors, other genetic variants, age, sex or other variables may, if substantial, also diminish apparent effect sizes, so detailed analyses of interaction with validated variants may be informative. Finally, other than the MC4R coding region, these loci have not yet been explored thoroughly for additional rare (or common) variants. As such, it is not known whether additional variants at these loci (those causal for the index association or those representing independent causal events) could explain a greater fraction of BMI variation. There are a growing number of examples, including at MC4R, where genes containing common variants associated with a particular phenotype also harbor lower-frequency, higher-penetrance variants with more severe phenotypic consequences16,43-46. Comprehensive sequencing studies in these and other loci (perhaps in individuals with extreme obesity) may represent a path to finding such variants and beginning to explore the relative contributions of common and rare variation to BMI. Discovering additional variants will slowly increase predictive power. However, a greater immediate impact of these studies is the identification of previously unsuspected loci that participate in the biology of body weight regulation, and which may help guide the development of new therapies.
METHODS
Study design
This study is comprised of two stages. Stage 1 is a meta-analysis of GWA studies comprised of 32,387 individuals of European ancestry. This meta-analysis allowed us to select 35 loci for detailed examination in stage 2, which included direct genotyping in 45,018 European-origin individuals from nine studies and in silico comparisons with results from 14,064 European-origin individuals from five studies with GWA data (Supplementary Fig. 1 and Supplementary Table 1).
Stage 1 samples and genotyping
The GIANT consortium currently encompasses 15 study cohorts with 32,387 individuals of European ancestry informative for adult BMI (Supplementary Fig. 1 and Supplementary Table 1). The 15 study cohorts, including between 1,094 and 5,433 individuals each, were genotyped using the Affymetrix 500K Mapping Array Set (11 cohorts, n = 25,394), Illumina HumanHap300 BeadChip (2 cohorts, n = 2,385), Illumina HumanHap300+240 (1 cohort, n = 2,235) or Illumina HumanHap 550 BeadChip (1 cohort, n = 2,265) (Supplementary Tables 2 and 3). To allow for meta-analysis across different marker sets and to improve coverage of the genome, we performed imputation of polymorphic HapMap CEU SNPs (Supplementary Note and Supplementary Table 3) using either MACH (Y. Li, C.J.W., J. Ding, P.S. and G.R.A., unpublished data) or IMPUTE47.
Association analysis with BMI and meta-analysis of association results
First, each study performed GWA analyses for BMI assuming an additive model implemented in either MACH2QTL (Y. Li, C.J.W., J. Ding, P.S. and G.R.A., unpublished data), Merlin48,49 or SNPTEST47. Covariates, trait transformation and strategies for excluding outliers or accounting for family relatedness varied according to each study's original design (Supplementary Tables 2 and 3), but the main results were essentially unchanged when we repeated meta-analysis after imposing a uniform set of analyses and procedures across the 15 study cohorts. For those samples based around case-control designs (such as those from FUSION and from the type 2 diabetes, coronary artery disease, and hypertension components of the Wellcome Trust Case Control Consortium), cases were analyzed separately from controls. To allow for relatedness in the SardiNIA and FUSION samples, regression coefficients were estimated in the context of a variance component model that modeled background polygenic effects49.
Next, we carried out meta-analysis using a weighted z-score method, which accounts for the direction of association relative to a consistent reference allele. In this method, P values for each study are first converted to z scores. Then, a weighted sum of z scores is calculated where each statistic is weighted by the square root of the sample size for each study. The resulting sum is divided by the square root of the total sample size to obtain an overall z statistic, which can be used to evaluate the overall evidence for association. The method takes direction of effect across studies into account by reversing the sign of the z score for a study if the effect is in the opposite direction. We obtained similar results when we analyzed each cohort using a uniform protocol (which involved a quantile transformation to approximate normality and adjusting for age and age2 in men and women separately) and combined the results using the regression coefficients and standard errors estimated from each study (Supplementary Fig. 2; Pearson correlation r = 0.91). Both meta-analysis procedures were implemented in the freely available METAL software package. The genomic control parameter λ was 1.10 in our initial meta-analysis without using genomic control correction in any study except SardiNIA, which, given our large sample size, suggests only a modest impact of unmodeled relatedness or population stratification in our results. The P values we report have all subsequently been corrected for this unmodeled relatedness or population stratification by application of a genomic control correction to all input studies as well as to the meta-analysis results.
Selection of SNPs for follow-up
For follow-up analyses (stage 2), we genotyped 35 SNPs drawn from the most significantly associated independent loci. We defined signals at two SNPs to be independent of each other if the SNPs were in low LD (r2 < 0.3) or if they were >1 Mb apart. In some cases, the SNP with the strongest signal of association at a locus could not be genotyped for technical reasons, and we substituted another SNP that was strongly correlated with the original SNP in the HapMap CEU sample (Supplementary Note). Because SNP selection was based on an earlier version of the meta-analysis and because some SNPs failed primer design, not all of the top signals were represented among the 35 SNPs. Among the SNPs that were followed up, the highest stage 1 P value was 6.9 × 10−4.
Stage 2 samples and genotyping
We genotyped 35 SNPs in a total of 45,018 individuals of European ancestry from nine study cohorts using Sequenom iPLEX or TaqMan (Supplementary Note). Individuals were eliminated from analysis if <80% of SNPs were called successfully. Among successfully typed individuals, genotype frequencies were in Hardy-Weinberg equilibrium (P > 10−6), call rates were >94%, and concordance of duplicate genotypes was >99% in each of the follow-up study cohorts.
For in silico replication, we also obtained association results for 35 SNPs from 14,064 individuals of European ancestry from five studies (Supplementary Table 1). The five study cohorts, each including 856 to 5,373 individuals, were genotyped using the Illumina HumanHap 550, 300 or Illumina Human CNV370 DUO (Supplementary Tables 2 and 3). To allow for meta-analysis across different marker sets and to improve coverage of the genome, we carried out imputation of polymorphic (minor allele frequency >1%) autosomal HapMap SNPs (Supplementary Note and Supplementary Table 3) using either MACH or IMPUTE with the HapMap CEU sample as a reference panel. We accounted for uncertainty in each genotype prediction in the analysis of imputed genotype data by using either the dosage information from MACH or the genotype probabilities from IMPUTE. Stage 1 and 2 results for FTO and MC4R are not presented directly in the main text but are shown for comparison in Supplementary Table 5 and Supplementary Figure 5 online.
Association analyses with BMI and secondary analyses
Association with BMI was tested as in stage 1, assuming an additive model. Logistic regression analysis was used to test for association with the risk of being overweight (defined as BMI ≥ 25 kg/m2) or obese (BMI ≥ 30 kg/m2), with adjustment for age, age2, and sex, testing for SNP effects in an additive genetic model. Evidence for association between our replicating SNPs and type 2 diabetes15, lipidlevels16 and coronary artery disease17,18 was extracted from publicly available datasets. The effect of the replicating SNPs on expression of nearby genes was determined from publicly available eQTL GWA studies from lymphocytes42 and brain tissue27.
Gene expression studies
Adult human RNA samples were obtained from Clontech either as poly(A) purified RNA (hypothalamus and adipocyte) or as total RNA (cerebellum, cortex, spleen, pancreas, lung, kidney, liver, testes and total brain). The total RNAs were purified to poly-A RNA using the Micro-Poly(A)Purist kit (Ambion) according to manufacturer's instructions. We used 20 ng of poly(A) RNA in a random-primed first-strand cDNA synthesis using SuperScript II (Invitrogen) according to manufacturer's instructions. The resulting cDNAs were diluted fourfold, and 5 μl of each sample were used in 12 μl reaction with SYBR Green PCR Master Mix kit (Applied Biosystems). Quantitative PCR reactions were done in triplicate on an ABI 7900HT (Applied Biosystems). We calculated expression levels from their average crossing points and expressed relative to the control gene EEF2 (elongation factor 2) and normalized to levels of gene-specific expression in total brain.
CNV analyses
We previously typed 1,350 copy number polymorphisms (CNPs) in the HapMap analysis panels; 360 of these CNPs were found to be common (minor allele frequency >5%) in individuals with European ancestry (HapMap CEU), explaining more than 80% of the copy number differences between any two individuals. 323 common CNPs seemed to be diallelic, and of these 261 were in strong LD with HapMap SNPs that are close to, but do not overlap, the CNPs20. For the current work, for each of these common, diallelic CNPs, we identified (from among the SNPs successfully typed or imputed in the GIANT meta-analysis) the SNP that best captured each CNP via LD in HapMap CEU. This formed the set of 261 ‘CNP-tagging SNPs’ that were used for analysis here; we used the GIANT meta-analysis P values for these SNPs.
At the NEGR1 locus, we found that the 10-kb deletion, the 45-kb deletion and the reference structural allele at NEGR1 each have perfect tagging SNPs (r2 = 1.0) in the HapMap CEU sample. In constructing Figure 4c, we colored each SNP according to which of these structural-allele-tagging SNPs it showed the strongest LD with in HapMap CEU. Locations of conserved elements were obtained from the phastConsElements17way track of the UCSC Genome Browser. A threshold score of 300 was set for inclusion in this figure.
URLs
MaCH, http://www.sph.umich.edu/csg/abecasis/mach/. METAL, http://www.sph.umich.edu/csg/abecasis/metal/.
Supplementary Material
ACKNOWLEDGMENTS
We are extremely grateful to all of the participants in each of the studies contributing to this effort. Full acknowledgments can be found in the Supplementary Note.
Support for this research was provided by: US National Institutes of Health grants CA65725, CA87969, CA49449, CA67262, CA50385, DK062370, DK072193, DK075787, HG02651, HL084729, HL087679 (through STAMPEED, 1RL1MH083268), 5UO1CA098233, 1Z01 HG000024, 1RL1MH083268, T32 DK07191, F32 DK079466, K23 DK080145, K23 DK067288, CIDR NIH Contract Number N01-HG-65403, NIA contract NO1-AG-1-2109; the Intramural Research Program of the Division of Cancer Epidemiology and Genetics; contracts from the Division of Cancer Prevention, National Cancer Institute and EU FP6 funding (contract no LSHM-CT-2003-503041); GlaxoSmithKline; the Faculty of Biology and Medicine of Lausanne, Switzerland; the Intramural Research Program of the National Institute on Aging (NIA); Cancer Research United Kingdom; the UK Medical Research Council (including grants G0000649, G0000934 and G0601261); the Wellcome Trust (including Strategic Award 076113, grants 068545/Z/02 and 076467/Z/05/Z); the NIHR through the Biomedical Research Centres at Oxford, King's College London; Guys and St. Thomas' Foundation Hospitals' Trust; the British Heart Foundation (including grant FS/05/061/19501), European Community's Seventh Framework Programme (ENGAGE:HEALTH-F4-2007-201413); Diabetes UK; Unilever Corporate Research; American Diabetes Association including a Smith Family Foundation Pinnacle Program Project Award #7-03-PPG-04R; the Academy of Finland (grants 118065 and 124243); National Genome Research Net Germany; Munich Center of Health Sciences (MC Health) as part of LMUinnovativ; the Helmholtz Center Munich; the Sigrid Juselius Foundation; University of Bristol; Linné grant from Swedish Research Council; Wallenberg Foundation; Folkhälsan Research Foundation; University of Southampton; Netherlands Organisation of Scientific Research NWO (nr. 175.010.2005.011); Erasmus Medical Center and Erasmus University, Rotterdam; Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Netherlands Ministry of Education, Culture and Science; the Netherlands Ministry for Health, Welfare and Sports; the European Commision (DG XII) and the Municipality of Rotterdam. G.R.A. and K.L.M. are Pew Scholars for the Biomedical Sciences; A.L.E. is supported by a Sarnoff Cardiovascular Research Foundation Fellowship; C.M.L. is a Nuffield Department of Medicine Scientific Leadership Fellow; S.A.M. is supported by a Life Sciences Research Fellowship; M.K. is supported by the Finnish Cultural Foundation; N.J.S. holds a BHF Chair; M.N.W. is a Vandervell Foundation Research Fellow; C.J.W. is supported by an American Diabetes Association postdoctoral fellowship; and E.Z. is a Wellcome Trust-RD Fellow (grant number 079557).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
Reprints and permissions information is available online at http://npg.nature.com/ reprintsandpermissions/
References
- 1.Flegal KM, Graubard BI, Williamson DF, Gail MH. Cause-specific excess deaths associated with underweight, overweight, and obesity. J. Am. Med. Assoc. 2007;298:2028–2037. doi: 10.1001/jama.298.17.2028. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein EA, et al. The lifetime medical cost burden of overweight and obesity: implications for obesity prevention. Obesity (Silver Spring) 2008;16:1843–1848. doi: 10.1038/oby.2008.290. [DOI] [PubMed] [Google Scholar]
- 3.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 4.Atwood LD, et al. Genomewide linkage analysis of body mass index across 28 years of the Framingham Heart Study. Am. J. Hum. Genet. 2002;71:1044–1050. doi: 10.1086/343822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farooqi IS. Genetic aspects of severe childhood obesity. Pediatr. Endocrinol. Rev. 2006;3(Suppl. 4):528–536. [PubMed] [Google Scholar]
- 6.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 9.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers JC, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 11.Benzinou M, et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 12.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2008 Dec 14; doi: 10.1038/ng.274. advance online publication, doi:10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 13.Herbert A, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 14.Liu YJ, et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum. Mol. Genet. 2008;17:1803–1813. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freathy RM, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–1426. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 21.Ren D, et al. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J. Clin. Invest. 2007;117:397–406. doi: 10.1172/JCI29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grinberg M, et al. Mitochondrial carrier homolog 2 is a target of tBID in cells signaled to die by tumor necrosis factor alpha. Mol. Cell. Biol. 2005;25:4579–4590. doi: 10.1128/MCB.25.11.4579-4590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marg A, et al. Neurotractin, a novel neurite outgrowth-promoting Ig-like protein that interacts with CEPU-1 and LAMP. J. Cell Biol. 1999;145:865–876. doi: 10.1083/jcb.145.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafer M, Brauer AU, Savaskan NE, Rathjen FG, Brummendorf T. Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol. Cell. Neurosci. 2005;29:580–590. doi: 10.1016/j.mcn.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Rankinen T, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 27.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 28.Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog. Neurobiol. 2005;76:99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Unger TJ, Calderon GA, Bradley LC, Sena-Esteves M, Rios M. Selective deletion of Bdnf in the ventromedial and dorsomedial hypothalamus of adult mice results in hyperphagic behavior and obesity. J. Neurosci. 2007;27:14265–14274. doi: 10.1523/JNEUROSCI.3308-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huszar D, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 31.Marsh DJ, et al. Response of melanocortin-4 receptor-deficient mice to anorectic and orexigenic peptides. Nat. Genet. 1999;21:119–122. doi: 10.1038/5070. [DOI] [PubMed] [Google Scholar]
- 32.Stratigopoulos G, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Zhou Y, Carter-Su C, Myers MG, Jr, Rui L. SH2B1 enhances leptin signaling by both Janus kinase 2 Tyr813 phosphorylation-dependent and -independent mechanisms. Mol. Endocrinol. 2007;21:2270–2281. doi: 10.1210/me.2007-0111. [DOI] [PubMed] [Google Scholar]
- 35.Fritschy JM, Mohler H. GABAA-receptor heterogeneity in the adult rat brain: differential regional and cellular distribution of seven major subunits. J. Comp. Neurol. 1995;359:154–194. doi: 10.1002/cne.903590111. [DOI] [PubMed] [Google Scholar]
- 36.Fehr C, et al. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr. Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- 37.Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 38.Edenberg HJ, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am. J. Hum. Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthoud HR, Morrison C. The brain, appetite, and obesity. Annu. Rev. Psychol. 2008;59:55–92. doi: 10.1146/annurev.psych.59.103006.093551. [DOI] [PubMed] [Google Scholar]
- 40.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon AL, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 43.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lettre G, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winckler W, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 46.Bonnycastle LL, et al. Common variants in maturity-onset diabetes of the young genes contribute to risk of type 2 diabetes in Finns. Diabetes. 2006;55:2534–2540. doi: 10.2337/db06-0178. [DOI] [PubMed] [Google Scholar]
- 47.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 48.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 49.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.