Abstract
Recent data has implicated the Ski protein as being a physiologically relevant negative regulator of signaling by Retinoic Acid (RA). The mechanism by which Ski represses RA signaling is unknown. Co-immunoprecipitation and immunofluorescence assay showed that Ski and RARα are in the same complex in both the absence and presence of RA, which makes Ski different from other corepressors. We determined that Ski can stabilize RARα and HDAC3. These results suggest that Ski represses RA signaling by stabilizing corepressor complex.
Keywords: Ski, RA, RARα, HDAC3, Proteasome, nuclear hormone signaling
Introduction
Recently, Ski was shown to act as a transcriptional corepressor by multiple direct and indirect interactions with several distinct repression complexes. These include complexes containing histone deacetylases (HDAC), N-CoR/SMRT/Sin3A corepressors and SMAD proteins. Ski's ability to interact with these various complexes result in transcriptional repression of distinct signaling pathways, which are important for cell growth and proliferation [1; 2]. Expression of Ski can induce immortalization and self-renewal of primary multipotential myeloid progenitor cells from avian bone marrow [3] This latter property of Ski is reminiscent of the effects of expressing a dominant-negative form of the retinoic acid receptor in hematopoietic cells [4], and thus may reflect Ski's ability to repress retinoic acid signaling.
Retinoic acid (RA) is essential for mammalian development. It functions by activating transcription involving the nuclear hormone receptor family of retinoic acid receptors (RARs). In the absence of ligand, retinoid receptors are primarily in the nucleus bound to RA responsive elements (RARE) on DNA. These complexes contain various corepressors and repress transcription in the absence of ligand [5]. RA binds to RAR and triggers a cascade of events, which favor the interactions between RAR and RXR and causes corepressors release, followed by coactivators recruitment, chromatin decompaction, and transcription initiation [6]. Tight regulation of the RA signaling pathway is essential for physiological responses. There is accumulating evidence that Ski is important in the regulation of transcription induced by RA. Analysis of mice genetically engineered to be null for the Ski gene revealed defects similar to those associated with excess intake of retinoids in humans. In humans the Ski gene maps to chromosome 1p36.3 and terminal deletions of chromosome 1 gives rise to monosomy 1p36 syndrome [7]. Children with this syndrome have craniofacial defects and digit defects, which are virtually identical to those in the Ski-/- mice; implying that these defects may be due to reduced levels of Ski and reflect a hypersensitivity to retinoids [8]. These data indicate that Ski plays a role in regulating RA signaling during development.
Recent experiments have shown that stabilization of inactive Smad complexes on DNA is a critical event in Ski-mediated inhibition of TGFβ signaling [9]. In this report, we show that expression of Ski leads to stabilization of proteins in the RA-repression complex, namely HDAC3 and RARα. These data indicate that, like repression of TGFβ signaling by Ski, repression of RA signaling by Ski also involves stabilization of repression complexes and may indicate a common mechanism by which Ski represses transcription.
Materials and methods
Cell Culture, Reagents, and Transfection
COS-1, HeLa, QT6, MEF Ski-/- (a gift from Dr. C. Colmenares. Lerner Research Institute, Ohio, USA) and MG63 cells were maintained in Dulbecco's modified Eagle's medium (Gibco/Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, penicillin G (100 units/ml), and streptomycin (100μg/ml). For QT6 cells, medium was supplemented with 1% chicken serum (Sigma, St Louis, MO, USA). Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). Expression plasmids were introduced into the cells using FuGENE 6 (Roche Applied Science, Indianapolis, IN, USA).
Plasmid Constructs
T7-wt-cSki, Flag-RARα, Myc-RARα, RXR, Myc-HDAC3, 6His-Ub and Flag-N-CoR-C were described previously [10; 11; 12; 13]. Flag-HDAC3 was constructed by cloning mouse HDAC3 cDNA into pCMV-Tag2B vector (Stratagene, La Jolla, CA, USA). GFP-Ski was made by cloning hSki cDNA into pEGFP-C1 vector (Clontech). Flag-Ski was constructed by inserting human c-Ski fragment into pCMV-Tag2C vector (Stratagene). For the LMP-Ski-shRNA, PCR-amplified DNA insert for expression of hSki specific small hairpin RNA (target sequence: gtactcggcccagatcgaa) was cloned into LMP retroviral vector (a gift from Dr. S.W. Lowe, Cold Spring Harbor Laboratory, NY, USA). Inducible hSki expression plasmid was generated by inserting hSki fragment into pRVYtet vector [14]. All cloning procedures were verified by DNA sequencing.
Co-Immunoprecipitation and Western Blot Analysis
Co-immunoprecipitation and western blot were performed as described [10] Antibodies used were anti-Myc (9E10, Santa Cruz Biotechnology, CA, USA), anti-Flag mouse monoclonal antibody (Sigma), anti-T7 mouse monoclonal antibodies (Novagen, Madison, WI, USA), anti-Ski (H-329, Santa Cruz Biotechnology), anti-HDAC3 (3G6, Upstate, Charlottesville, VA, USA), anti-Ski (G8, Cascade Bioscience, Winchester, MA, USA), anti-RARα (Santa Cruz Biotechnology), anti-β-Actin (Sigma), and anti-α-Tubulin (Sigma). Proteins were detected by chemiluminescence (Perkin Elmer, Shelton, CT, USA).
Luciferase Reporter Assays
Luciferase reporter assay was performed as described [10]. The luciferase reporter construct CRBPII-Luc is a gift from Dr. Vimla Band, Tufts University School of Medicine, Boston, MA, USA). For ligand stimulation, cells were treated with RA (1μM) or appropriate solvent 24h after transfection.
Indirect Immunofluorescence and microscopy
Indirect Immunofluorescence and microscopy were carried out as described previously [12]. Cells were visualized with an Axiovert 200M (Zeiss, Thornwood, NY, USA) using a 63X oil DIC lens and the images were analyzed using the Axiovision software (Zeiss).
Proteasome inhibition and in vivo ubiquitination assays
The proteasome inhibitor MG132 (Sigma) was used at a concentration of 25μM or 5μM as indicated. Ubiquitinated intermediates were purified and detected as described previously [12].
Retrovirus-mediated Gene Transfer
The amphotropic retroviral packaging cells Phoenix A were transfected and grown for 48h to confluence prior to harvesting the viral supernatant. MEF Ski-/- cells were infected with the supernatant supplemented with Polybrene (10μg/ml) and incubated for 6h. The infected cells were cultured for 48h and then selected for 1 week in the presence of appropriate antibiotics.
Results
RA-induced RARα degradation is important for optimal RARα-mediated transactivation
RA induces the degradation of its receptor RARα [15] and this is thought to be a resetting mechanism for efficient RA target gene expression [16]. Therefore we considered it possible that Ski may inhibit RA signaling by influencing this degradation. We first demonstrated that in our system RA did increase RARα turnover. Cycloheximide (CHX) was used to inhibit protein synthesis. RA clearly increased the turnover of RARα (Fig1A). Next we treated the transfected cells with proteasome inhibitor MG132. Addition of MG132 inhibited RARα turnover, indicating that degradation involved the ubiquitin proteasome pathway (Fig1B). To determine the affect of RARα turnover on RA signaling, we performed a luciferase reporter assay using a RA-responsive reporter, CRBPII-Luc. Addition of RA resulted in an approx 4-fold increase of luciferase activity (Fig1C). The addition of MG132 inhibited this induction in a time-dependent manner. Western Blot analysis showed MG132 treatment inhibited Flag-RARα degradation, and simultaneously decreased RA-dependent transcriptional activity. To rule out the possibility that the effects of MG132 were non-specific, we determined the effects of MG132 on another reporter. MG132 had no effect on the transactivation of a TK-luc reporter gene (Fig1D), indicating that the MG132 effect on the CRBPII-Luc reporter gene expression was to some extent RA specific. These data confirm that RA can induce RARα degradation, and this degradation is important for optimal RA signaling.
Fig 1. RA-induced RARα degradation is important for optimal RARα-mediated transactivation.
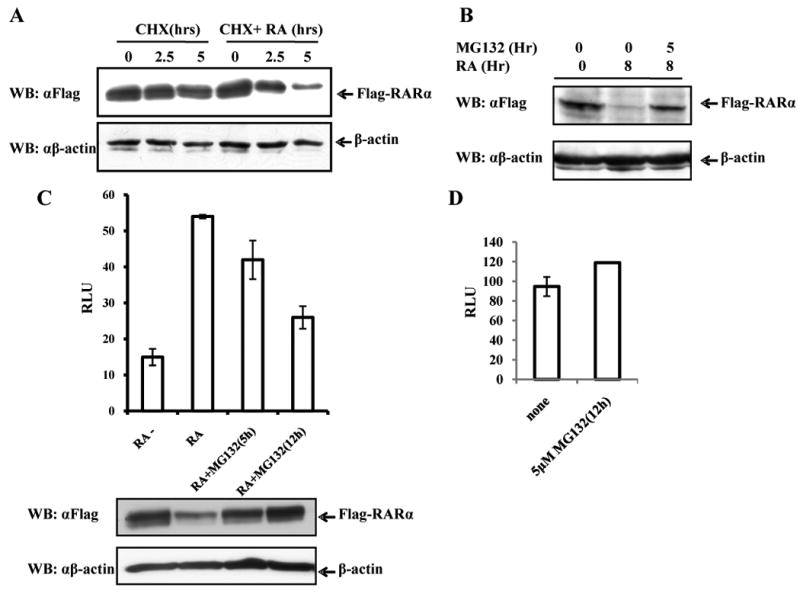
(A) COS-1 cells, transfected with plasmid encoding Flag-RARα, were treated without or with 1 μM RA for different times in the presence of CHX as indicated. (B) COS-1 cells were transfected with plasmid encoding Flag-RARα. 24h after transfection, cells were treated without or with RA for 8h in the absence or presence of 5μM MG132 as indicated. (C) QT6 cells were transfected with CRBPII-Luc, Flag-RARα and RXR, and after 24h were left untreated or treated with RA (1μM) for 12h. MG132 (5μM) treatment is as indicated. RLU, relative light units (arbitrary activity). (D) QT6 cells were transfected with Tk-luc reporter and 24h after transfection the cells were treated with or without MG132 (5μM) for 12h. The results are expressed as means ±S.D. from three independent experiments.
Ski expression inhibits RA-induced RARα degradation
Since Ski inhibits Ra-signaling we determined the effects of expressing Ski on RARα turnover. To exclude the possibility that Ski may affect transcriptional or translationally regulated levels of RARα, we used CHX to inhibit new protein synthesize. Expression of Ski slowed down the RA induced turnover of RARα (Fig2A). We quantified this effect and found the expression of Ski increased the half-life of RARα by more than two-fold (Fig2B). Since the degradation of RARα involves the proteasome, we determined the effect of Ski expression on RARα ubiquitination by using an in vivo ubiquitination assay. There was a marked reduction of ubiquitinated forms of RARα in the presence of Ski (Fig2C), indicating that the ubiquitination and subsequent ubiquitin-dependent degradation of RARα is inhibited by the presence of Ski.
Fig 2. Ski expression inhibits RA-induced RARα degradation.
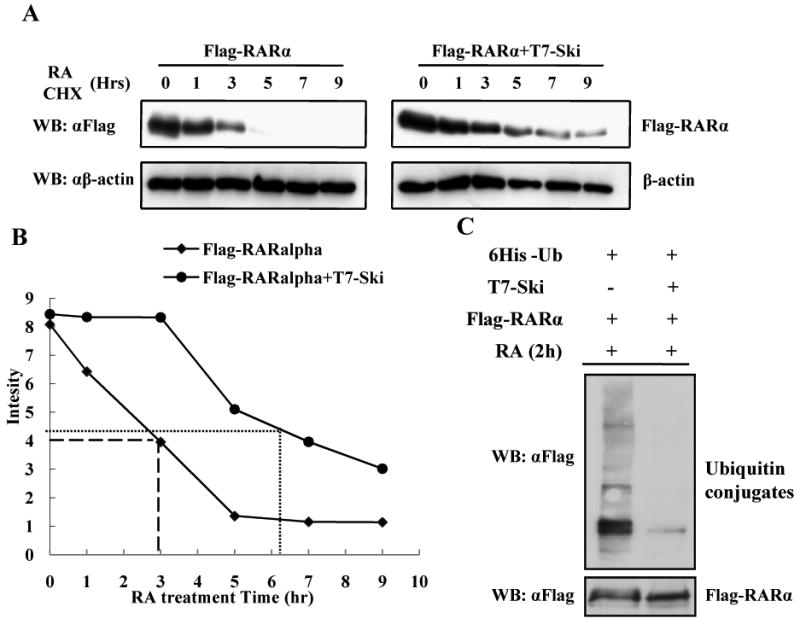
(A) COS-1 cells were transfected with Flag-RARα with or without T7-Ski. CHX and RA were added to cells after 24h transfection. The cell lysates were collected at different time points. (B) The quantification of results from (A). (C) QT6 cells were transfected with different combinations of three plasmids: Flag-RARα, T7-Ski and 6His-Ub as indicated for 24h, and then treated with RA for 2h. The ubiquitinated forms of Flag-RARα were detected by Western Blot.
Ski and RARα are in the same complex in both the presence and absence of RA
Ski has been reported to interact with RARα [11; 17], therefore we looked at the interaction of RARα and Ski in the absence and presence of RA. Ski and RARα interaction was determined by co-immunoprecipitation and was unaffected by 6h RA treatment (Fig3A). Similarly immunofluorescence studies showed that Ski and RARα were both present in nuclear dot structures (Fig3B), and this co-localization was unaffected by the addition of RA. The above data indicate Ski is complexed with RARα in both the absence and presence of RA treatment.
Fig 3. Ski and RARα are in the same complex in both the presence and absence of RA.
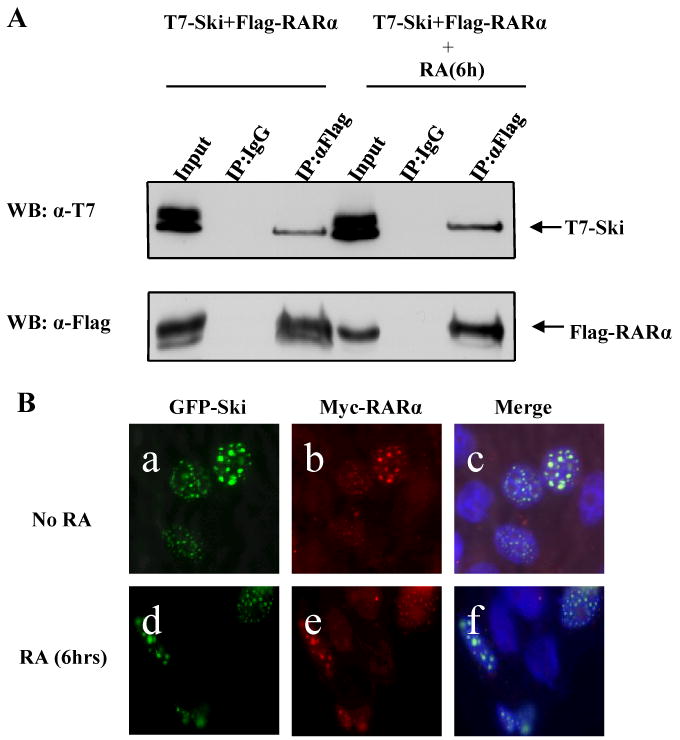
(A) T7-Ski was co-immunoprecipitated with Flag-RARα using an anti-Flag antibody, and a mouse normal IgG as a control. The immunocomplexes and 10% input were analyzed by Western Blot using an anti-T7 antibody (the upper panel) or an anti-Flag antibody (the lower panel). (B) Representative fluorescent images of HeLa cells expressing Myc-RARα and GFP-Ski without (a-c) or with (d-f) 6h RA (1μM) treatment. Myc-RARα is shown in red, GFP-Ski in green and Hoechst staining of DNA in blue.
Ski associates with HDAC3 and inhibit HDAC3 degradation
The nuclear receptor corepressor complex, which is necessary for the nuclear receptor-mediated repression, consists of N-CoR,/SMRT, HDAC3, transducin β-like 1 (TBL1), TBLR1 and GPS2 [18] and probably several more proteins [19]. This complex mediates repression and its degradation by the proteasome system is essential for efficient RA signaling [16] as it is necessary for coactivator proteins to be recruited to the RAR/RXR complex. We hypothesized that Ski might be associating with the corepressor complex and inhibiting its degradation. A key functional member of this complex is the histone deacetylase HDAC3. Thus we determined if Ski can interact with HDAC3 and influence its turnover. Co-immunoprecipitation experiments demonstrated Ski-HDAC3 interaction in both the absence and presence of RA (Fig4A). We demonstrated that RA induces HDAC3 degradation which can be inhibited by MG132 (Fig4B). Hence we assessed the effect of expressing Ski on HDAC3 stability. We found that HDAC3 turnover is inhibited by the expression of Ski (Fig4C). The effects of Ski expression on endogenous HDAC3 levels were also examined. We utilized mouse embryonic fibroblasts (MEF) isolated from Ski -/- animals in which Ski expression was now driven by a Doxycycline (Dox)-regulated promoter. As shown in Fig4D, Ski expression can be induced by the withdrawal of Dox (lane1-3) whereas the addition of Dox turned off Ski expression (lane4-6). Expression of Ski clearly inhibited RA-induced endogenous HDAC3 turnover. In the presence of CHX, Ski was also degraded. To complement these studies, we used the MG63osteosarcoma cell line, which express multiple forms of endogenous Ski. These cells were infected with a retrovirus expressing a short hairpin-type RNA (shRNA) directed against Ski to knockdown the expression of Ski. Fig4E showed that the shRNA reduced the levels of Ski expression (lane2) and this reduction correlated with reduced levels of HDAC3. Cells expressing control shRNA, lane1& 3, maintained Ski expression and this correlated with increased levels of HDAC3. These data show that Ski can stabilize not only exogenously expressed HDAC3 but also endogenous HDAC3 and imply that expression of Ski is capable of stabilizing the corepressor complex and by doing so Ski can repress transcription induced by RA.
Figure 4. Ski associates with HDAC3 and inhibit HDAC3 degradation.

(A). Flag-HDAC3 was co-immunoprecipitated with an anti-Ski antibody (rabbit). The immunocomplexes and 10% input were detected by an anti-Flag antibody. (B) COS-1 cells transfected with Flag-HDAC3 were treated without or with RA for 18h in the absence or presence of 5μM MG132. (C) COS-1 cells were transfected with Flag-HDAC3 with or without T7-Ski, and CHX was added to the cells after 24h transfection. Flag-HDAC3 levels were checked by Western Blot using an anti-Flag antibody (middle panel), Ski levels were determined by anti-T7 antibody (the upper panel) and the α-tubulin level was used as a loading control (the lower panel). (D) The MEF Ski-/- cells were maintained in the presence or absence of Dox for 48h, then untreated or treated with CHX as indicated. The endogenous HDAC3 levels were determined by an anti-HDAC3 antibody using 50ug of cell lysate for each sample. (E) The osteosarcoma cell line MG63 was infected with a retrovirus expressing the short hairpin-type RNA (shRNA) directed against Ski in order to knockdown the expression of Ski (lane2) or control shRNA (lanes 1 &3) and the endogenous HDAC3 levels were detected by an anti-HDAC3 antibody.
Discussion
Ski expression is up-regulated by two- three fold in a subset of human acute myeloid leukemia, AML, and this increased expression correlates with bad prognosis and resistance to RA-induced differentiation therapy [11], implying that repression of RA signaling plays a role in this subset of AML patients. In addition, several reports suggest that Ski plays a physiological role in regulating the repression of RA signaling [7; 8; 20; 21; 22]. However, the molecular mechanism of how Ski can repress RA signaling is still unclear. In this report we examined the effects of Ski expression on the stability of two key members of the RA corepressor complex and found that Ski expression stabilizes these proteins. Previous reports have indicated that nuclear hormone-mediated nuclear hormone receptor (NR) degradation is necessary for efficient target genes activation. This has been reported for several NRs [23; 24; 25; 26]. Furthermore, it has been reported that corepressor/coactivator complex exchange is required for transcriptional activation by RARα [18]. We found that Ski can inhibit RA-induced RARα degradation and also stabilize HDAC3. These data are consistent with the hypothesis that corepressor/coactivator complex exchange is blocked by the presence of Ski and thus results in inhibition of RA signaling.
The nuclear receptor corepressor complex consists of N-CoR,/SMRT, HDAC3, transducin β-like 1 (TBL1), TBLR1 and GPS2 [18] and probably several more proteins[19]. We tested whether Ski regulates the protein level of HDAC3, which provides enzymatic activity and promotes transcription repression by deacetylating histones [27]. Our data showed that Ski and HDAC3 are associated in both the absence and presence of RA (Fig4A). We found that RA indeed also induces HDAC3 proteasome degradation (Fig4B). Similarly to the effects on RA-induced RARα degradation, Ski can also inhibit HDAC3 proteasome degradation. We demonstrated that the endogenous levels of HDAC3 could also be stabilized by the regulated Ski expression. To complement this analysis, we used shRNA directed against Ski to reduce the levels of endogenous Ski protein in MG63 osteosarcoma cell line. As predicted, reduction in the level of expression of Ski led to a parallel reduction of HDAC3 levels. Together these data indicate that Ski levels can influence the levels of expression of HDAC3 and RARα.
Ski stabilizes the inactive Smad complex on the Smad binding element (SBE) of TGFβ target genes, and this stabilization inhibits the access of newly activated Smad proteins to the SBE [9]. Thus the stabilization of the inactive complex by Ski results in inhibition of TGFβ signaling. In a mechanistically similar manner we report that Ski might maintain the repressed state of the RA target genes by stabilizing the HDAC3 containing corepressor complex. The Ski-related protein SnoN has been reported to positively regulate the protein levels of the corepressor mSin3A [28]. These data indicate that, like Ski, SnoN can also regulate the levels of corepressors. These findings point to similar mechanistic functions of these family members by stabilizing corepressors to repress transcription.
We hypothesize that the mechanism of Ski's repressive effects on RA signaling is primarily at the corepressor/coactivator exchange level by stabilizing proteins in the corepressor complex in the presence of ligand. The presence of RA induces active corepressors release and degradation through the ubiquitin proteasome pathway. When Ski protein is present it associates with the corepressor complex by virtue of its ability to interact, directly or indirectly, with RARα, N-CoR/SMRT and HDAC3, and prevents the efficient dissociation and/or degradation of the corepressor complex. Since the degradation of the corepressor complex is inhibited, the coactivator complex cannot be recruited. This inhibition of corepressor degradation means that efficient transcription does not occur and, since RARα degradation takes places after the transcription of the target genes, the degradation of RARα is inhibited indirectly. Thus we hypothesize that Ski's primary mode of action is to stabilize the corepressor complex and this indirectly results in the stabilization of RARα.
In summary, we propose that Ski protein inhibits the RA signaling pathway through maintaining the basal repressed state of the RA target genes by stabilizing the corepressor complex. Our findings reveal a novel mechanism of Ski's repressive effect on RA signaling pathway which might also apply to the other signaling pathways inhibited by the Ski protein.
Acknowledgments
We thank all the members of the Hayman laboratory for helpful discussions and criticisms of the manuscript. This work was supported by U.S. Public Health Service Grant CA42573 from the National Cancer Institute to MJH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nomura T, Khan MM, Kaul SC, Dong HD, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–23. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13:2196–206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beug H, Dahl R, Steinlein P, Meyer S, Deiner EM, Hayman MJ. In vitro growth of factor-dependent multipotential hematopoietic cells is induced by the nuclear oncoprotein v-Ski. Oncogene. 1995;11:59–72. [PubMed] [Google Scholar]
- 4.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831–41. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 5.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 6.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. Embo J. 2000;19:1045–54. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slavotinek A, Shaffer LG, Shapira SK. Monosomy 1p36. J Med Genet. 1999;36:657–63. [PMC free article] [PubMed] [Google Scholar]
- 8.Colmenares C, Heilstedt HA, Shaffer LG, Schwartz S, Berk M, Murray JC, Stavnezer E. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski-/- mice. Nat Genet. 2002;30:106–9. doi: 10.1038/ng770. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H, Yagi K, Kondo M, Kato M, Miyazono K, Miyazawa K. c-Ski inhibits the TGF-beta signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene. 2004;23:5068–76. doi: 10.1038/sj.onc.1207690. [DOI] [PubMed] [Google Scholar]
- 10.Ueki N, Hayman MJ. Signal-dependent N-CoR requirement for repression by the Ski oncoprotein. J Biol Chem. 2003;278:24858–64. doi: 10.1074/jbc.M303447200. [DOI] [PubMed] [Google Scholar]
- 11.Ritter M, Kattmann D, Teichler S, Hartmann O, Samuelsson MK, Burchert A, Bach JP, Kim TD, Berwanger B, Thiede C, Jager R, Ehninger G, Schafer H, Ueki N, Hayman MJ, Eilers M, Neubauer A. Inhibition of retinoic acid receptor signaling by Ski in acute myeloid leukemia. Leukemia. 2006 doi: 10.1038/sj.leu.2404093. [DOI] [PubMed] [Google Scholar]
- 12.Marcelain K, Hayman MJ. The Ski oncoprotein is upregulated and localized at the centrosomes and mitotic spindle during mitosis. Oncogene. 2005;24:4321–9. doi: 10.1038/sj.onc.1208631. [DOI] [PubMed] [Google Scholar]
- 13.Ueki N, Zhang L, Hayman MJ. Ski can negatively regulates macrophage differentiation through its interaction with PU.1. Oncogene. 2008;27:300–7. doi: 10.1038/sj.onc.1210654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweasy JB, Lang T, Starcevic D, Sun KW, Lai CC, Dimaio D, Dalal S. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102:14350–5. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopf E, Plassat JL, Vivat V, de The H, Chambon P, Rochette-Egly C. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin-proteasome pathway. J Biol Chem. 2000;275:33280–8. doi: 10.1074/jbc.M002840200. [DOI] [PubMed] [Google Scholar]
- 16.Andela VB, Rosier RN. The proteosome inhibitor MG132 attenuates retinoic acid receptor trans-activation and enhances trans-repression of nuclear factor kappaB. Potential relevance to chemo-preventive interventions with retinoids. Mol Cancer. 2004;3:8. doi: 10.1186/1476-4598-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl R, Kieslinger M, Beug H, Hayman MJ. Transformation of hematopoietic cells by the Ski oncoprotein involves repression of retinoic acid receptor signaling. Proc Natl Acad Sci U S A. 1998;95:11187–92. doi: 10.1073/pnas.95.19.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–26. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 19.Tabata T, Kokura K, Ten Dijke P, Ishii S. Ski co-repressor complexes maintain the basal repressed state of the TGF-beta target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells. 2009;14:17–28. doi: 10.1111/j.1365-2443.2008.01246.x. [DOI] [PubMed] [Google Scholar]
- 20.Berk M, Desai SY, Heyman HC, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–39. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atanasoski S, Notterpek L, Lee HY, Castagner F, Young P, Ehrengruber MU, Meijer D, Sommer L, Stavnezer E, Colmenares C, Suter U. The protooncogene Ski controls Schwann cell proliferation and myelination. Neuron. 2004;43:499–511. doi: 10.1016/j.neuron.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 22.McGannon P, Miyazaki Y, Gupta PC, Traboulsi EI, Colmenares C. Ocular abnormalities in mice lacking the Ski proto-oncogene. Invest Ophthalmol Vis Sci. 2006;47:4231–7. doi: 10.1167/iovs.05-1543. [DOI] [PubMed] [Google Scholar]
- 23.Lin HK, Altuwaijri S, Lin WJ, Kan PY, Collins LL, Chang C. Proteasome activity is required for androgen receptor transcriptional activity via regulation of androgen receptor nuclear translocation and interaction with coregulators in prostate cancer cells. J Biol Chem. 2002;277:36570–6. doi: 10.1074/jbc.M204751200. [DOI] [PubMed] [Google Scholar]
- 24.Lonard DM, Nawaz Z, Smith CL, O'Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–48. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 25.Lange CA, Shen T, Horwitz KB. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc Natl Acad Sci U S A. 2000;97:1032–7. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dace A, Zhao L, Park KS, Furuno T, Takamura N, Nakanishi M, West BL, Hanover JA, Cheng S. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc Natl Acad Sci U S A. 2000;97:8985–90. doi: 10.1073/pnas.160257997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor-beta-mediated transcription repression. Mol Cell Biol. 2008;28:1988–98. doi: 10.1128/MCB.01442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


