Abstract
We have studied the role of the basic helix–loop–helix–PAS transcription factor EPAS-1/hypoxia-inducible factor 2α in vascular development by gene targeting. In ICR/129 Sv outbred background, more than half of the mutants displayed varying degrees of vascular disorganization, typically in the yolk sac, and died in utero between embryonic day (E)9.5 and E13.5. In mutant embryos directly derived from EPAS-1−/− embryonic stem cells (hence in 129 Sv background), all embryos developed severe vascular defects both in the yolk sac and embryo proper and died between E9.5 and E12.5. Normal blood vessels were formed by vasculogenesis but they either fused improperly or failed to assemble into larger vessels later during development. Our results suggest that EPAS-1 plays an important role at postvasculogenesis stages and is required for the remodeling of the primary vascular network into a mature hierarchy pattern.
The genesis of a functional vascular system requires the successful execution of a complex set of developmental programs. First, vascular channels arise from nonendothelial progenitors through vasculogenesis, a process requiring differentiation, proliferation, migration, and cell/cell interactions. Once formed, some primary vascular channels, often small in size, interact with one another to form large blood vessels. Such interactions appear to be rather specific because typically only certain subsets of microvessels at defined locations would actively merge to form large vessels, whereas others maintain their discrete small tubular structures. This process, often referred to as vascular remodeling, reshapes the morphology of the vascular system from one that consists of uniformly sized small vessels to one that displays a hierarchy of large and small vessels. Thus, like other processes such as vasculogenesis and angiogenesis, remodeling is a critical aspect of vascular development.
Many components important for vascular development have been discovered. For instance, it is now well established that the vascular endothelial growth factor (VEGF/VEGF-A) (1, 2) and its receptors, Flt-1(3, 4), Flk-1/KDR (5–7), and neuropillin-1 (8), play essential roles in vasculogenesis and angiogenesis (9–13). Other important examples include angiopoietins (14–17), Tie-1 and Tie-2/Tek tyrosine kinases (18–20), VEGF-C and its major receptor Flt-4 (21, 22), ephrin-B2 and its receptor Eph-B4 (23, 24).
Recently, two basic helix–loop–helix (bHLH)-PAS transcription factors, ARNT-1 (25, 26) and hypoxia-inducible factor 1α (27–29), have been shown to play important roles in embryonic vascularization. Mouse embryos deficient in these proteins exhibit dilated and fused blood vessels and die in utero. The bHLH-PAS protein EPAS-1 (hypoxia-inducible factor 2α, HLF, HRF, or MOP2) (30–34) is closely related to hypoxia-inducible factor 1α, interacts with the common heterodimerization partner ARNT-1 (35), and is predominantly expressed in endothelial cells. However, the role of EPAS-1 in vascular development remains elusive, although previous studies have demonstrated its requirement in regulating catecholamine homeostasis (36). Here we report that in our independently generated EPAS-1 knockout mice, a significant percentage of homozygous mutants displayed hemorrhaging and failed to maintain discrete vascular tubular structures or assemble larger vessels from smaller precursors. These observations indicate that EPAS-1 plays an important role in controlling vascular remodeling.
Materials and Methods
DNA Manipulation.
A 407-bp EPAS-1 cDNA fragment (nucleotides 99–505) (30) was generated by reverse transcription–PCR from mouse lung total RNA by using two primers (AATGACAGCTGACAAGGAGAAAAA and GAGTGAAGTCAAAGATGCTGTGTC), and a genomic clone was isolated from 129 Sv genomic library (Stratagene). Construction of the targeting vector is described in Fig. 1. The following primers were used for genotyping by PCR: AGCTCAGAGCTGAGGAAGGAGAAA and CTTATGTGTCCGAAGGAAGCTGA for the wild-type allele, amplifying a 174-bp band; and TGGCCACAACCATGGAACAGGG and AAAGCGCCATTCGCCATTCAGGCTGCGCA for the targeted allele, amplifying a 246-bp fragment (Fig. 1C). To detect the expression of EPAS-1 mRNA, primers AGGCCGACCAGCAAATGGA (exon 3) and GAGTGAAGTCAAAGATGCTGTGTC (exon 4) were used to amplify a 163-bp fragment of EPAS-1 cDNA, or CTTCTATGGGAGGCAGATCCAACACG and CAACAGGTAAGGCTCGAACGATGGCC were used to amplify a 633-bp fragment. The exact locations for the latter two primers are unknown on the mouse gene, but they correspond to exons 15 and 16 in the human homologue (30).
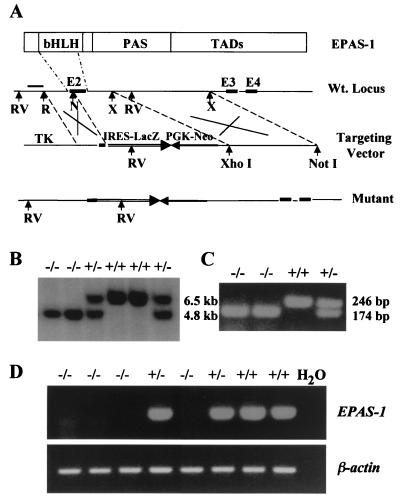
Figure 1.
Targeting of the EPAS-1 gene. (A) Targeting construct. A 1.9-kb EcoRI (R)/NaeI (N) fragment was used as the 5′ arm and a 6.2-kb XmnI (X) fragment as the 3′ arm. Homologous recombination results in the replacement of most of exon 2 (E2) and a region of intron after it (≈ 1.0 kb) by IRES-lacZ and PGK-Neo sequences. EcoRV (RV) digestion generates a 6.5-kb fragment for the wild-type allele and a 4.8-kb fragment for the targeted allele, both of which hybridize with the external probe (bar). (B) A Southern blot of DNA samples from embryos resulting from heterozygotes mating. (C) Genotyping by PCR. (D) Reverse transcription–PCR analysis of total RNA of E11.5 embryos for the expression of EPAS-1. The amount of template used and PCR efficiency for each reaction was normalized by reverse transcription–PCR for β-actin.
Embryonic Stem (ES) Cell Cultures.
The targeting construct was linerized by NotI and introduced into R1 ES cells (gift from A. Nagy and J. Rossant, University of Toronto) by electroporation. ES cell clones double-resistant to G418 and gancyclovior were selected and expanded, and genomic DNA was isolated for Southern blotting, according to the protocols of Wurst et al. (37). To select for EPAS-1−/− ES cells, EPAS-1+/− ES cells were cultured for 9 days in the presence of 2.0 mg/ml of G418 at 2 × 105 cell per 10-cm plate (GIBCO/BRL). Surviving colonies were expanded, duplicated, and screened by Southern blotting.
Mice.
Chimeric mice were constructed by in vitro aggregation of targeted ES cells (EPAS-1+/−) with ICR morulae (Harlan Sprague-Dawley) (38). Male chimeras were mated with ICR females to obtain germ-line transmissions of the targeted allele. The heterozygotes were identified by 5-bromo-4-chloro-3-indolyl β-d-galactoside staining of tail specimen, because IRES-lacZ was expressed strongly in tail blood vessels despite its low levels of expression in embryos. The validity of this procedure was confirmed by Southern blotting. Genotyping of embryos was done by Southern blotting or PCR.
Completely ES cell-derived embryos were produced as described by Nagy et al. (39). Briefly, wild-type embryos were electrically fused at the two-cell stage, resulting in tetraploidity. Each single-celled tetraploid embryo developed to four cells after overnight culturing and was used for aggregation with ES cells. The aggregates formed blastocysts after overnight incubation and were transferred into the uteri of pseudopregnant females. In our hands, each aggregation experiment had the capacity of generating 0–10 embryos. Therefore, multiple aggregation experiments were performed for both +/− and −/− ES cells, and cumulative numbers are reported.
Histology.
Immunohistochemical staining with anti-CD31 (Mec13.3, PharMingen) was performed essentially as described by Sato et al. (19). Briefly, embryos were fixed at 4°C with 4% paraformaldehyde and rinsed with PBS. The fixed embryos were treated with 5% H2O2, rinsed with PBS, and incubated overnight in PBSMT-NGS (PBS containing 3% instant milk, 0.1% Triton X-100, and 2% normal goat serum). Embryos then were incubated with Mec13.3 in PBSMT-NGS overnight, rinsed, incubated with goat-anti-rat IgG-horseradish peroxidase, and subjected to color development after extensive washing. The stained embryos and yolk sac membranes were photographed and processed for paraffin-based sections. Sections were cut to 5 μm and counterstained with nuclear fast red.
Results
Generation of a Null Mutation in the EPAS-1 Gene.
To introduce a targeted mutation in the EPAS-1 gene, we deleted most of the second exon and about 1.0 kb of the adjacent intron (Fig. 1A), disrupting the basic and helix–loop–helix domains essential for DNA binding and heterodimer formation. The first exon encodes only nine amino acid residues without known function and so is unlikely to have partial EPAS-1 activity. Independent germ lines carrying the targeted allele were obtained from two different ES cell clones. Examples are shown in Fig. 1 B and C. To determine whether there was any cryptic EPAS-1 mRNA in EPAS-1−/− mutants (abbreviated as −/− from here on), we did carefully controlled reverse transcription–PCR analyses of total RNA extracted from embryonic day (E)11.5 embryos. To ensure the reliability of these experiments, two independent pairs of primers were used that corresponded to two different regions downstream from the targeted sequence. Both sets of primers demonstrated specific absence of signals in −/− samples, and an example is shown in Fig. 1D. Therefore, cryptic EPAS-1 mRNA, if any, was at levels undetectable by PCR.
Hemorrhage in Homozygous Mutants.
We bred male chimeras to ICR females to obtain germ-line transmissions of the targeted allele, resulting in F1 progenies in 129 Sv/ICR background. Among F2 progenies resulting from heterozygotes mating, about one-third of −/− mice were born alive, but they were much smaller than normal littermates and had relatively short lifespans, typically a few weeks. The remaining two-thirds of the homozygotes died in utero. The −/− embryos fell into four classes when examined at various stages between E9.5 and E17.5: (i) dead and severely necrotic (completely pale or even partially resorbed); (ii) dead but not yet necrotic (or only slightly necrotic), often hemorrhagic; (iii) viable but hemorrhagic; and (iv) viable without obvious defects. The relative abundance of different classes of mutants varied at different stages, with E9.5 having the lowest number of dead embryos (three of 29 −/− embryos, or about 10%). By 15.5 days postcoitum, −/− embryos either belonged to class i or iv, indicating that embryonic lethality occurred much earlier than E15.5. In fact, about 60% of all −/− embryos died at or before E13.5, with the peak of embryonic death occurring at E11.5. Fig. 2 shows some examples of class ii and iii mutants, displaying hemorrhaging in the yolk sac as well as embryo proper.
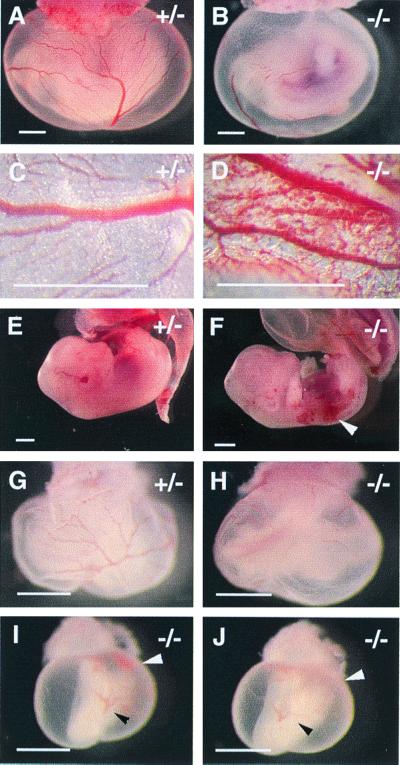
Figure 2.
Hemorrhaging in EPAS-1−/− mutants. (A and B) E11.5. (A) Normal morphology of a mouse conceptus. (B) The yolk sac membrane appears pale, most likely because of hemorrhaging significantly before dissection, rather than failure in hematopoiesis. This is supported by data shown in D and F, both photographed at the time of active hemorrhaging. (G–J) Embryos dissected at E10.0. The littermate shown in I and J appears to have been arrested at an earlier stage, approximately at E9.5. Interestingly, pools of blood cells (white arrow) were found to be deposited at the bottom of the yolk sac cavity, indicating leakage of blood vessels. (I) Photographed immediately after the original bottom side was flipped up. (J) Photographed 15 min after flipping (black arrow shows the reference point). The blood pool had diffused, indicating that these cells were indeed outside blood vessels. (Bars in A, B, E, and F, 2 mm; C, D, and G–J, 600 μm.)
Subtle Vascular Disorganization in −/− Embryos in 129 Sv/ICR Background.
To analyze vascular development in more detail, we visualized the blood vessels by immunohistochemical staining with anti-CD31 antibody (40). [The lacZ insert was expressed only weakly at E11.5, perhaps because of low efficiency of translational initiation from the IRES element, although the level of expression gradually increased at later stages. However, lacZ expression was mostly limited to endothelial cells, consistent with the findings of Tian et al. (30) and Ema et al. (32).] Fig. 3 shows examples of vascular anomalies in yolk sac membranes at E11.5. Such anomalies, however, were very subtle at the whole-mount level even in most severe cases (compare Fig. 3 A and B, where the specimen in B was from a severely hemorrhagic yolk sac). Nonetheless, many endothelial cells were misarranged to form extended lines at the expense of intact ring structures as revealed in two-dimensional sections (Fig. 3 D and F), indicative of extended endothelial sheets rather than tubular structures in the yolk sac. The proportion of endothelial cells involved in such disorganized structures varied from one yolk sac to another, with a maximum level of about one-third. Yolk sac membranes with moderate hemorrhaging showed normal-looking vascular patterns at both whole-mount and tissue-section levels (Fig. 3 G and H), perhaps because the number of lesions were too few to detect.
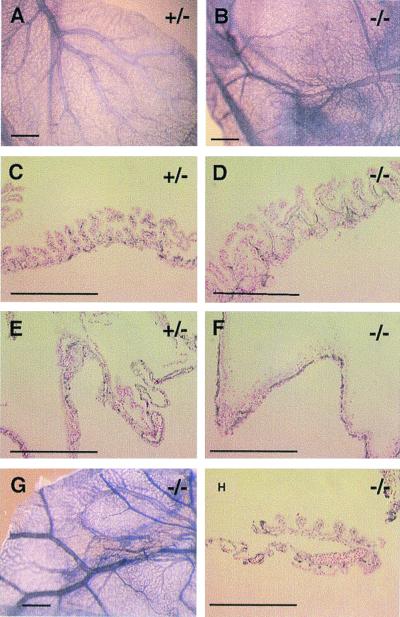
Figure 3.
Immunohistochemical staining of normal and mutant yolk sac membranes (all at E11.5) with anti-CD31 antibody. (A) Normal yolk sac membrane, showing a highly organized vascular tree pattern. (B) The vascular pattern from a severely hemorrhaging mutant differs from the normal control only slightly when viewed at the whole-mount level. (C–F) Histological sections of anti-CD31-stained specimens. (D and F) In most severe mutants, about a third of endothelial cells form extended endothelial sheets without proper lumen structures. (G and H) Moderately hemorrhaging mutants had no significant vascular disorganization, when viewed both at the whole-mount level and in histological sections. (Bars: A, B, and G, 600 μm; C–F and H,100 μm.)
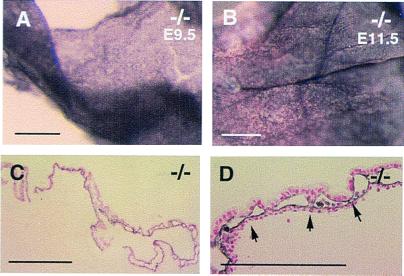
At E9.5, only about 10% of the mutants were hemorrhagic. When examined by immunohistochemical staining with anti-CD31, even the most severely hemorrhagic yolk sacs showed essentially normal vascular patterns at the whole-mount level (Fig. 4 A and B). But in histological sections, an estimated 2–10% of the vessels in hemorrhagic yolk sacs (varying from one embryo to the next) appeared disorganized (Fig. 4 D and F). Sporadically, sections were found that contained unsealed endothelium (Fig. 4F), through which blood could leak out readily. The reduced frequency and severity of vascular defects at E9.5 indicated that this was perhaps the earliest stage at which vascular defects began to develop. Indeed, when more than 30 −/− embryos were examined at E8.5, none had vascular defects, indicating that vasculogenesis was normal.
Figure 4.
Subtle vascular disorganization and hemorrhaging at E9.5. (A and B) Whole-mount anti-CD31 staining of yolk sac membranes, where B was taken from one of the embryos similar to that shown in Fig. 2 I and J. Sporadic lesions were found in histological sections (D and F). The very long endothelial linings shown in D were never found in sections from normal controls. (E and F) In areas where active remodeling was occurring to form larger vessels, large openings such as shown in F, were occasionally found in mutants only, which allowed blood cells to leak out (arrow). Note that there is no sign of tissue tearing or cracking in nearby areas, arguing that the opening was not an artifact of poor specimen preparation. (Bars: A and B, 500 μm; C–F, 50 μm.)
We also observed hemorrhaging in the embryo proper in freshly dissected mutants but not in normal littermates (Fig. 2 E and F). However, we were unable to find obvious crevices in the blood vessels. One possibility is that there are very few vascular lesions in the embryo proper, rendering them difficult to find. Nevertheless, they are perhaps responsible for the leakage of blood contents.
Severe Vascular Defects in Embryos Directly Derived from EPAS-1−/− ES Cells.
We speculated that variation in vascular defects was caused by nonidentical strain background in individual embryos, because of their outbred origin. To further address the role of EPAS-1 in vascular development, we constructed embryos in 129 Sv background by the “tetraploid aggregation” technique (39). In this technique, ES cells (or their knockout derivatives) are aggregated in vitro with tetraploid morulae. Because the tetraploid cells cannot contribute to the embryo proper and the mesoderm-derived components of the yolk sac, the entire embryo and the mesoderm-derived tissues of the yolk sac, such as the vascular system, are contributed only by ES cells (129 Sv background).
To make such −/− embryos, we first isolated −/− ES cells from two independent +/− ES cell lines (data not shown), and both −/− and +/− ES cells were used for aggregation. In more than 50 EPAS-1−/− embryos analyzed, approximately 20% developed severely disorganized vasculature at E9.5. Transient states were found where adjacent vessels were fusing to one another to form large cavities lined by endothelial sheets (Fig. 5). By E11.5, −/− embryos were either dead or at least had moderate hemorrhaging if still alive. Typically, however, the yolk sac vasculature was grossly disorganized by this stage (Fig. 5). By E12.5, all −/− embryos died of vascular defects. In aggregation experiments using +/− cells, 15 embryos were examined between E10.5 and birth, and none had vascular defects and all were viable at the stage of analysis. With both +/− and −/− ES cells, a large number of aggregates did not undergo successful embryogenesis for technical reasons. Such nonviable “embryos” (which have been excluded from the numbers reported above), however, were easily recognizable because they died early (at preimplantation or early postimplantation stages) and underwent rapid disintegration, leaving empty decidua by the time dissections were performed. Thus the technical limitation did not interfere with our analyses.
Figure 5.
Severe vascular disorganization in the yolk sac membranes of mutant embryos in 129 Sv strain background. (A) E9.5. About 20% of the E9.5 mutants showed such pattern. (B) E11.5. Vascular patterns derived from +/− ES cell lines were identical to those shown in Figs. 3A and 4A and are not duplicated here. (C and D) Histological sections. About half of the E9.5 yolk sac membranes demonstrated disorganization. (C) Elongated endothelial linings and enlarged vascular lumen (compare with Fig. 4C). (D) Lumen enlargement appeared to be caused by merging of adjacent vessels. Arrows indicate sites of vascular merging. (Bars: A and B, 500 μm; C, 150 μm; and D, 75 μm.)
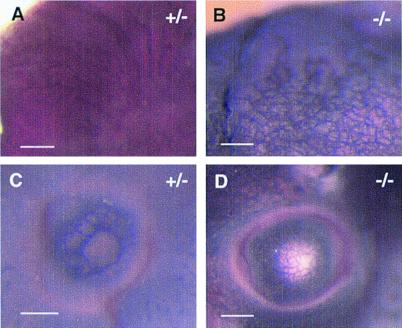
In 129 Sv background, vascular defects were observed within the embryo proper in addition to the yolk sac. In normal embryos, the primary vascular beds undergo extensive remodeling to form vessels of distinct sizes starting from E9.5. In −/− embryos, however, such a hierarchy failed to develop in some organs. Instead, a collection of small blood vessels persisted. This defect was most clearly manifested in the head and eyes at E11.5 (Fig. 6). Other blood vessels such as the dorsal aorta and intersomitic arteries, however, appeared normal (not shown), indicating that EPAS-1 is required for the development of a subset of blood vessels.
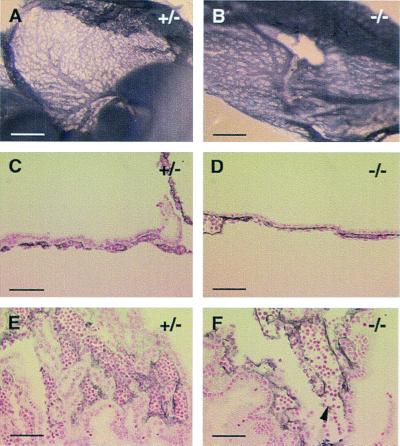
Figure 6.
Vascular defects in the embryo proper in 129 Sv background (E11.5). (A) An area of perineural vasculature in +/− embryos, showing an organized hierarchy, where large vessels branch into smaller ones. (B) Perineural vessels in −/− embryos failed to evolve into a vascular tree consisting of large and small vessels. (C and D) A similar defect is also observed in the developing eyes. (Bars: 300 μm.)
Discussion
The nascent vasculature in developing embryos is a dynamic structure that gradually evolves into a vascular tree by continuous vascular remodeling and branching angiogenesis. Although many angiogenic regulators are known, the process of vascular remodeling is much less understood. In this paper, we present evidence that the bHLH-PAS transcription factor, EPAS-1, plays an important role in vascular remodeling. In EPAS-1-deficient embryos, small blood vessels in the optical vesicle and cephalic tissues fail to form larger vessels by combining to one another. In the yolk sac, smaller blood vessels do merge to one another; however, they do so without proper regulation, often leading to the formation of extensive endothelial sheets. Furthermore, when large vessels do form in −/− yolk sacs, they sometimes fail to seal completely, leaving openings where blood contents leak out. Thus, in EPAS-1 mutant mice, although vasculogenesis occurs normally, there appears to be improper interactions among blood vessels once they are formed, resulting in aberrant vascular remodeling. Our analyses at the cellular level thus far have revealed no defects in endothelial cell survival, proliferation, differentiation, and recruitment of vascular smooth muscle cells (data not shown). Therefore, as a future direction, it will be of particular interest to investigate the potential alterations in endothelial cell migration, cell/cell, and cell/matrix interactions.
Interestingly, in an independent gene targeting experiment, Tian et al. (36) found that EPAS-1 was important for catecholamine homeostasis instead of vascularization. It is unlikely that the difference in the mutant phenotypes has arisen from construct designs, because both groups disrupted the bHLH domain and demonstrated that the mutations were null alleles. Therefore, we suggest that the discrepancy is a result of the use of different mouse strains. In addition, subtle differences in the ES cells used by the two groups also may have contributed to the apparent disagreement. This is possible because the 129 Sv mice, from which ES cells are derived, include several closely related strains and so different ES cell lines may have nonidentical strain background.
The mechanisms leading to the phenotypic differences remain elusive but several possibilities are discussed below. In one scenario, there may exist, among different strains, proteins whose functions overlap with EPAS-1. One interesting candidate for investigation of potential overlapping functions is hypoxia-inducible factor α, which shares extensive sequence identity with EPAS-1. In addition, instead of functional overlaps at the level of EPAS-1, functional overlap also could occur at the level of downstream genes regulated by EPAS-1. By analogy, genes that are not directly controlled by EPAS-1, but lie further downstream or even in other signaling pathways, also might modify the mutant phenotypes.
Tian et al. (36) demonstrated a defect in catecholamine homeostasis in EPAS-1-deficient embryos, which suffered from reduced heart rate (25%) and died between E12.5 and E16.5. However, embryonic lethality was rescued by supplementing mother's diet with d,l-threo-3,4-dihydroxyphenylserine (DOPS), a precursor to norepinephrine. The rescued pups died within 24 h of birth, because of a lack of continued supply of DOPS through mother's circulation. In our study, more than half of the mutant embryos died from vascular defects before E13.5. Those that were born alive survived for several weeks, and no spontaneous death was observed within 24 h after birth. When mothers were fed with DOPS, however, three of 11 live-born −/− pups (from a total of 84 pups of all genotypes) died within 24 h. We hypothesize that dietary supplement of DOPS rescued these three pups from embryonic lethality. This finding implies that some of our mutants also suffered from reduced level of catecholamine, although we were unable to directly demonstrate a statistically significant reduction in the heart rate possibly because of the infrequent occurrence of this defect.
Together, our data and those of Tian et al. (36) indicate that EPAS-1 may have at least two distinct roles: one to regulate the production of catecholamine and the other to regulate vascular remodeling. The relative importance of these functions, however, may depend on strain background. In ICR/129 Sv outbred background, whereas most of the mutants carried vascular defects, the severity varied greatly among individual embryos. On the other hand, even those that were born alive also had disorganized blood vessels in various organs (to be described elsewhere), explaining the runt phenotype. That EPAS-1 plays an important role in vascular development is further supported by experiments with completely −/− ES cell-derived embryos. Vascular defects were greatly amplified in these mutants and all mutants died at or before E12.5.
The regulation of the expression and function of EPAS-1 is complex. The protein level of EPAS-1 is regulated by oxygen tension where hypoxia induces stabilization of the protein (41) and the functionality of the protein is in part controlled by its redox states (42). In our knockout mice, we demonstrated that there is no functional EPAS-1 mRNA templates, suggesting that the complexity of these protein-level regulations should not influence our mutant phenotype because the protein per se cannot be produced.
Several target genes have been previously reported whose expression was activated by EPAS-1 in cultured cells, including vascular endothelial growth factor, flk-1, and tie-2 (30, 41, 43). However, we were unable to find alterations in the expression of these genes (unpublished data), suggesting that EPAS-1 may regulate the expression of novel target molecules. The characterization of the vascular defects in EPAS-1 knockout mice presents an opportunity for the identification of such genes and opens an avenue for the investigation of the controlling mechanisms of vascular development.
Acknowledgments
This work was supported by the Medical Research Council of Canada. G.-H.F. is a recipient of the Medical Research Council Scholarship of Canada.
Abbreviations
- bHLH
basic helix–loop–helix
- ES
embryonic stem
- En
embryonic day n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140087397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140087397
References
- 1.Leung D W, Cachianes G, Kuang W J, Goeddel D V, Ferrara N. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 2.Senger D R, Connolly D T, Van de Water L, Feder J, Dvorak H F. Cancer Res. 1990;50:1774–1778. [PubMed] [Google Scholar]
- 3.Shibuya M, Yamaguchi S, Yamane A, Ikeda T, Tojo A, Matsushime H, Sato M. Oncogene. 1990;5:519–524. [PubMed] [Google Scholar]
- 4.de Vries C, Escobedo J A, Ueno H, Houck K, Ferrara N, Williams L T. Science. 1992;255:989–991. doi: 10.1126/science.1312256. [DOI] [PubMed] [Google Scholar]
- 5.Matthews W, Jordan C T, Gavin M, Jenkins N A, Copeland N G, Lemischka I R. Proc Natl Acad Sci USA. 1991;88:9026–9030. doi: 10.1073/pnas.88.20.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millauer B, Wizigmann-Voos S, Schnurch H, Martinez R, Moller N P, Risau W, Ullrich A. Cell. 1993;72:835–846. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- 7.Quinn T P, Peters K G, De Vries C, Ferrara N, Williams L T. Proc Natl Acad Sci USA. 1993;90:7533–7537. doi: 10.1073/pnas.90.16.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soker S, Takashima S, Miao H Q, Neufeld G, Klagsbrun M. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 9.Fong G-H, Rossant J, Gertsenstein M, Breitman M L. Nature (London) 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 10.Fong G-H, Zhang L, Bryce D-M, Peng J. Development (Cambridge, UK) 1999;126:3015–3025. doi: 10.1242/dev.126.13.3015. [DOI] [PubMed] [Google Scholar]
- 11.Shalaby F, Ho J, Stanford W L, Fischer K D, Schuh A C, Schwartz L, Bernstein A, Rossant J. Cell. 1997;89:981–990. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 12.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X F, Breitman M L, Schuh A C. Nature (London) 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 13.Kawasaki T, Kitsukawa T, Bekku Y, Matsuda Y, Sanbo M, Yagi T, Fujisawa H. Development (Cambridge, UK) 1999;126:4895–4902. doi: 10.1242/dev.126.21.4895. [DOI] [PubMed] [Google Scholar]
- 14.Davis S, Aldrich T H, Jones P F, Acheson A, Compton D L, Jain V, Ryan T E, Bruno J, Radziejewski C, Maisonpierre P C, Yancopoulos G D. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 15.Maisonpierre P C, Suri C, Jones P F, Bartunkova S, Wiegand S J, Radziejewski C, Compton D, McClain J, Aldrich T H, Papadopoulos N, et al. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 16.Suri C, Jones P F, Patan S, Bartunkova S, Maisonpierre P C, Davis S, Sato T N, Yancopoulos G D. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 17.Suri C, McClain J, Thurston G, McDonald D M, Zhou H, Oldmixon E H, Sato T N, Yancopoulos G D. Science. 1998;282:468–471. doi: 10.1126/science.282.5388.468. [DOI] [PubMed] [Google Scholar]
- 18.Dumont D J, Gradwohl G, Fong G-H, Puri M C, Gertsenstein M, Auerbach A, Breitman M L. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 19.Sato T N, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Nature (London) 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 20.Puri M C, Rossant J, Alitalo K, Bernstein A, Partanen J. EMBO J. 1995;14:5884–5891. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumont D J, Jussila L, Taipale J, Lymboussaki A, Mustonen T, Pajusola K, Breitman M, Alitalo K. Science. 1998;282:946–949. doi: 10.1126/science.282.5390.946. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Linden P, Farnebo J, Cao R, Eriksson A, Kumar V, Qi J H, Claesson-Welsh L, Alitalo K. Proc Natl Acad Sci USA. 1998;95:14389–14394. doi: 10.1073/pnas.95.24.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H U, Chen Z F, Anderson D J. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- 24.Adams R H, Wilkinson G A, Weiss C, Diella F, Gale N W, Deutsch U, Risau W, Klein R. Genes Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes H, Reisz-Porszasz S, Hankinson O. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 26.Maltepe E, Schmidt J V, Baunoch D, Bradfield C A, Simon M C. Nature (London) 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 27.Wang G L, Semenza G L. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 28.Iyer N V, Kotch L E, Agani F, Leung S W, Laughner E, Wenger R H, Gassmann M, Gearhart J D, Lawler A M, Yu A Y, Semenza G L. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan H E, Lo J, Johnson R S. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 31.Jain S, Maltepe E, Lu M M, Simon C, Bradfield C A. Mech Dev. 1998;73:117–123. doi: 10.1016/s0925-4773(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 32.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 34.Flamme I, Frohlich T, von Reutern M, Kappel A, Damert A, Risau W. Mech Dev. 1997;63:51–60. doi: 10.1016/s0925-4773(97)00674-6. [DOI] [PubMed] [Google Scholar]
- 35.Crews S T. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 36.Tian H, Hammer R E, Matsumoto A M, Russell D W, McKnight S L. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wurst W, Joyner A L. In: Gene Targeting. Joyner A L, editor. New York: Oxford Univ. Press; 1993. pp. 33–61. [Google Scholar]
- 38.Wood S A, Allen N D, Rossant J, Auerbach A, Nagy A. Nature (London) 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]
- 39.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldwin H S, Shen H M, Yan H C, DeLisser H M, Chung A, Mickanin C, Trask T, Kirschbaum N E, Newman P J, Albelda S M, et al. Development (Cambridge, UK) 1994;120:2539–2553. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- 41.Wiesener M S, Turley H, Allen W E, Willam C, Eckardt K U, Talks K L, Wood S M, Gatter K C, Harris A L, Pugh C W, et al. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- 42.O'Rourke J F, Tian Y M, Ratcliffe P J, Pugh C W. J Biol Chem. 1999;274:2060–2071. doi: 10.1074/jbc.274.4.2060. [DOI] [PubMed] [Google Scholar]
- 43.Kappel A, Ronicke V, Damert A, Flamme I, Risau W, Breier G. Blood. 1999;93:4284–4292. [PubMed] [Google Scholar]