Abstract
Transmembrane chemoreceptors, also known as methyl-accepting chemotaxis proteins (MCPs), translate extracellular signals into intracellular responses in the bacterial chemotaxis system. MCP ligand binding domains control the activity of the CheA kinase, situated ∼200 Å away, across the cytoplasmic membrane. The 2.15 Å resolution crystal structure of a T. maritima soluble receptor (Tm14) reveals distortions in its dimeric four-helix bundle that provide insight into the conformational states available to MCPs for propagating signals. A bulge in one helix generates asymmetry between subunits that displaces the kinase-interacting tip, which resides over 100 Å away. The maximum bundle distortion maps to the adaptation region of transmembrane MCPs where reversible methylation of acidic residues tunes receptor activity. Minor alterations in coiled-coil packing geometry translate the bulge distortion to a >25 Å movement of the tip relative to the bundle stalks. The Tm14 structure discloses how alterations in local helical structure, which could be induced by changes in methylation state and/or by conformational signals from membrane proximal regions, can reposition a remote domain that interacts with the CheA kinase.
Keywords: Chemotaxis, Conformational change, Four-helix bundle, Receptor, Signal transduction
The bacterial chemotaxis system has served as an important model for understanding transmembrane and intracellular signal transduction (1-3). The molecular mechanisms underlying chemotaxis allow bacteria to sense chemical gradients with high sensitivity, wide dynamic range, memory, and signal integration (2). Central to the chemotaxis system is the histidine autokinase, CheA, which phosphorylates CheY, a diffusible regulator of the direction of flagellar rotation (1-3). Clusters of transmembrane chemoreceptors, also known as MCPs (for methyl-accepting chemotaxis proteins), engage CheA and an adaptor protein, CheW, to regulate CheA activity in response to ligand binding to the extracellular domain of the receptor (4-6). In addition, the MCPs undergo methylation and demethylation of specific glutamate residues in a feedback loop that modifies receptor properties in accordance with the level of kinase activity (5, 6). Despite considerable structural and biochemical characterization of MCPs, details are lacking for how the extracellular ligand binding domain affects the membrane-distal cytoplasmic regions where CheA interacts.
MCPs are broadly represented in Bacteria and Archea with multiple paralogs present in a given organism: E. coli has four well-studied MCPs (Tar, Tsr, Trg and Tap) (5, 6), whereas the pathogen Vibrio cholera has 45 identifiable MCP sequences (1). All biochemically and structurally characterized MCP domains have a dimeric architecture that likely holds for all members due to a universally conserved sequence of repeating hydrophobic residues that composes the C-terminal dimerization domain. The structural elements of MCPs include an N-terminal transmembrane helix (TM1), an extracellular ligand-binding domain (which can be variable), a second transmembrane helix (TM2), a cytoplasmic HAMP domain and a C-terminal domain (MCPC) that folds into a long anti-parallel 4-helix bundle, with two helices supplied from each subunit. Crystallographic structures for the E. coli Tar and Tsr extracellular domains show a dimer of two antiparallel four helix bundles that bind ligands at their interface (7). In contrast, MCP extracellular domains from receptors found in other organisms are expected to have quite different folds (8). An NMR structure for a naturally isolated HAMP domain of unknown function reveals an unusual parallel helical bundle structure, with two helices supplied from each subunit (9). Recent biochemical and genetic data strongly suggest that this structure is relevant for the HAMP domains within the E. coli MCPs (10, 11).
Conformational changes in the extracellular and HAMP domain translate into conformational changes in the cytoplasmic domain (MCPC) that affect the activity of CheA. MCPC can be further broken down into functional subdomains. Most proximal to the membrane is the adaptation region, which contains glutamine and glutamate residues that undergo covalent modification (6). Methylation of the Glu (and deamidation of Gln residues in certain receptors) affects ligand binding and CheA activation (12-15). The region with the highest sequence conservation is the membrane distal tip of the receptor that binds CheA and CheW. The subdomain between the adaptation region and signaling region has been defined as the flexible bundle region because it contains Gly residues important for function, exhibits higher thermal (B)-factors in crystal structures, and less canonical coiled-coil packing (8). There is considerable data for how conformational signals propagate from the ligand binding site, through TM2 to the HAMP domain for the E. coli MCPs Tar, Tsr, and Trg. Crosslinking studies, solid-state NMR, spin-label measurements, and replacement of membrane interfacial residues all indicate a piston-like motion of TM2 when ligands bind the extracellular domains (16-20) (5, 6). Less is known about how these signals affect MCPC and CheA. Furthermore, the length of MCPC can vary greatly among receptors, which can be classified based on the number of heptad repeats present in the 4-helix coiled-coil (28−44)(8). Crystal structures have been determined for a truncated form of MCPC from E. coli Tsr (class 36) (21, 22) and MCPC 1143 from T. maritima (Tm1143C class 44) (23). Both structures depict similar 4-helix bundles, although the C-terminal ends of TsrC are frayed due to truncation of the N-terminal helices (21). TsrC and Tm1143C also form very different packing interactions within their respective crystal lattices: TsrC forming a trimer of dimers (21), and Tm1143C an aligned hedgerow of dimers (23). There is strong evidence in E. coli that the MCPs do form trimers in the higher order structures that constitute the receptor arrays (4, 6, 24), although less is known about how the trimers associate with each other, CheA, and CheW. Whole cell tomography in several bacteria has revealed a hexagonal lattice for the arrays, which is fit well by a trimer-of-dimers (24-26). Nonetheless, Tm1143C does not form a trimeric structure in the crystal, although contacts between the molecules may be influenced by the low pH at which the crystals were grown (23). Direct contacts between neighboring molecules in the lattice are mediated by protonated Glu residues. Additional structures, crystallized under different conditions would be helpful to better explore the conformational states and detailed interactions possible among MCPC domains.
In addition to the transmembrane MCPs, there are related proteins in many bacteria that have an MCPC domain, but no transmembrane regions and are hence predicted to be soluble receptors (MCPS). MCPSs have CheA-interacting regions and N-terminal or C-terminal extension beyond the four-helix bundle that can be as small as a positively charged peptide, or as large as an entire domain(s) (e.g. P. aeruginosa McpS or R. sphaeroides TlpC) (27, 28). Herein, we report the structure of MCPS from T. maritima (Tm14). Tm14 has a small positively charged N-terminal peptide that extends beyond the bundle and no known modification sites. Tm14 does have sequence and overall structure similarity to Tm1143C; however its conformation is strikingly different in ways that provide insight into the conformational states available to cytoplasmic domains of chemoreceptors.
Materials/Methods
Gene Manipulation
The gene encoding Tm14 was PCR cloned into vector pET28a (Novagen) and expressed with a 6-Histidine tag in E. coli strain BL21 (RIL DE3) (Novagen). The cells were grown in Luria Broth (US Biological Sciences) with kanamycin (50 μg/mL) and the proteins were purified using Ni-NTA chelation chromatography as previously described (38). The purified protein was run on a Superdex200 26/60 sizing column prior to concentration (15 mg/mL) in 50 mM Tris, pH 7.5/150 mM NaCl. The Asn217Ile mutant was constructed with Quickchange mutagensis (Novagen) and expressed as described above.
Crystallization and Data Collection
Crystals of Tm14 fragment grew by vapor diffusion against a reservoir of 25% Dioxane after 4 days at room temperature. Pb derivatives were produced by soaking the crystal with 8.7 mM lead trimethyl acetate for 1 hour. Native diffraction data were collected under a 100 K nitrogen stream using a rotating anode X-ray generator with an R-AXIS IV detector (Rigaku). Anomalous diffraction data were collected at 13.1 KeV on the Pb derivative under a 100 K nitrogen stream at the Cornell High Energy Synchrotron Source beamline (F2) on an ADSC Quantum 315 CCD. In both cases 20% ethylene glycol was used as a cryoprotectant. The crystals belong to the space group P21 and contain one Tm14 dimer per asymmetric unit. Data were processed by HLK2000 (39) and XDS (40).
Structure Determination and Refinement
Diffraction data from both the native and Pb-derivatized crystals were processed with SOLVE and RESOLVE(41-43) to generate initial electron density maps based on anomalous diffraction from Pb and isomorphous differences between the Pb and native data (figure-of-merit = 0.4 to 2.6 Å resolution). A partial structure was built into the initial maps and then helices from the model were used as probes for molecular replacement with PHASER (44) to place the missing helical regions. Positional and thermal parameters were refined with CNS amidst cycles of manual model building and solvent molecule placement (45).
Graphics
Molecular representations were made with Molscript (46) and SPOCK (46). Solvent accessible (molecular) surfaces were calculated with SPOCK (46) using a solvent probe of radius 1.4 Å.
Results
Tm14 (278 residues, MW = 31,660 kD, and pI = 5.23) has a positively charged 15 residue peptide that extends beyond the predicted 4-helix bundle. When expressed in E. coli, the purified protein inhibits the autophosphorylation activity of T. maritima CheA in a manner that is augmented by the presence of CheW (Supplemental Fig. 1). The full-length protein had a tendency to degrade over time and thus a more stable, symmetric variant (residues 41−254) was produced for crystallization.
Crystals of the truncated Tm14 (space group P21, a = 68.7 Å b = 25.8 Å c = 119.6 Å β = 93.81) grew in 20% dioxane and diffracted to 2.15 Å resolution, which far exceeds that of the other two known MCPC structures (PDB codes 1QU7 and 2CH7). The Tm14 structure was determined by single wavelength anomalous diffraction (SAD) of Pb-soaked crystals and refined to R = .254 and Rfree = .280 (Table 1).
Table 1.
Data collection and Refinement Statistics for Tm14
| Diffraction Statistics | |||
|---|---|---|---|
| Space group = P21 (β = 93.81) | a = 68.71 | b = 25.75 | c = 119.61 |
| Native Tm14 | Pb Tm14 | Tm14 Asn217Ile | |
| Resolution (Å) | 2.16 (2.22 − 2.16)g | 2.15 (2.21 − 2.15)g | 3.00 (3.11 − 3.00) |
| Number of unique reflections | 24221 | 45726 | 8972 |
| Number of observations | 70197 | 168739 | 31676 |
| % Completeness | 96.5 | 97.5 | 99.8 |
| 〈I/σI〉a | 12.3 (4.6)g | 16.9 (6.1)g | 22.5 (6.3) |
| RSymb (%) | 6.1 (23.6)g | 6.2 (20.3)g | 16.4 (47.6) |
| SAD structure solution statistics | |
|---|---|
| Resolution cut-off (Å) | 2.50 |
| Number of Anomalous sites found | 2 |
| Mean figure of merit | 0.38 |
| Overall Z-score | 6.59 |
| Refinement Statistics | Native Tm14 | Asn217Ile |
|---|---|---|
| Number of residues | 426 | 426 |
| Resolution | 2.17 (50 − 2.17) | 3.00 (20.0 − 3.00) |
| Wilson B | 39.3 | 52.6 |
| Number of water molecules | 424 | 416 |
| Rc (%) | 24.2( 38.5 )g | 25.9 (26.4) |
| Rfreed (%) | 27.8 ( 41.4 )g | 30.5 (32.8) |
| Overall 〈B〉e (Å2) | 45.6 | 58.1 |
| Mainchain 〈B〉 (Å2) | 40.8 | 52.5 |
| Sidechain 〈B〉 (Å2) | 46.0 | 59.3 |
Intensity of the signal to noise ratio.
for all reflections (no σ cutoff).
Rfree calculated against 10% of reflections removed at random.
Overall model average thermal 〈B〉 factor.
Root mean square deviations from bond and angle restraints.
Highest resolution bin for compiling statistics.
Similar to Tm1143C and TsrC, the structure of Tm14 forms a dimeric antiparallel four-helix bundle (Fig. 1), with each helix having a heptad repeat of hydrophobicity that is commonly found in coiled-coils (i.e. a-b-c-d-e-f-g with the a and d residues mostly hydrophobic and buried inside the core of the dimer interface). However, unlike the structures of Tm1143C and TsrC, Tm14 has an unusual distortion in its middle and shows deviations from the standard helical packing patterns (Figs. 1 and 2). This “bulge” separates the helices and allows a water molecule to penetrate the hydrophobic core. The bulge, which resides at a position analogous to that of MCP modification sites in the adaptational regions, affects the overall shape of the helix bundle (Figs. 2 and 3).
Fig. 1.
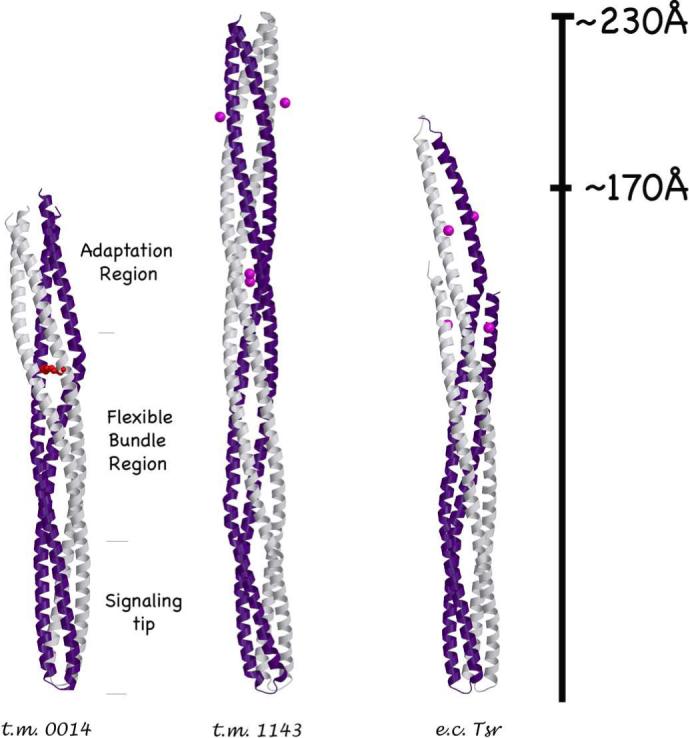
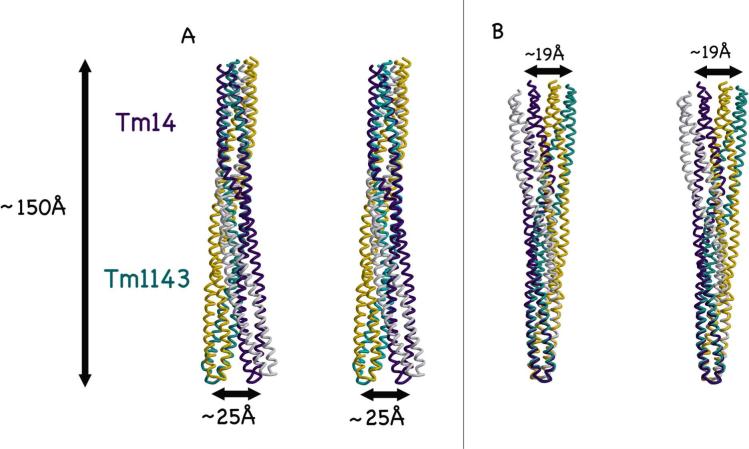
Tm14 compared to other known MCPC structures. Chain A (gray) and Chain B (purple). Methylation sites on Tm1143C and E. coli TsrC (magenta spheres) are found in the same region as the Tm14 bulge (red).
Fig. 2.
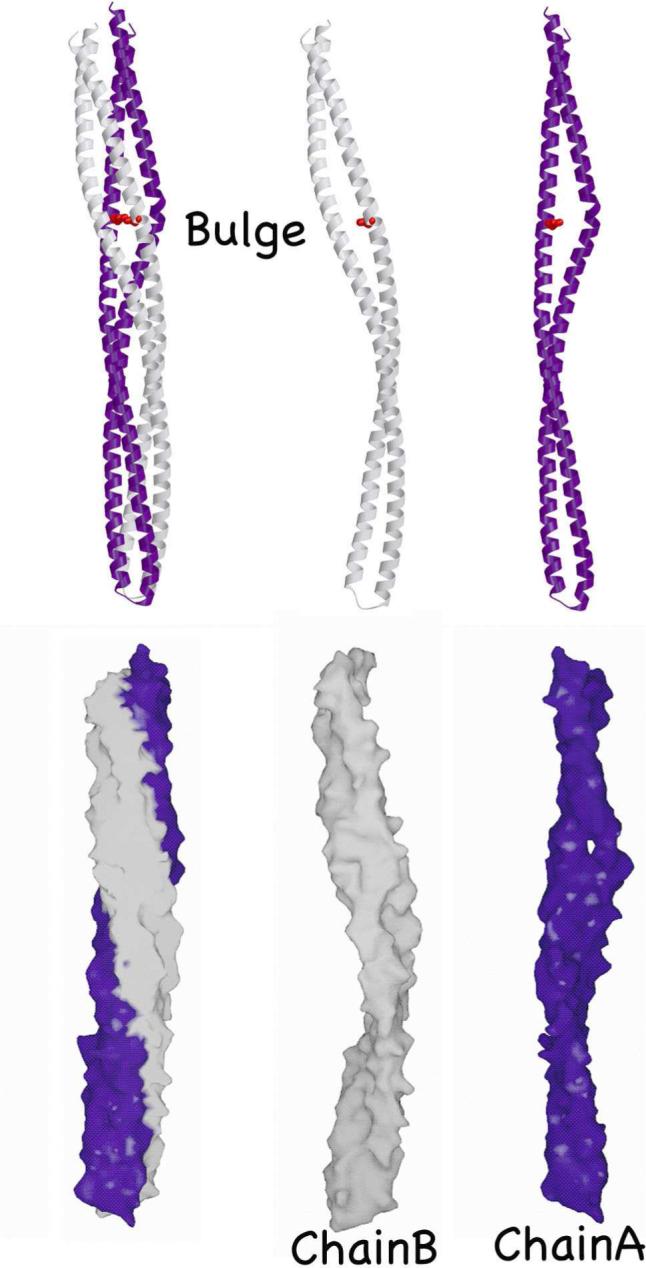
Asymmetry between subunits A and B in Tm14. Subunit B is kinked at the bulge, but elsewhere quite linear, whereas subunit A is curved through out its length. The bulge in subunit B makes a solvent accessible gap in the molecular surface.
Fig. 3.
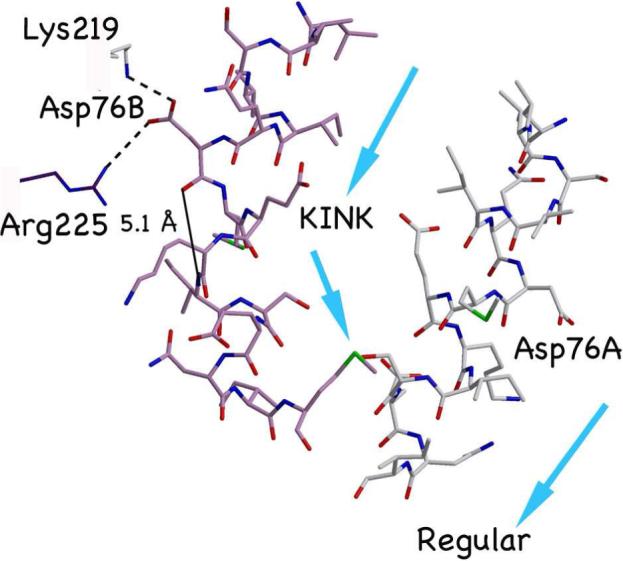
Close-up of the bulge distortion. The kink at Asp76B generates a 45 degree change in the direction of the helical axis. Interactions of Asp76 correlate with local helix unwinding in subunit B. Asp76B breaks peptide hydrogen bonding within the helix, but the side chain forms salt bridges with two positively charged residues on different dimers in the crystal lattice. Arrows show directions of the individual helical axes.
There are two general packing arrangements found in anti-parallel 4-helix coiled-coils. In one class, the relative offset of the heptad repeats on N- and C-terminal helices is half of one heptad, which would place the 4 interior residues in roughly the same plane perpendicular to the supercoil axis. However, Tm14 and the other chemoreceptors have an offset of 0.25/heptad, which interdigitates the hydrophobic sites on N- and C-terminal helices. This smaller offset is typical of a ferritin-like alacoil such as the Lac repressor ((29) and PDB code: 1LBH) and shifted from that found in the coiled-coil modules of other chemotaxis proteins, such as the CheA dimerization domain. The structures of the three known receptors (TsrC, Tm1143C, Tm14) are all very similar in the highly conserved, membrane distal tip that interacts with CheA. However, there is more structural variation among the receptors in the adaptation and flexible bundle regions that compose the helical stalks.
Helical bundle parameters, calculated with HELANAL (30), reveal substantial distortions and asymmetry in Tm14. HELANAL calculates vectors fit to four successive C-alpha positions and defines a local bending angle between neighboring vectors along the helix. If the local bending angle is more than 20 degrees the helix is classified as kinked. Furthermore, the degree to which the origins of the vectors trace a line or a circle classify the helix as linear or curved (30). This analysis shows that both subunits (A and B) of Tm1143C and TsrC have kinks throughout their structure (Supplemental Fig. 2). However in Tm14, subunit A curves for its entire extent, but subunit B kinks at the bulge and then is mostly linear moving down towards the tip (Fig. 2). This asymmetry is also reflected in the local helical bending angle of subunit A (Supplemental Fig. 2), which does not change substantially, unlike that of subunit B and both subunits of either TsrC or Tm1143C. Pulling the two subunits apart reveals that subunit B must become straight after the kink in order to maintain close contact with the continually curved subunit A (Fig. 2). Not only is there asymmetry among subunits A and B but also among the N-terminal and C-terminal helices. Taking the pitch between parallel sets of helices as an indicator for supercoiling, the C-terminal helices supercoil to a much greater degree (93 Å pitch) compared to the N-terminal helices and typical coiled-coils (150−200 Å pitch) (31). Viewed another way, the C-terminal helices have more left-handed twist than the N-terminal helices. To our knowledge, the degree of structural asymmetry in Tm14 is unprecedented for a long 4-helix bundle structure.
The net result of subunit asymmetry resulting from the helical bulge is to shift the conserved signaling tip relative to the flexible bundle region (Fig. 4). Packing within the conserved region of the tip is regular and very similar to that found in the structures of Tm1143C and TsrC. However, superimposing the helical stalks of Tm14 with either Tm1143C or TsrC reveals that in Tm14 the tip is displaced ∼ 25 Å away from its position in the more symmetric receptors. In contrast, if one superimposes he conserved tips, the stalks spread out by about 20 Å. In either case, large motions of the regions known to interact with CheA and CheW, or the modification enzymes may have functional relevance (see below).
Fig. 4.
The bulge causes displacement of the Tm14 signaling tip relative to the helical stalk. A) Stereoview of the superposition of Tm1143 (light seagreen and gold) and Tm14 (dark purple and light grey) reveal the repositioning of the tip by ∼25 Å as a result of a distortion in the helical stalks. Tm1143 represents how Tm14 would appear if both subunits were more symmetric. The C-alpha trace of residues 217−252 of Tm14 superimposed with a RMS of 1.7 Å . B) Stereoview of the 19 Å movement of the stalks if the conserved tips are superimposed (residues 149−164).
The greatest degree of asymmetry between the Tm14 subunits localizes in the aforementioned bulge 108 Å from the signaling tip. Here, the side chains of Met77A and Met77B in the d positions on the N-terminal helices move away from the center of the bundle, whereas the corresponding d residues on the C-terminal helix, Asn217A and B, direct their side chains at each other and hydrogen bond across the bundle core. Although most d positions of chemoreceptors hold hydrophobic residues, nearly all MCP sequences contain at least one hydrophilic d residue. In Tm14, the Asn217 interaction forms an “x-layer”, as defined by (32), in which the two Cα-Cβ bonds point at each other and are nearly in the same plane (Fig. 5). The polar Asn side chains also likely facilitate the penetration of ordered solvent into the bundle core.
Fig. 5.
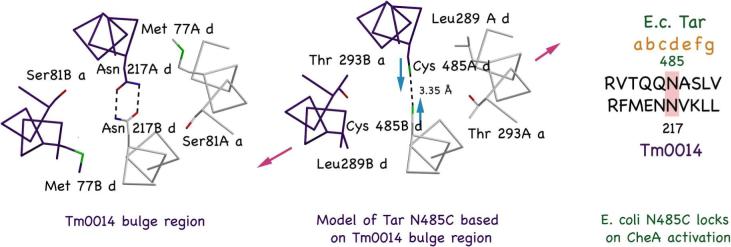
Relevance of Tar lock-on disulfide bonds to the bulge distortion of Tm14. Sequence similarity between the heptads in Tar where an engineered Cys cross-link locks on CheA activation and the heptad in Tm14 that forms the bulge (center). Left: Helical packing of Tm14 showing the two internally hydrogen-bonded Asn217 residues. Middle: Model of Tar adaption region where disufide crosslinking locks-on CheA activity. A disulfide bond would generate an even shorter distance between helices and a more prismatic distortion of the bundle core. Right: Sequence alignment of the Tar heptad model with the Tm14C distortion region.
The pinching together of Asn217 A and B displaces the N-terminal helices outward and forms a prismatic structure in cross-section (Fig. 5). At the periphery of the bulge, Asp76B breaks from helical geometry to the greatest degree and has larger thermal (B) factors than nearby residues. The Asp76B carbonyl oxygen points out from the helix and is 5.1 Å from what would be its i+4 hydrogen-bonding partner, the amide nitrogen of Ile 80 (Fig. 3). Notably, the charge on Asp76B is at least partially neutralized through salt bridges with Lys 219A and Arg 225B on neighboring dimers in the crystal lattice (Fig 3). In contrast, Asp 76A does not salt bridge with symmetry related dimers, maintains regular helical packing, and unlike Asp76B, has low B factors consistent with the rest of the structure. Charge compensation at an aspartate is interesting because Asp76 is positioned analogously to Glu residues in other MCPs that are known to undergo neutralization by the methylation reactions of the adaptation response. Thus, the crystal contacts provided by Lys219A and Arg225B may be a chemical analogy to methylation and have thereby serendipitously promoted a distortion normally caused by the biologically relevant mechanism of charge neutralization. It is worth noting that because the interactions between molecules in the lattice are extensive (as they also are for the other MCPC structures), we cannot rule out additional influences of crystal packing on the Tm14C conformation. Nonetheless, these domains are situated close to each other in the receptor arrays and thus their modes of interaction, as demonstrated in the crystal lattice are of interest.
To test the importance of hydrogen-bonding within the helix core for stabilizing the Tm14 distortion, the Asn217 residues were mutated to Ile, a common d position residue. Crystals of the Asn217Ile Tm14C grew under similar conditions and were isomorphous with those derived from the wild-type (wt) sequence, but diffracted to much lower resolution (3.0 Å, Table 1). The structure of the variant is similar to that of the wt in the region of the bulge with the Ile217 residues forming van der Waals contacts across the bundle core (data not shown). However, increased thermal factors and less discernible electron density indicate considerably more disorder in the Ile variant structure. Thus, under this crystallization condition and in this lattice environment, buried Asn residues are not necessary to form a bulge. Nonetheless, the energetic penalty for not satisfying the Asn hydrogen bonds in the bundle core makes it likely that these residues promote the distorted conformation in the context of the wt sequence.
Crystal packing interactions provide some information for how MCPCs will interact with each other when at high concentration. For one dimension of the crystal lattice, the Tm14 receptors stack in hedgerows, much like those found in the structure of Tm1143C. However, the hedgerow stacking includes the tip distortion, such that all of the aligned receptors bend in the same direction. Neighboring hedgerows associate head-to-tail with interactions between neighboring dimers mediated by salt bridges between highly conserved residues that flank the signaling tip. Arg131A and Arg146B on one dimer interact with Glu149B and Glu160B on another, respectively. Arg146, Glu149, and Glu160 are strictly conserved in chemoreceptors (8) and their salt bridges bring two receptor tips into a close-packed interface of 633 Å2 of buried surface area per dimer (Supplemental Fig. 3). Arg131 lies at the beginning of the highly conserved region and this residue position mediates receptor-to-receptor contacts in the other two MCP structures. In TsrC this residue is a Phe involved in the trimer interface (21, 33) whereas in Tm1143C this residue is a Glu (presumably protonated) which interacts with its symmetry mate in an aligned neighboring dimer (23). All the other T. maritima transmembrane receptors including Tm1143 have a Glu in this position, only the soluble Tm14 has an Arg. In the Tm14 structure, the conserved Glu and Arg residues align the respective tips in an anti-parallel configuration, which would be a permitted association mode for a soluble receptor, and also for a transmembrane receptor in the context of a membrane invagination (26).
Discussion
The overall conformation of the receptor dimer is much more distorted in Tm14 than in Tm1143C. These structural differences may derive from the higher crystallization pH of Tm14 compared to Tm1143C, which produced crystals at low pH where most of the surface acid groups were likely neutralized (23). Structures of TsrC that had two Glu residues replaced with Gln to mimic methylation had lower thermal (B)-factors in the adaptation region (22). Furthermore, studies of E. coli Tar corroborate that removal of negative charge on the receptor surface by residue substitution reduces flexibility; this in turn enhances CheA activation (34). Thus, the greater distortions of Tm14 may reflect its increased dynamics under conditions where more surface anionic groups are ionized. Notably, the largest distortion in the Tm14 bundle occurs at a position where a surface Asp (76) salt-bridges to other positively charged residues in adjacent molecules. Thus, neutralization of surface charge in Tm14 correlates with the local unwinding of a helix and disruption of the bundle packing, thereby imparting flexibility to the entire molecule.
Engineering disulfide bridges into the Tar receptor can dramatically affect its ability to regulate CheA. In particular, cross-links at some d positions, lock on CheA activity (35). These sites are contained in heptads very similar in sequence and position to the Tm14 heptad that forms the bulge containing Asn217 (Fig. 6). A disulfide bond at the 217 position would bring d residues on adjacent subunits even closer together than is achieved by the hydrogen-bonding Asn residues. For a disulfide to form, the C-terminal helices will have to pinch in, and a bulge of the N-terminal helices, as found in Tm14, will likely result (Fig. 6). Thus, a distortion, not unlike that observed for Tm14 may contribute to the lock-on phenotype.
Fig. 6.
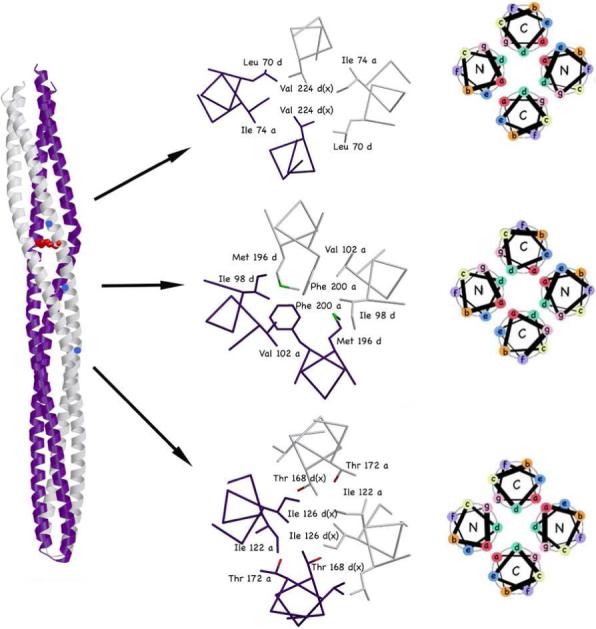
Variations in helical packing in different positions throughout the Tm14 structure. Subunits A and B shown in gray and purple, respectively. The heptad is represented by sites a-f in the helical wheel formation. The helical packing in the Tm14 structure is not regular but varies significantly across the length of the receptor. Top: mixed x-da layers packing; Center: da-layer packing; Bottom: x-layer packing.
How then could such local distortions affect the CheA kinase, which binds to the conserved tip over 100 Å away? The kink in Tm14 does not translate to irregular helical packing in the kinase-binding region and the conserved tip is very similar in structure among all of the characterized MCPCs. However, what does differ among Tm14, TsrC and Tm1143C is the position of the tip relative to the stalks. The kink in subunit B generates asymmetry between the subunits that manifests over their entire length. As a consequence, the tip swings out, > 25 Å from its comparable position in the other receptors if the stalks are aligned above the adaptation region (Fig. 4). Conversely, if the tips are taken as a fixed point and superimposed, the stalks spread ∼20 Å. Thus, the Tm14C structure demonstrates one way a local conformational change in the flexible region high-up in the helical stalks can influence a more static kinase-binding region downstream.
Movement of the receptor tip relative to the stalks in Tm14C derives directly from helical packing irregularities in the bulge. Charge neutralization of Asp76B in the crystal lattice suggests how packing distortions may result from changes in methylation state within the adaptation region, but could such distortions also be propagated from the membrane proximal regions of the receptor in response to ligand binding? It has been proposed that signaling through the HAMP domain involves switching between two nearly isoenergetic helical packing modes related by relative rotations of the helices (9). Instead of “knobs-into-holes” packing typical of coiled coils, the HAMP domain structure has “knobs-into-knobs” packing that generates so-called complementary x-da layers (Fig. 5). In a canonical coiled-coil, the a and d residues pack into the core symmetrically and equally. Nonetheless, most coiled coils of length greater than a few heptads show some kind of discontinuity, which can be classified as either a stutter (4-residue insertion into the heptad repeat) or a stammer (3-residue insertion) (32). In a stutter, a residues point into the center of the core and push the d and e residues out to form a ring around the core. In a stammer, d residues point into the center of the core and the a and g residues form the ring around the core. An “x-layer” is formed from residues that point into the core, whereas a “da-layer” forms from residues in the peripheral ring ((32) and Fig 5). Such discontinuities can impart flexibility and may generate the structural specificity needed to discriminate subunit interactions among different types of receptors
The HAMP domain is thought to toggle between knobs-into-holes packing and complementary x-da packing through a rotation induced from signals sent through the membrane (9). Surprisingly the structure of Tm14 assumes a variety of packing modes at different positions along its length, including the two proposed for the signaling states of the HAMP (Fig. 5). In general most of the C-terminal helices are in the x position where in the N-terminal helices the da position dominates. However, the C-terminal helices become more x-like moving away from the tip, whereas the N-terminal helices become more x-like down toward the tip. In the bulge, a da configuration of the N-terminal helices allows the C-terminal helices to form a highly prismatic x-layer in which the Asn217 residues make close contact. Outside the flexible bundle region of Tm14 and in all of Tm1143C the packing is mostly symmetric x-layers, staggered by 0.25 of a heptad. Such pure x-layer packing is incompatible with the bulge distortion of Tm14 because of steric hindrance from the core residues in the layers above and below. However, it seems likely that in solution the helical packing in Tm14 may fluctuate to a more symmetric x configuration. The different packing arrangements observed in the MCPC structures over their length, despite very similar sequence contexts suggests that their structures can readily change. Thus, forces that influence local helix interactions within the coiled-coil, perhaps exerted by conformational change within the HAMP domain or transmembrane region could have a substantial impact on the overall receptor conformation.
The Tm14 structure indicates that changes in local helical packing, mediated by x-da layers, can be readily translated into bending of the stalks and translation of the conserved tip. This amplification of spatial displacement at the tip derives directly from the length of the receptor, the position of the distortion and coupling among the main structural parameters that ultimately determine coiled-coil conformation. FRET studies of Tsr-YFP fusions suggest that activating ligands induce substantial bending motions in the receptor stalks (36). Furthermore, genetic studies of Tsr that introduced mutations into the signaling region showed that prolines (as opposed to Trp or Ala residues) were the most devastating for clustering, kinase activation and ternary complex assembly (33). Proline residues generate kinks in helices and may thereby distort the position and/or conformation of the tip. Similarly, replacements at conserved Gly residues in the helical stalks of the E. coli receptors have dramatic effects on CheA activation (37). The inherent flexibility of Gly residues may facilitate tip-bending distortions. Indeed, distortions in from regular helical packing in Tm1143C often localize at Gly residues.
Overall, the structure of Tm14 reveals a new MCPC structural state that provides possible explanations for how changes in packing and surface properties within the receptor stalk can reposition the kinase-activating tip (Fig. 4A). Such local distortions could arise from modification in the adaptation region and/or packing rearrangments induced by the HAMP domain. If the distortions observed in Tm14C are important for receptor function we are left with the possibilities of either the stalks or tips remaining fixed and the other moving, or some combination of both. Because CheA and CheW bind the tip, it seems likely that this interaction region must in some way change structure or position to relay signals to the kinase. If the tips were to remain fixed in all signaling states of the receptor and the major conformational changes were in the stalk region, it is not evident how the kinase would be impacted given that our structures do not support a conformational change within the tip itself. The structure within the region of high sequence conservation at the tip does not change between the two conformational states, i.e. the subunit asymmetry and distortions that differentiate the receptors manifest “above” the kinase-interacting region. A recent study of cryo-electron tomograms of overexpressed Tsr chemoreceptors finds two primary structural states: 1) an expanded trimer of dimers (CheA inhibiting) and 2) a compact trimer-of-dimers (CheA activating). These states differ mainly by movement of the HAMP domains by 25 Å in a plane roughly perpendicular to the trimer axis. If we superimpose the distortion in Tm14 onto Tm1143 this time with the assumption that the tips always remain associated by the trimer contact then the difference in distance between the start of the adaptation region is about 11 Å (Fig. 4B), which on extrapolation up toward the membrane would be consistent with a 25 Å difference at the HAMP domains (26). This study also observed that the expanded form results in an 8% reduction in the height of the trimers (26). It seems plausible that this height reduction might result from a bending or tilting of the individual dimers in the trimer formation. Thus although the precise mechanism by which conformational changes in the receptor trimers effect CheA remain unresolved, flexing of the helical stalks and repositioning of the receptor tips are likely to play an important role.
Finally we note that the Tm14C structure is highly asymmetric. If the receptors contained within the signaling arrays are in contact with each other, there may be a strong tendency for them to distort together in one direction. Thus, the swinging motion of the tip could reorganize interactions among receptors, CheA, and CheW in a highly cooperative manner.
Supplementary Material
Acknowledgements
We would like to thank Andy Arvai for assistance with the structure determination, the Cornell High Energy Synchrotron Source (CHESS) for access to data collection facilities.
Abbreviations
- HAMP domain
domain found in Histidine kinases, Adenylyl cyclases, Methyl-accepting chemotaxis proteins and Phosphatases
- MCP
methyl-accepting chemotaxis protein
- Tar
E. coli aspartate receptor
- Tm14
T. maritima soluble receptor
- Tm1143
T. maritima transmembrane receptor
- Tm1143C
Tm1143 cytoplasmic domain
- Tsr
E. coli serine receptor
- wt
wild-type
Footnotes
This work was supported by NIH Grant GM066775 (B.R.C)
Supporting Information
Affect of Tm14 on CheA activity. Structural parameters of three receptor signaling domains. Head-to-tail crystal packing of Tm14. Electron density for Tm14. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 4.Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr Opin Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends In Biochemical Sciences. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh JI, Biemann HP, Prive GG, Pandit J, Koshland DE, Kim SH. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. Journal Of Molecular Biology. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 8.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 10.Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: A disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. Journal Of Bacteriology. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bornhorst JA, Falke JJ. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: Development of a heterogeneous two-state model. Journal Of Molecular Biology. 2003;326:1597–1614. doi: 10.1016/s0022-2836(03)00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai WC, Beel BD, Hazelbauer GL. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Molecular Microbiology. 2006;61:1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x. [DOI] [PubMed] [Google Scholar]
- 14.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 15.Park CY, Dutton DP, Hazelbauer GL. Effects Of Glutamines And Glutamates At Sites Of Covalent Modification Of A Methyl-Accepting Transducer. Journal Of Bacteriology. 1990;172:7179–7187. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller A. S. a. F., J.J. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 18.Danielson MA, Biemann HP, Koshland DE, Falke JJ. Attractant-Induced And Disulfide-Induced Conformational-Changes In The Ligand-Binding Domain Of The Chemotaxis Aspartate Receptor - A F-19 Nmr-Study. Biochemistry. 1994;33:6100–6109. doi: 10.1021/bi00186a009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottemann KM, Thorgeirsson TE, Kolodziej AF, Shin YK, Koshland DE. Direct measurement of small ligand-induced conformational changes in the aspartate chemoreceptor using EPR. Biochemistry. 1998;37:7062–7069. doi: 10.1021/bi980305e. [DOI] [PubMed] [Google Scholar]
- 20.Ottemann KM, Xiao W, Shin Y-K, Koshland DE., Jr. A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 21.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 22.Kim SH, Wang WR, Kim KK. Dynamic and clustering model of bacterial chemotaxis receptors: Structural basis for signaling and high sensitivity. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor:kinase assembly. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 24.Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Molecular Microbiology. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang PJ, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khursigara CM, Wu XW, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. Journal Of Bacteriology. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardy SL, Maddock JR. Polar localization of a soluble methyl-accepting protein of Pseudomonas aeruginosa. J Bacteriol. 2005;187:7840–7844. doi: 10.1128/JB.187.22.7840-7844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wadhams GH, Martin AC, Porter SL, Maddock JR, Mantotta JC, King HM, Armitage JP. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Molecular Microbiology. 2002;46:1211–1221. doi: 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- 29.Gernert KM, Surles MC, Labean TH, Richardson JS, Richardson DC. The Alacoil - A Very Tight, Antiparallel Coiled-Coil Of Helices. Protein Science. 1995;4:2252–2260. doi: 10.1002/pro.5560041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bansal M, Kumar S, Velavan R. HELANAL: A program to characterize helix geometry in proteins. Journal Of Biomolecular Structure & Dynamics. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- 31.Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. Journal Of Structural Biology. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 32.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Fibrous Proteins: Coiled-Coils, Collagen And Elastomers. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 33.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: Electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winston SE, Mehan R, Falke JJ. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: Cysteine and disulfide scanning studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. Journal Of Molecular Biology. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W. Automatic Processing Of Rotation Diffraction Data From Crystals Of Initially Unknown Symmetry And Cell Constants. Journal Of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 41.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallographica Section D-Biological Crystallography. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallographica Section D-Biological Crystallography. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallographica Section D-Biological Crystallography. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal Of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Christopher JA, Baldwin TO. SPOCK: Real-time collaborative molecular modelling. Journal Of Molecular Graphics & Modelling. 1998;16:285–285. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.