Abstract
Tetraspanins are multiple membrane-spanning proteins that likely function as the organizers of membrane microdomains. Tetraspanins associate with other membrane-bound molecules such as cell-adhesion proteins, growth factor receptors, and Ig superfamily members and regulate key cellular processes such as adhesion, migration, and fusion. Tetraspanins are widely expressed in vascular and haematopoietic cells and are involved in both physiological and pathological processes related to angiogenesis, vascular injury, thrombosis, and haemostasis. A wide body of evidence suggests that tetraspanins directly regulate the development and functions of the vascular system and the pathogenesis of vascular diseases. This article reviews current understanding of the roles of tetraspanins in vascular functions.
KEYWORDS: Tetraspanins, CD9, CD151, CD63, Vascular system, Smooth muscle cells, Endothelial cells, Platelets
1. Introduction
Tetraspanins comprise a group of integral membrane proteins that contain four conserved hydrophobic transmembrane domains (TM1-TM4), intracellular N- and C-termini, and small (EC1) and large (EC2) extracellular domains.1–5 A schematic representation of tetraspanin structure is shown in Figure 1. The EC2 domain of most tetraspanins features four–six cysteine residues, glycosylation sites, and a highly conserved ‘CCG’ motif. The ‘CCG’ motif and conserved strong polar residues in the TM domains are considered to be the signature structural elements for all tetraspanins. The EC2 region, together with transmembrane regions, confers structural and functional specificities to tetraspanins and is also important in mediating tetraspanin interactions with other membrane-bound proteins such as integrins1–5 and Ig superfamily proteins.6 Tetraspanin-containing multimolecular complexes are designated as ‘tetraspanin web’ or ‘tetraspanin enriched microdomains’ (TEM) as the isolation of a given tetraspanin typically yields a range of other tetraspanins and associated transmembrane proteins.2,7,8 However, with the exception of CD151-integrin α3 binding, most tetraspanin-transmembrane protein interactions are probably not based on the direct amino-acid-residue–amino-acid-residue interactions at the EC regions because these interactions are relatively weak and readily disrupted under the so called ‘high stringency’ detergent conditions used for cell lysate preparation (e.g. 1% Triton X-100).
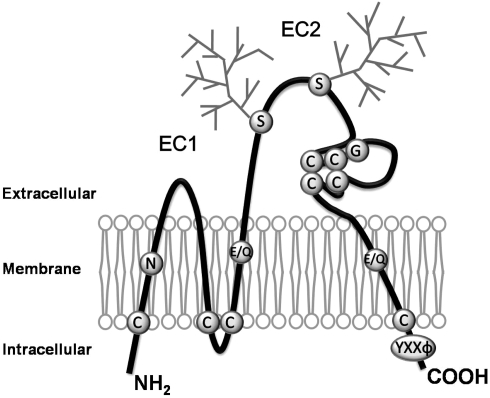
Figure 1.
Schematic drawing of the structure of tetraspanins. Tetraspanins are composed of four transmembrane, an intracellular N- and C-termini, and two extracellular (one shorter, EC1, and one longer, EC2) domains. The conserved motifs or residues featured by tetraspanins are denoted. Most tetraspanins contain four or six cysteine residues in EC2, and two of those are in a highly conserved ‘CCG’ motif. Most of tetraspanins have glycosylation sites in EC2 as indicated as the squares, while CD9 is glycosylated in EC1. In each tetraspanin, there are several cysteine residues proximal to the interface of the inner leaflet and cytosol, which are the sites for palmitoylation and characteristic strong polar residues in the transmembrane domains. Tetraspanins also contains ‘YXXΦ’ sorting motif in the C-terminal cytoplasmic domain. C, Cysteine; E, Glutamic acid; G, Glycine; Q, Glutamine; N, Aspargine; S, Serine; Y, Tyrosine; X, any aminoacid; Φ, Hydrophobic aminoacid.
As the organizers of membrane microdomains, tetraspanins regulate vital cellular events such as adhesion, spreading, migration, and fusion (Figure 2). The biochemical characteristics of tetraspanin microdomains and the regulatory roles of tetraspanins in phenomena such as malignancy, immune cell regulation, and viral infections have been thoroughly reviewed in several articles.9,10–13 Recent studies underline that tetraspanins actively traffic between plasma membrane and intracellular vesicular compartments. The trafficking of tetraspanins is accompanied by the vesicular release and trafficking of other membrane proteins.14,15 Another emerging theme is that tetraspanins appear to regulate pericellular proteolysis near the plasma membrane leading to the altered cell motility and adhesiveness.16–19 Although the biochemical and biophysical nature of tetraspanin activities remains unknown, existing evidence has expanded the function of tetraspanins as molecular ‘facilitators’ or ‘organizers’ at the plasma membrane to both intracellular vesicles and the extracellular micro-environment.
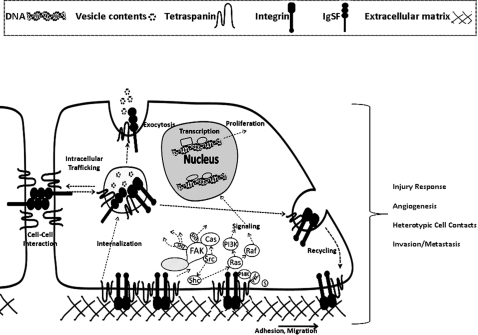
Figure 2.
Schematic drawing showing cellular functions of tetraspanins. Tetraspanins associate with integrins, Ig superfamily proteins, and other transmembrane proteins. Additionally, tetraspanins are in molecular complex with cytoplasmic proteins, such as syntenin-1 (s) and signalling molecules. Tetraspanin containing complexes are designated as ‘tetraspanin web’ or ‘tetraspanin enriched microdmians’ (TEM). In addition to serving as organizers at the plasma membrane, tetraspanins are also enriched in the intracellular vesicles. Trafficking of tetraspanins between various cellular compartments tightly regulates exocytosis and trafficking of their associated partners. Tetraspanins in conjunction with other membrane proteins regulate cell–extracellular matrix adhesion, cell–cell interactions, cell migration, and modulate intracellular signaling events. Tetraspanins regulate phenomena such as vascular injury response, angiogenesis, heterotypic cell contacts, and tumour invasion and metastasis.
Tetraspanins exhibit diverse tissue distribution patterns. For example, CD9, CD63, and CD151 are widely expressed in a variety of tissue including vascular and haematopoietic cells, whereas others such as TSSC6 are expressed only in the haematopoietic cells. For the 33 identified human tetraspanins, the expression and function of most newly identified tetraspanins in the vascular system remain to be elucidated. Because vascular events such as neointimal formation, angiogenesis, and thrombosis are tightly regulated by cell adhesion proteins, e.g., integrins, and tetraspanins are key players in adhesion and migration, tetraspanins may regulate important pathophysiological phenomena of the vascular system. Indeed, the roles of tetraspanins in thrombosis, vascular morphogenesis, and vascular remodelling are beginning to be increasingly understood. Considering the rapidly growing interest in vascular tetraspanin functions, we discuss the biology of various vascular tetraspanins and their functional role in regulating pathophysiological processes related to the cardiovascular system in this review, which is distinct from earlier reviews focusing on other functional aspects of tetraspanins such as malignancy, immune response, and viral infection.
2. Specific roles of tetraspanins in vascular functions
2.1. CD9
Human CD9 (Tspan29) was first detected by its reactivity with the monoclonal antibody (mAb) BA-1 raised against a leukaemia cell line NALM-6.20 Using specific DNA probes, the gene encoding CD9 was localized to the short arm of chromosome 12.21 The primary structure of CD9 was elucidated in 1991 when CD9 cDNA was isolated from a megakaryocytic library. The CD9 gene consists of eight coding regions (exons) that span about a 20 kb region of genome. Transcription of the CD9 gene generates a 1.4 kb mRNA that encodes a protein of 228 amino acids with an apparent molecular weight of a 24 kDa on SDS–PAGE.22 This report also confirmed that the mature protein contained four hydrophobic sequences representative of four transmembrane spanning domains. CD9 expression has been demonstrated on several cells such as macrophages, eosinophils, basophils, fibroblasts, epithelial cells, neuronal cells, oocytes, and various tumours and tumour cell lines.23 CD9 regulates phenomena such as cell morphology, migration, proliferation, cell fusion, and tumour cell metastasis.2,24 CD9 is the first tetraspanin for which knock out (KO) mice were generated, and oocytes derived from CD9 KO mice have drastically reduced fertility rates.25 In various cell systems, CD9 exists in molecular complexes with β1 and β3 integrins,26,27 Ig superfamily proteins,6 membrane-anchored heparin-binding epidermal growth factor (HB-EGF)-like growth factor,28 and other tetraspanins.2
Smooth muscle cells (SMC) constitute a major component of muscular arteries. Endothelial damage in the vessel wall elicits phenotypic changes in the SMC leading to the formation of neointimal layer. Neointimal hyperplasia directly contributes to the pathogenesis of restenosis after angioplasty and predisposes vessels to occlusion by platelet-rich thrombi.29 Therefore, membrane proteins regulating SMC phenotypes can profoundly influence the progression of atherothrombosis culminating in vessel occlusion. CD9 is expressed in cultured SMC as well as in the SMC of the vessel wall following vascular injury.30,31 Specific effects of tetraspanin CD9 in the regulation of SMC phenotypes and in neointima formation following vascular injury have been studied using various model systems.30–33 Earlier reports clearly established an association between CD9 and β1 integrins in SMC and suggested that CD9 levels are upregulated in SMC undergoing switch from the synthetic to proliferative phenotype. In addition, a peri-vascular electrical injury to the femoral artery suggested that the extent of neointima formation was not significantly altered in CD9 KO mice when compared with that of the wild-type controls.32 More recently, antibody perturbation and adenoviral CD9 gene delivery studies in a carotid ligation injury model conclusively established a connection between CD9 and neointimal hyperplasia.30 In vitro experimental results using human coronary artery SMC indicated that CD9 is important in regulating both proliferative and migratory phenotypes of SMC.30 CD9 in SMC associates with various β1 integrins including the fibronectin receptor α5β1.30,31 Since vascular injury is associated with tissue remodelling involving extracellular matrix (ECM) reorganization, we predict that CD9 and its associated integrins play a critical role in the overall healing response of the injured vessel.
CD9 in other model systems associates with certain growth factors belonging to epidermal growth factor (EGF) family. In this regard, CD9 specifically associates with the membrane anchored HB-EGF-like growth factor receptors and regulates its juxtacrine growth factor activity.28 Such findings warrant future investigations to study the relative importance of other CD9 associated proteins in the vascular injury response. When exogenously expressed, CD9 significantly upregulates PI-3K/Akt signalling pathway in SMC as well as in other model systems.23,30 As PI-3K/Akt plays an important pivotal role in the modulation of SMC cell migration and proliferation,34,35 it can be speculated that augmentation of PI3-K/Akt activity by CD9 may play a critical role in CD9-induced SMC phenotypic changes. Future studies are required to delineate upstream molecular signalling mechanisms that lead PI-3K/Akt enhanced activation by CD9 and to define the downstream events that affect SMC phenotypes upon PI-3K activation.
Within the vascular tissue CD9 expression is not limited to the SMC of the medial layer. As studies have shown endothelial cells (ECs) derived from various tissue sources express CD9. For example, EC derived from bovine retina,36 human umbilical vein,37,38 saphenous vein grafts,39 and lymphatic vessels40 all express significant amounts of CD9. CD9 in EC is attributed to be important in several cellular events. Using anti-CD9 mAbs, Klein-Soyer et al.39 showed a regulatory role for CD9 in EC migration. In cultured human umbilicial vein cells (HUVEC), CD9 is predominately localized to EC junctions.41 Furthermore, anti-CD9 mAbs inhibited transwell haptotactic migration of EC to fibronectin as well as EC migration into wound areas in an in vitro scratch assay. However, in contrast with SMC, the effects of anti-CD9 mAbs appeared to be limited to the regulation of EC migration as EC proliferation was unaltered upon anti-CD9 mAb treatment.39 However, anti-CD9 mAb treated HUVEC were reported to have a diminished platelet-induced proliferative response.42
In addition to EC migration, CD9 is implicated to modulate leucocyte transendothelial migration.38 Both neutralizing anti-CD9 antibody and recombinant CD9 protein corresponding to the EC2 domain inhibited leucocyte migration.38 During the process of leucocyte transendothelial cell migration, CD9 redistributed and clustered near the regions of EC contacts with leukocytes. Thus, by differentially clustering with the adhesion molecules such as intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1), CD9 enriched microdomains in the plasma membrane of EC may take part in regulating the endothelial barrier function.
In addition, the regulatory role of CD9 in mediating cell–cell interactions is not limited to homotypic aggregation events as evidence suggests that CD9 indeed can regulate heterotypic cell contacts. For example, very early observations suggested that CD9 mAbs can enhance neutrophil adhesion to the endothelium.43 Furthermore, anti-CD9 mAbs inhibited transendothelial migration of melanoma cells suggesting that CD9 modulates tumor cell–EC interactions.44 More recent evidence indicates that in activated EC, CD9 containing tetraspanin microdomains take part in the formation of specialized structures designated as endothelial adhesion platforms (EAP) that are biochemically distinct from lipid rafts.45 Ligation of CD9 with antibodies resulted in tetraspanin clustering and recruitment of ICAM-1 and VCAM-1 to the docking structures providing the first direct evidence for the role of tetraspanins in heterotypic cellular interactions.45
As CD9 is abundantly expressed in platelets as well as in the endothelium, it remains to be determined whether similar heterotypic cell contact regulatory mechanisms are operational in platelet interactions with the vascular endothelium. A putative role for CD9 in the regulation of EC matrix metalloproteinase (MMP) activity has been investigated.19 In mouse lung endothelial cells (MLEC), a specific knockdown of CD9 does not appear to interfere with the MMP-2 activity, whereas CD151 knockdown in these cells significantly enhances MMP2 activity and its collagenolytic activity. Thus, it appears that tetraspanins differentially regulate EC functions and this feature may be due to the ability of tetraspanins to reorganize membrane protein complexes. Other than SMC and EC, the involvement of CD9 in cardiac myocyte growth and heart muscle hypertrophy has also been reported.46 Adenoviral-mediated exogenous CD9 expression in mouse cardiac myocytes inhibited cardiac muscle hypertrophy and reduced mortality following myocardial infarction.46
2.2. CD63
CD63 or Tspan30 is primarily an intracellular tetraspanin and is mainly localized to late endosomal and lysosomal compartments. In the vascular system, CD63 is relatively well studied in platelets and EC. In EC, CD63 was identified as a component of Weibel-Palade bodies,47,48 which store and secrete von Willebrand factor (vWF). Together with P-selectin, CD63 is one of the few proteins known to reside in the membrane of Weibel-Palade bodies.48–50 Although it is not well understood how CD63 is targeted to Weibel-Palade bodies, vWF clearly plays a role in CD63 trafficking into these structures.51,52 CD63 and P-selectin are initially recruited to the vWF storage granules that further develop into Weibel-Palade bodies, suggesting that the luminal vWF contains a sorting signal that targets CD63 and P-selectin.47 During the exocytosis of vWF, the surface expressions of CD63 and P-selectin in EC are transiently enhanced due to the fusion between the Weibel-Palade body membrane and the plasma membrane,47 which likely contributes to the leucocyte attachment to the endothelium.
Several lines of evidence suggest that CD63 is functionally involved in vascular cell adhesion. First, CD63 antibodies inhibit monocyte and neutrophil adhesion to serum-coated substratum and EC layer, respectively.53,54 Second, CD63 physically associates with several integrins such as α3β1, α6β1, and αLβ2,55,56 which are found in the vascular system. Third, a CD63 mAb induces integrin αLβ2-dependent neutrophil adhesion to HUVEC, and integrin αLβ2 is upregulated in CD63-activated neutrophils so that these neutrophils become competent to bind HUVEC.56 Moreover, CD63 likely regulates cell migration based on the observations of altered cell motility upon CD63 antibody treatment and CD63 overexpression.57,58
Mechanistically, CD63 appears to regulate exocytosis at the cellular level. For example, the Weibel-Palade bodies in EC undergo exocytosis upon inflammatory stimulation while an antibody to CD63 inhibits IgE-mediated release of histamine in basophils.59 Although the activities of protein phosphatases (such as receptor-linked tyrosine phosphatase α), protein kinases (such as Src),56,60–62 and lipid kinases (such as type II PI-4kinase) were detected in CD63 immunoprecipitates, little knowledge has been gained in understanding the CD63-mediated signalling pathways. Recently, CD63 was found to bind syntenin-1, a PDZ domain-containing adapter protein.63 CD63 C-terminal cytoplasmic domains and syntenin-1 PDZ domains are needed for the interaction while the C-terminus of syntenin-1 stabilizes the interaction.63 Syntenin-1 retards CD63 internalization and the deletion of the N-terminal 100 residues of syntenin-1 completely blocks CD63 internalization.63 In addition, CD63 associates with membrane-type 1 MMP (MT1-MMP) to facilitate its lysosomal degradation,64 functions as a cellular receptor of tissue inhibitor of metalloproteinase-1 (TIMP-1),65 and interacts with H, K-ATPase β-subunit to enhance its internalization.66
2.3. CD151
CD151 or Tspan24, originally identified as platelet-EC tetra-span antigen 3 (PETA-3), is widely expressed in ECs, SMCs, and epithelial cells, and in haematopoietic cells such as platelets and megakaryocytes.67–69 CD151 shows stable and stoichiometric association with laminin-binding integrins α3β1, α6β1, α6β4, and α7β1 in a number of cell systems and functions together with the integrins in a variety of cellular processes.70–72 Much of the knowledge on biological properties of CD151 and its associated integrins is gained in the areas of cell motility,73–75 cell–cell adhesion,76–78 cell–ECM adhesion75,79 and tumour metastasis.74,80 In the vessel wall, CD151 is expressed in the SMC-enriched medial layer, fibroblast-enriched adventitial layer, and endothelium. Although the expression of CD151 in atherosclerotic arteries appears to be increased when compared with the normal arteries,81 precise regulatory role of CD151 in SMC functions has so far not been investigated.
In cultured HUVEC, intracellular CD151 accounts for up to 66% of the total CD151 and is mostly localized to the endosomal/lysosomal vesicles. The cell surface CD151 in ECs is localized at cell–cell junctions,41 a pattern also seen with tetraspanins CD9 and CD81, which are the primary tetraspanins associating with CD151.41,82 Remarkably, the tetraspanin web formed by CD9, CD81, and CD151 on EC are critical for proper adhesive function of ICAM-1 and VCAM-1, and appropriate distribution of CD151 in the webs on EC is required for lymphocyte transendothelial migration and can strengthen the firm adhesion of lymphocytes during extravasation.38 In addition, CD151 has been implicated to regulate several essential functions of EC including migration, invasion, and spreading.41,82,83
Accumulating evidence reveals that CD151 is an important regulator of vasculogenesis and angiogenesis. First, CD151 regulates EC cable formation in in vitro Matrigel assays.82–84 Second, CD151 expression via viral vectors substantially increases microvessel density in the rat ventricular myocardium undergoing ischaemic infarction.85 Last, CD151 KO results in mice defective in pathological angiogenesis as evidenced by in vivo Matrigel plug assays, corneal micropocket and tumour implantation assays, and ex vivo aortic ring assays.83 Despite these striking vascular abnormalities, the physiological vascular morphogenic process appears to be unaffected in CD151 KO mice.83
Mechanistically, studies in epithelial cells indicate that CD151 associates with α3β1 integrin and regulates cell–cell contact through organizing junctional complexes containing cadherin, β-catenin, PKC, and PTPμ.76,77 Also, CD151, like its partner integrin α6β4, is a constitutive component of hemidesmosomes,78 a major cell–matrix adhesion machinery in epidermis. At the molecular level, CD151 promotes cell adhesion strengthening mediated by integrin α6β1 and potentiates the ligand-binding activity of integrin α3β1 by stabilizing the activated conformation of this integrin.79,86 Intracellularly, CD151 expression leads to the PKC- and Cdc42-dependent actin cytoskeletal reorganization, a process critical for both adhesion and migration.76,77 In EC, both overexpression and knockout studies indicate that CD151 can upregulate eNOS, Akt, and Rac activities, which are apparently needed for endothelial cell–cell adhesion and angiogenesis.83,85 Endothelial CD151 also promotes the collagenolytic activity and TEM association of MT1-MMP, an ECM remodelling enzyme involved in angiogenesis.41 However, the precise mechanisms of CD151 controlling EC adhesion, migration, and angiogenesis remain largely unknown.
Within haematopoietic cells, CD151 expression is mostly restricted to platelets, megakaryocytes, erythrocytes, and activated T lymphocytes.69,87,88 A C-terminal deletion mutation of human CD151 presents with severe defects of erythropoiesis,89 suggesting the connection between CD151 and the proper functioning of haematopoietic cells. However, CD151-null mice are normal in the development of haematopoietic cells and proliferation of T lymphocytes,90–92 CD151 appears to modulate cell adhesion in haematopoietic cells such as the homotypic cell–cell adhesion of erythroleukaemia and megakaryoblastic cells.87,91 In human T lymphocytes, CD151 expression is transactivated by Tax in response to human T cell leukaemia virus type 1 (HTLV-1) infection where CD151 promotes α5β1 integrin-dependent adhesion to fibronectin in HTLV-1-positive T cells.93,94 In addition, CD151 may also play a role in human immunodeficiency virus type 1 (HIV-1) entry into macrophages. Recombinant soluble forms of the EC2 domains of CD151 as well as tetraspanins CD9, CD63, and CD81 are capable of inhibiting HIV-1 virion uptake perhaps by altering the organization of CD4-HIV complexes within tetraspanin web that are required for membrane fusion events.95
3. Function of tetraspanins in haemostasis and thrombosis
3.1. CD9
Platelets express at least five different tetraspanin members, CD9, CD151, CD63, TSSC6,11 and Tspan9,96 of which CD9 has the highest surface expression. At 50 000–80 000 copies per platelet, CD9 is approximately equimolar with that of fibrinogen receptor integrin GPIIb/IIIa.97 CD9 in platelets is in a molecular complex with the GPIIb/IIIa and other platelet membrane glycoproteins CD36,98 GPIb/V/IX complex, the integrin-associated protein (IAP),26 and with CD63.99 CD9 interactions with GPIIb/IIIa are disrupted by strong non-ionic detergents such as Triton-X100, and with the detergents such as digitonin that disrupt tetraspanin–tetraspanin interactions.
All described anti-human CD9 monoclonal antibodies (mAbs) bind in the EC2 region of CD9 and induce platelet aggregation via FcγRII-mediated crosslinking mechanism. Antibody ligation to CD9 results in activation of G proteins leading to phosphoinositide hydrolysis and inhibition of adenylate cyclase. These events mediated by phospholipase C, required neither secreted ADP nor thromboxane generation.100 Due to the propensity of CD9 mAbs to actuate FcγRII-dependent mechanisms, employing intact anti-CD9 IgG in functional assays has made understanding the specific contribution of CD9 in platelets a challenge.101,102 Soluble F(ab’)2 fragments to CD9 derived from intact mAbs do not elicit such pro-aggreagtory response.103 However, immobilized F(ab’)2 are capable of inducing dense granule release and platelet aggregation, suggesting that immobilization or direct clustering of CD9 contributes to initiation of a platelet signal.104
Recent studies performed using monovalent Fab fragments of CD9 mAbs reveal that engagement of CD9 with Fab promotes aggregate stability and enhanced fibrinogen binding to GPIIb/IIIa under threshold concentrations of agonists.105 Similarly, recombinant protein corresponding to the CD9 EC2 domain inhibits low-dose agonist induced platelet aggregation (J. Kotha et al., unpublished observations). Scanning electron microscopy with immunogold labelling with CD9 mAb suggest that CD9 is enriched at the regions of platelet–platelet contacts and is localized with the GPIIb/IIIa in the alpha granules and pseudopodial process of activated platelets.106 Confocal laser scanning microscopy studies on platelets at various stages of spreading reveal that in addition to high-density localization of CD9 to platelet–platelet contact sites, filopodial extensions show markedly enhanced CD9 staining.105
Significance of CD9 in platelet function and haemostasis has also been studied using CD9 KO mice model. Despite being the most abundant tetraspanin on platelets, CD9 deficiency does not appear to alter agonist-induced platelet aggregation or expression of the platelet activation marker, P-selectin (F. Lanza, personal communication). Tail bleeding assays suggest that CD9 KO mice have a decreased bleeding tendency that is in total contrast with that of other tetraspanin KO models (CD151 and TSSC6). However, CD9 KO platelets display an increased αIIbβ3 activation without changes in total levels of platelet αIIbβ3. These results point to CD9 as a repressor of integrin αIIbβ3 activation in platelets (F. Lanza, personal communication). The exact role of CD9 in platelet function and haemostasis remains elusive, and it is yet to be determined whether CD9 Fabs or other agents that functionally perturb CD9 carry a potential therapeutic value in regulating thrombosis and haemostasis.
3.2. CD63
CD63 is a tetraspanin expressed abundantly in the platelets. In contrast with CD9, CD63 is mostly localized to the dense granules and lysosomes of resting platelets and redistributes to platelet surface only upon platelet activation. Thus, CD63, along with P-selectin, is routinely used as a surface marker for platelet activation. CD63 is also expressed in several megakaryocytic cell lines.107 As a constitutively expressed protein in the platelet granules, CD63 was found to be associated with the platelet storage pool deficiency disorders such as the Hermansky–Pudlak Syndrome.108 Biochemical studies show that the surface translocated CD63 preferentially complexes with the CD9- and GPIIb/IIIa- containing multimeric complexes.109 A direct role for CD63 in the platelet granule release and in altering GPIIb/IIIa-fibrinogen interactions remain to be determined. Immunoelectron microscopy studies demonstrate that CD63 and type II PI-4 kinase colocalize at both internal membranes of resting platelets and filopodia of thrombin-activated platelets.110
Antibody perturbation of platelets with anti-CD63 mAbs exhibits incomplete spreading on fibrinogen and impairs tyrosine phosphorylation of focal adhesion kinase (FAK).99 CD63 also modulates platelet spreading on the fibrinogen-immobilized substratum in an integrin GPIIb/IIIa-dependent manner.99 Despite these characteristics, perturbation of CD63 function in platelets via mAbs does not alter platelet adhesion and activation per se. Recent evidence from CD63-null mice indicated CD63 is not required for platelet development, activation, and adhesion to collagen under shear flow and thrombus formation both in vitro and in vivo. However, when compared with the wild-type, CD63-deficient platelets consistently displayed slightly but statistically insignificantly stronger responses in standard aggregation assay and reduced reversibility of aggregation at the low and intermediate concentrations of agonist.111
3.3. CD151
CD151 is expressed in human as well as murine platelets. Like CD9, CD151 also complexes with GPIIb/IIIa.91 However, unlike CD9-GPIIb/IIIa complexes, CD151-GPIIb/IIIa interactions are retained under stringent detergent conditions such as 1% Triton X-100.91 Similar to CD9, certain mAbs against human CD151 induce platelet aggregation via FcRγII-mediated cross-linking mechanism.112 Despite having no changes in GPIIb/IIIa expression, platelets from CD151 KO mice have impaired GPIIb/IIIa outside-in signalling and exhibit diminished spreading on fibrinogen and reduced formation of filopodia.91 Other haemostatic anomalies in CD151 KO mice include delayed clot retraction and reduced agonist-induced platelet aggregation response to collagen, ADP, and PAR-4 agonist peptide.91 However, platelet adhesion to immobilized fibrinogen and the alpha and dense granules secretion by platelets appear to be unaffected by CD151 deficiency.91
3.4. TSSC6 and Tspan9
TSSC6 or Tspan32 was shown to be expressed in murine platelets. Platelets derived from the TSSC6 KO mice exhibit unstable haemostasis (including increased tail bleeding time, blood lost, rebleeding, and emboli), delayed clot retraction, reduced spreading on fibrinogen, and reduced aggregation to low dosages of PAR-4 agonist peptide and collagen.113 Whether or not TSSC6 is expressed in human platelets and has a functional role in human thrombosis and haemostasis is yet to be determined. Tspan9 is a recently identified platelet tetraspanin that selectively complexes with the platelet collagen receptor, GPVI, and α6β1 but not with adhesion receptors GPIb and GPIIb-IIIa.100 The role of Tspan9 has not been determined.
4. Concluding remarks
Evolutionarily, tetraspanin expression is primarily restricted to multicellular organisms,114,115 suggesting that their primordial function is in modulating intercellular interactions. Functional association of tetraspanins with cell motility is evident even in organisms such as fungi where a tetraspanin homolog directs the formation of cellular protrusive structure that is important for fungal cell invasive activity.116 In vascular tissue and platelets, tetraspanins clearly regulate cell migration and intercellular interactions (Table 1). Thus, a common theme for tetraspanin function in vascular tissue appears to be the modulation of the adhesive and migratory properties of cells. Another characteristic feature for vascular tetraspanins is their localization in cellular vesicles, suggesting that they regulate vesicle trafficking and release events. For example, all four tetraspanins discussed are either largely or partially localized in intracellular granules or vesicles of platelets. Their translocation to the cell surface upon activation correlates with the platelet aggregation. In addition, the vesicular trafficking of CD151 regulates cell migration while EC movement is likely involved in CD151-mediated neovascularization.15,83
Table 1.
The distribution, partners, and functions of vascular tetraspanins
| Tetraspanin | Cell type | Protein interactions | Specific functions |
|---|---|---|---|
| CD9 | Smooth muscle cells | α5β1 | Migration, proliferation, signalling |
| α2β1 | |||
| α3β1 | |||
| Endothelial cells | ICAM-1 | Transendothelial migration of leucocytes | |
| VCAM-1 | |||
| α3β1 | |||
| Platelets | GPIIb/IIIa | Aggregation, signalling | |
| GPIb/V/IX | |||
| CD151 | Smooth muscle cells | Not done | Not done |
| Endothelial cells | Tetraspanins such as CD9 and CD81 | Cell–ECM adhesion | |
| Integrins such as α3β1, α6β1, and α5β1 | Cell–cell adhesion | ||
| IgSF proteins such as ICAM-1 and VCAM-1 | Cell migration | ||
| MT1-MMP | Angiogenesis | ||
| Platelets | GPIIb/IIIa | Aggregation, spreading, adhesion to fibrinogen | |
| Tetraspanins such as CD9 | |||
| CD63 | Smooth muscle cells | Not done | Not done |
| Endothelial cells | vWF | Vesicle trafficking | |
| Platelets | GPIIb/IIIa | Spreading, signalling | |
| Type II PI-4K |
Earlier studies have highlighted the importance of tetraspanins in the regulation of various vascular events. Several critical questions emerge from the earlier observations. For example, how do tetraspanins coordinate cell–matrix and cell–cell adhesions in vascular events? How do tetraspanins coordinate motogenic and mitogenic activities during vascular morphogenesis, remodelling, and vascular injury response? What are the precise roles that vascular tetraspanins play in vesicle trafficking and release? How are the tetraspanins in vesicular compartments connected to cell adhesion and migration? How important are the intricate interactions within tetraspanin webs for fine-tuning specific phenotypes of platelets, SMC, and EC? Could tetraspanins serve as prognostic indicators for vascular disease or as targets for drug therapy?
To answer these outstanding questions, the challenges lie in unraveling the functional associations of the tetraspanin web, elucidating the specific signalling pathways that regulate cell phenotypes, and understanding the trafficking mechanism of tetraspanins in vascular cells. Another key issue to be addressed is the interpretation of the functional data obtained from tetraspanin KO models as other tetraspanins can provide functional compensation.117 Whether or not such mutual functional compensatory phenomena contribute significantly to the overall cell phenotypes observed in these models must be determined before a specific function can be assigned to a tetraspanin.
Based on the fundamental nature of these outstanding questions and key issues, we believe that the future progress largely depends on the integrated approaches that combine biophysical and high resolution imaging, signalling and genetic, or in vitro and in vivo studies. Also, because of the functional versatility of tetraspanins, their specific effector functions may be unique to the vascular system. Future directions will entail the extensive use of vascular models to determine the exact mechanism of vascular tetraspanin function. Furthermore, given the importance of VSMC in the pathogenesis of multiple vascular diseases, functional analysis of tetraspanins in this understudied cell system is predicted to be fruitful. Moreover, besides the tetraspanin members discussed in this review, the expression characteristics and functional roles of many novel and less studied human tetraspanins in vascular system are largely unclear and will become a major exploring area in the coming years. Finally, translational research linking tetraspanins to vascular pathology will provide useful insights into the discovery of preventive measures, diagnostic markers, and therapeutic agents for vascular diseases.
Conflict of interest: none declared.
Funding
This work was supported by the Vascular Biology Center of Excellence, University of Tennessee Health Science Center and an American Heart Association Southeast Affiliate Grant-in-Aid to L.K.J.; American Heart Association Pre-doctoral Fellowship Award and The Patricia Kouns Fellowship Award to J.K.; and the National Institutes of Health Grant CA-96991 and American Heart Association Southeast Affiliate Grant-in-Aid 0855307E to X.A.Z.
References
- 1.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 3.Horejsi V, Vlcek C. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett. 1991;288:1–4. doi: 10.1016/0014-5793(91)80988-f. [DOI] [PubMed] [Google Scholar]
- 4.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 5.Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 6.Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem. 2001;276:40545–40554. doi: 10.1074/jbc.M107338200. [DOI] [PubMed] [Google Scholar]
- 7.Rubinstein E, Le Naour F, Lagaudriere-Gesbert C, Billard M, Conjeaud H, Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur J Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 8.Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005;20:218–224. doi: 10.1152/physiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 9.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 10.Lazo PA. Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 2007;98:1666–1677. doi: 10.1111/j.1349-7006.2007.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goschnick MW, Jackson DE. Tetraspanins-structural and signalling scaffolds that regulate platelet function. Mini Rev Med Chem. 2007;7:1248–1254. doi: 10.2174/138955707782795656. [DOI] [PubMed] [Google Scholar]
- 12.Wright MD, Moseley GW, van Spriel AB. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens. 2004;64:533–542. doi: 10.1111/j.1399-0039.2004.00321.x. [DOI] [PubMed] [Google Scholar]
- 13.Martin F, Roth DM, Jans DA, Pouton CW, Partridge LJ, Monk PN, et al. Tetraspanins in viral infections: a fundamental role in viral biology? J Virol. 2005;79:10839–10851. doi: 10.1128/JVI.79.17.10839-10851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, He B, Liu WM, Zhou D, Cox JV, Zhang XA. Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J Biol Chem. 2007;282:31631–31642. doi: 10.1074/jbc.M701165200. [DOI] [PubMed] [Google Scholar]
- 16.Sugiura T, Berditchevski F. Function of alpha3beta1-tetraspanin protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2) J Cell Biol. 1999;146:1375–1389. doi: 10.1083/jcb.146.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito Y, Tachibana I, Takeda Y, Yamane H, He P, Suzuki M, et al. Absence of CD9 enhances adhesion-dependent morphologic differentiation, survival, and matrix metalloproteinase-2 production in small cell lung cancer cells. Cancer Res. 2006;66:9557–9565. doi: 10.1158/0008-5472.CAN-06-1131. [DOI] [PubMed] [Google Scholar]
- 18.Arduise C, Abache T, Li L, Billard M, Chabanon A, Ludwig A, et al. Tetraspanins regulate ADAM10-mediated cleavage of TNF-alpha and epidermal growth factor. J Immunol. 2008;181:7002–7013. doi: 10.4049/jimmunol.181.10.7002. [DOI] [PubMed] [Google Scholar]
- 19.Yanez-Mo M, Barreiro O, Gonzalo P, Batista A, Megias D, Genis L, et al. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 2008;112:3217–3226. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- 20.Kersey JH, LeBien TW, Abramson CS, Newman R, Sutherland R, Greaves M. P-24: a human leukemia-associated and lymphohemopoietic progenitor cell surface structure identified with monoclonal antibody. J Exp Med. 1981;153:726–731. doi: 10.1084/jem.153.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benoit P, Gross MS, Frachet P, Frezal J, Uzan G, Boucheix C, et al. Assignment of the human CD9 gene to chromosome 12 (region P13) by use of human specific DNA probes. Hum Genet. 1991;86:268–272. doi: 10.1007/BF00202407. [DOI] [PubMed] [Google Scholar]
- 22.Lanza F, Wolf D, Fox CF, Kieffer N, Seyer JM, Fried VA, et al. cDNA cloning and expression of platelet p24/CD9. Evidence for a new family of multiple membrane-spanning proteins. J Biol Chem. 1991;266:10638–10645. [PubMed] [Google Scholar]
- 23.Kotha J, Longhurst C, Appling W, Jennings LK. Tetraspanin CD9 regulates beta 1 integrin activation and enhances cell motility to fibronectin via a PI-3 kinase-dependent pathway. Exp Cell Res. 2008;314:1811–1822. doi: 10.1016/j.yexcr.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert Rev Mol Med. 2001;2001:1–17. doi: 10.1017/S1462399401002381. [DOI] [PubMed] [Google Scholar]
- 25.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, et al. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 26.Longhurst CM, White MM, Wilkinson DA, Jennings LK. A CD9, alphaIIbbeta3, integrin-associated protein, and GPIb/V/IX complex on the surface of human platelets is influenced by alphaIIbbeta3 conformational states. Eur J Biochem. 1999;263:104–111. doi: 10.1046/j.1432-1327.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- 27.Rubinstein E, Le Naour F, Billard M, Prenant M, Boucheix C. CD9 antigen is an accessory subunit of the VLA integrin complexes. Eur J Immunol. 1994;24:3005–3013. doi: 10.1002/eji.1830241213. [DOI] [PubMed] [Google Scholar]
- 28.Higashiyama S, Iwamoto R, Goishi K, Raab G, Taniguchi N, Klagsbrun M, et al. The membrane protein CD9/DRAP 27 potentiates the juxtacrine growth factor activity of the membrane-anchored heparin-binding EGF-like growth factor. J Cell Biol. 1995;128:929–938. doi: 10.1083/jcb.128.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Meyer GR, Bult H. Mechanisms of neointima formation–lessons from experimental models. Vasc Med. 1997;2:179–189. doi: 10.1177/1358863X9700200304. [DOI] [PubMed] [Google Scholar]
- 30.Kotha J, Zhang C, Longhurst CM, Lu Y, Jacobs J, Cheng Y, et al. Functional relevance of tetraspanin CD9 in vascular smooth muscle cell injury phenotypes: A novel target for the prevention of neointimal hyperplasia. Atherosclerosis. 2008 doi: 10.1016/j.atherosclerosis.2008.07.036. Published online ahead of print on August 8. [DOI] [PubMed] [Google Scholar]
- 31.Scherberich A, Moog S, Haan-Archipoff G, Azorsa DO, Lanza F, Beretz A. Tetraspanin CD9 is associated with very late-acting integrins in human vascular smooth muscle cells and modulates collagen matrix reorganization. Arterioscler Thromb Vasc Biol. 1998;18:1691–1697. doi: 10.1161/01.atv.18.11.1691. [DOI] [PubMed] [Google Scholar]
- 32.Lijnen HR, Lupu F, Collen D, Le Naour F, Boucheix C. CD9 gene deficiency does not affect smooth muscle cell migration and neointima formation after vascular injury in mice. Thromb Haemost. 2000;83:956–961. [PubMed] [Google Scholar]
- 33.Scherberich A, Giannone G, Perennou E, Takeda K, Boucheix C, Rubinstein E, et al. FAK-mediated inhibition of vascular smooth muscle cell migration by the tetraspanin CD9. Thromb Haemost. 2002;87:1043–1050. [PubMed] [Google Scholar]
- 34.Goncharova EA, Ammit AJ, Irani C, Carroll RG, Eszterhas AJ, Panettieri RA, et al. PI3K is required for proliferation and migration of human pulmonary vascular smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2002;283:L354–L363. doi: 10.1152/ajplung.00010.2002. [DOI] [PubMed] [Google Scholar]
- 35.Oudit GY, Sun H, Kerfant BG, Crackower MA, Penninger JM, Backx PH. The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J Mol Cell Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 36.Deissler H, Kuhn EM, Lang GE, Deissler H. Tetraspanin CD9 is involved in the migration of retinal microvascular endothelial cells. Int J Mol Med. 2007;20:643–652. [PubMed] [Google Scholar]
- 37.Favaloro EJ. Differential expression of surface antigens on activated endothelium. Immunol Cell Biol. 1993;71:571–581. doi: 10.1038/icb.1993.63. [DOI] [PubMed] [Google Scholar]
- 38.Barreiro O, Yanez-Mo M, Sala-Valdes M, Gutierrez-Lopez MD, Ovalle S, Higginbottom A, et al. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood. 2005;105:2852–2861. doi: 10.1182/blood-2004-09-3606. [DOI] [PubMed] [Google Scholar]
- 39.Klein-Soyer C, Azorsa DO, Cazenave JP, Lanza F. CD9 participates in endothelial cell migration during in vitro wound repair. Arterioscler Thromb Vasc Biol. 2000;20:360–369. doi: 10.1161/01.atv.20.2.360. [DOI] [PubMed] [Google Scholar]
- 40.Erovic BM, Neuchrist C, Kandutsch S, Woegerbauer M, Pammer J. CD9 expression on lymphatic vessels in head and neck mucosa. Mod Pathol. 2003;16:1028–1034. doi: 10.1097/01.MP.0000089777.58000.B2. [DOI] [PubMed] [Google Scholar]
- 41.Yanez-Mo M, Alfranca A, Cabanas C, Marazuela M, Tejedor R, Ursa MA, et al. Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with alpha3 beta1 integrin localized at endothelial lateral junctions. J Cell Biol. 1998;141:791–804. doi: 10.1083/jcb.141.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko EM, Lee IY, Cheon IS, Kim J, Choi JS, Hwang JY, et al. Monoclonal antibody to CD9 inhibits platelet-induced human endothelial cell proliferation. Mol Cells. 2006;22:70–77. [PubMed] [Google Scholar]
- 43.Forsyth KD. Anti-CD9 antibodies augment neutrophil adherence to endothelium. Immunology. 1991;72:292–296. [PMC free article] [PubMed] [Google Scholar]
- 44.Longo N, Yanez-Mo M, Mittelbrunn M, de la Rosa G, Munoz ML, Sanchez-Madrid F, et al. Regulatory role of tetraspanin CD9 in tumor-endothelial cell interaction during transendothelial invasion of melanoma cells. Blood. 2001;98:3717–3726. doi: 10.1182/blood.v98.13.3717. [DOI] [PubMed] [Google Scholar]
- 45.Barreiro O, Zamai M, Yanez-Mo M, Tejera E, Lopez-Romero P, Monk PN, et al. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol. 2008;183:527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ushikoshi HTT, Takemura G, Li Y, Esaki M, Khai NC, Kawai T, et al. CD9 gene therapy inhibits cardiac hypertrophy and tachycardia, and attenuates the remodeling after myocardial infarction in mice. Mol Ther. 2005;11(Suppl. 1):S359. [Google Scholar]
- 47.Hannah MJ, Williams R, Kaur J, Hewlett LJ, Cutler DF. Biogenesis of Weibel-Palade bodies. Semin Cell Dev Biol. 2002;13:313–324. doi: 10.1016/s1084-9521(02)00061-7. [DOI] [PubMed] [Google Scholar]
- 48.Vischer UM, Wagner DD. CD63 is a component of Weibel-Palade bodies of human endothelial cells. Blood. 1993;82:1184–1191. [PubMed] [Google Scholar]
- 49.Bonfanti R, Furie BC, Furie B, Wagner DD. PADGEM (GMP140) is a component of Weibel-Palade bodies of human endothelial cells. Blood. 1989;73:1109–1112. [PubMed] [Google Scholar]
- 50.Schnyder-Candrian S, Borsig L, Moser R, Berger EG. Localization of alpha 1,3-fucosyltransferase VI in Weibel-Palade bodies of human endothelial cells. Proc Natl Acad Sci USA. 2000;97:8369–8374. doi: 10.1073/pnas.97.15.8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi T, Vischer UM, Rosnoblet C, Lebrand C, Lindsay M, Parton RG, et al. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol Biol Cell. 2000;11:1829–1843. doi: 10.1091/mbc.11.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michaux G, Cutler DF. How to roll an endothelial cigar: the biogenesis of Weibel-Palade bodies. Traffic. 2004;5:69–78. doi: 10.1111/j.1600-0854.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 53.Smith DA, Monk PN, Partridge LJ. Antibodies against human CD63 activate transfected rat basophilic leukemia (RBL-2H3) cells. Mol Immunol. 1995;32:1339–1344. doi: 10.1016/0161-5890(95)00113-1. [DOI] [PubMed] [Google Scholar]
- 54.Toothill VJ, Van Mourik JA, Niewenhuis HK, Metzelaar MJ, Pearson JD. Characterization of the enhanced adhesion of neutrophil leukocytes to thrombin-stimulated endothelial cells. J Immunol. 1990;145:283–291. [PubMed] [Google Scholar]
- 55.Berditchevski F, Bazzoni G, Hemler ME. Specific association of CD63 with the VLA-3 and VLA-6 integrins. J Biol Chem. 1995;270:17784–17790. doi: 10.1074/jbc.270.30.17784. [DOI] [PubMed] [Google Scholar]
- 56.Skubitz KM, Campbell KD, Iida J, Skubitz AP. CD63 associates with tyrosine kinase activity and CD11/CD18, and transmits an activation signal in neutrophils. J Immunol. 1996;157:3617–3626. [PubMed] [Google Scholar]
- 57.Mantegazza AR, Barrio MM, Moutel S, Bover L, Weck M, Brossart P, et al. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood. 2004;104:1183–1190. doi: 10.1182/blood-2004-01-0104. [DOI] [PubMed] [Google Scholar]
- 58.Radford KJ, Thorne RF, Hersey P. Regulation of tumor cell motility and migration by CD63 in a human melanoma cell line. J Immunol. 1997;158:3353–3358. [PubMed] [Google Scholar]
- 59.Kitani S, Berenstein E, Mergenhagen S, Tempst P, Siraganian RP. A cell surface glycoprotein of rat basophilic leukemia cells close to the high affinity IgE receptor (Fc epsilon RI). Similarity to human melanoma differentiation antigen ME491. J Biol Chem. 1991;266:1903–1909. [PubMed] [Google Scholar]
- 60.Carmo AM, Wright MD. Association of the transmembrane 4 superfamily molecule CD53 with a tyrosine phosphatase activity. Eur J Immunol. 1995;25:2090–2095. doi: 10.1002/eji.1830250743. [DOI] [PubMed] [Google Scholar]
- 61.Iida J, Skubitz AP, McCarthy JB, Skubitz KM. Protein kinase activity is associated with CD63 in melanoma cells. J Transl Med. 2005;3:42. doi: 10.1186/1479-5876-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin D, Kamsteeg EJ, Zhang Y, Jin Y, Sterling H, Yue P, et al. Expression of tetraspan protein CD63 activates protein-tyrosine kinase (PTK) and enhances the PTK-induced inhibition of ROMK channels. J Biol Chem. 2008;283:7674–7681. doi: 10.1074/jbc.M705574200. [DOI] [PubMed] [Google Scholar]
- 63.Latysheva N, Muratov G, Rajesh S, Padgett M, Hotchin NA, Overduin M, et al. Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol Cell Biol. 2006;26:7707–7718. doi: 10.1128/MCB.00849-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takino T, Miyamori H, Kawaguchi N, Uekita T, Seiki M, Sato H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem Biophys Res Commun. 2003;304:160–166. doi: 10.1016/s0006-291x(03)00544-8. [DOI] [PubMed] [Google Scholar]
- 65.Jung KK, Liu XW, Chirco R, Fridman R, Kim HR. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duffield A, Kamsteeg EJ, Brown AN, Pagel P, Caplan MJ. The tetraspanin CD63 enhances the internalization of the H,K-ATPase beta-subunit. Proc Natl Acad Sci USA. 2003;100:15560–15565. doi: 10.1073/pnas.2536699100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fitter S, Seldin MF, Ashman LK. Characterisation of the mouse homologue of CD151 (PETA-3/SFA-1); genomic structure, chromosomal localisation and identification of 2 novel splice forms. Biochim Biophys Acta. 1998;1398:75–85. doi: 10.1016/s0167-4781(98)00034-7. [DOI] [PubMed] [Google Scholar]
- 68.Fitter S, Tetaz TJ, Berndt MC, Ashman LK. Molecular cloning of cDNA encoding a novel platelet-endothelial cell tetra-span antigen, PETA-3. Blood. 1995;86:1348–1355. [PubMed] [Google Scholar]
- 69.Geary SM, Cambareri AC, Sincock PM, Fitter S, Ashman LK. Differential tissue expression of epitopes of the tetraspanin CD151 recognised by monoclonal antibodies. Tissue Antigens. 2001;58:141–153. doi: 10.1034/j.1399-0039.2001.580301.x. [DOI] [PubMed] [Google Scholar]
- 70.Berditchevski F, Gilbert E, Griffiths MR, Fitter S, Ashman L, Jenner SJ. Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis. J Biol Chem. 2001;276:41165–41174. doi: 10.1074/jbc.M104041200. [DOI] [PubMed] [Google Scholar]
- 71.Sterk LM, Geuijen CA, van den Berg JG, Claessen N, Weening JJ, Sonnenberg A. Association of the tetraspanin CD151 with the laminin-binding integrins alpha3beta1, alpha6beta1, alpha6beta4 and alpha7beta1 in cells in culture and in vivo. J Cell Sci. 2002;115:1161–1173. doi: 10.1242/jcs.115.6.1161. [DOI] [PubMed] [Google Scholar]
- 72.Yauch RL, Kazarov AR, Desai B, Lee RT, Hemler ME. Direct extracellular contact between integrin alpha(3)beta(1) and TM4SF protein CD151. J Biol Chem. 2000;275:9230–9238. doi: 10.1074/jbc.275.13.9230. [DOI] [PubMed] [Google Scholar]
- 73.Gesierich S, Paret C, Hildebrand D, Weitz J, Zgraggen K, Schmitz-Winnenthal FH, et al. Colocalization of the tetraspanins, CO-029 and CD151, with integrins in human pancreatic adenocarcinoma: impact on cell motility. Clin Cancer Res. 2005;11:2840–2852. doi: 10.1158/1078-0432.CCR-04-1935. [DOI] [PubMed] [Google Scholar]
- 74.Testa JE, Brooks PC, Lin JM, Quigley JP. Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res. 1999;59:3812–3820. [PubMed] [Google Scholar]
- 75.Winterwood NE, Varzavand A, Meland MN, Ashman LK, Stipp CS. A critical role for tetraspanin CD151 in alpha3beta1 and alpha6beta4 integrin-dependent tumor cell functions on laminin-5. Mol Biol Cell. 2006;17:2707–2721. doi: 10.1091/mbc.E05-11-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chattopadhyay N, Wang Z, Ashman LK, Brady-Kalnay SM, Kreidberg JA. Alpha3beta1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J Cell Biol. 2003;163:1351–1362. doi: 10.1083/jcb.200306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J Cell Biol. 2003;163:165–176. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sterk LM, Geuijen CA, Oomen LC, Calafat J, Janssen H, Sonnenberg A. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin alpha6beta4 and may regulate the spatial organization of hemidesmosomes. J Cell Biol. 2000;149:969–982. doi: 10.1083/jcb.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lammerding J, Kazarov AR, Huang H, Lee RT, Hemler ME. Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc Natl Acad Sci USA. 2003;100:7616–7621. doi: 10.1073/pnas.1337546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13:221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Liu Z, Shen X, Yao W, Qu H, Yang M, et al. Expression of CD151 in human atherosclerotic artery and its implication. J Huazhong Univ Sci Technolog Med Sci. 2005;25:629–631. doi: 10.1007/BF02896154. [DOI] [PubMed] [Google Scholar]
- 82.Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK. PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci. 1999;112:833–844. doi: 10.1242/jcs.112.6.833. [DOI] [PubMed] [Google Scholar]
- 83.Takeda Y, Kazarov AR, Butterfield CE, Hopkins BD, Benjamin LE, Kaipainen A, et al. Deletion of tetraspanin Cd151 results in decreased pathologic angiogenesis in vivo and in vitro. Blood. 2007;109:1524–1532. doi: 10.1182/blood-2006-08-041970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang XA, Kazarov AR, Yang X, Bontrager AL, Stipp CS, Hemler ME. Function of the tetraspanin CD151-alpha6beta1 integrin complex during cellular morphogenesis. Mol Biol Cell. 2002;13:1–11. doi: 10.1091/mbc.01-10-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng Z, Liu Z. CD151 gene delivery activates PI3K/Akt pathway and promotes neovascularization after myocardial infarction in rats. Mol Med. 2006;12:214–220. doi: 10.2119/2006-00037.Zheng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishiuchi R, Sanzen N, Nada S, Sumida Y, Wada Y, Okada M, et al. Potentiation of the ligand-binding activity of integrin alpha3beta1 via association with tetraspanin CD151. Proc Natl Acad Sci USA. 2005;102:1939–1944. doi: 10.1073/pnas.0409493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fitter S, Sincock PM, Jolliffe CN, Ashman LK. Transmembrane 4 superfamily protein CD151 (PETA-3) associates with beta 1 and alpha IIb beta 3 integrins in haemopoietic cell lines and modulates cell-cell adhesion. Biochem J. 1999;338:61–70. [PMC free article] [PubMed] [Google Scholar]
- 88.Sincock PM, Mayrhofer G, Ashman LK. Localization of the transmembrane 4 superfamily (TM4SF) member PETA-3 (CD151) in normal human tissues: comparison with CD9, CD63, and alpha5beta1 integrin. J Histochem Cytochem. 1997;45:515–525. doi: 10.1177/002215549704500404. [DOI] [PubMed] [Google Scholar]
- 89.Karamatic Crew V, Burton N, Kagan A, Green CA, Levene C, Flinter F, et al. CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood. 2004;104:2217–2223. doi: 10.1182/blood-2004-04-1512. [DOI] [PubMed] [Google Scholar]
- 90.Cowin AJ, Adams D, Geary SM, Wright MD, Jones JC, Ashman LK. Wound healing is defective in mice lacking tetraspanin CD151. J Invest Dermatol. 2006;126:680–689. doi: 10.1038/sj.jid.5700142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lau LM, Wee JL, Wright MD, Moseley GW, Hogarth PM, Ashman LK, et al. The tetraspanin superfamily member CD151 regulates outside-in integrin alphaIIbbeta3 signaling and platelet function. Blood. 2004;104:2368–2375. doi: 10.1182/blood-2003-12-4430. [DOI] [PubMed] [Google Scholar]
- 92.Wright MD, Geary SM, Fitter S, Moseley GW, Lau LM, Sheng KC, et al. Characterization of mice lacking the tetraspanin superfamily member CD151. Mol Cell Biol. 2004;24:5978–5988. doi: 10.1128/MCB.24.13.5978-5988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hasegawa H, Nomura T, Kishimoto K, Yanagisawa K, Fujita S. SFA-1/PETA-3 (CD151), a member of the transmembrane 4 superfamily, associates preferentially with alpha 5 beta 1 integrin and regulates adhesion of human T cell leukemia virus type 1-infected T cells to fibronectin. J Immunol. 1998;161:3087–3095. [PubMed] [Google Scholar]
- 94.Hasegawa H, Utsunomiya Y, Kishimoto K, Yanagisawa K, Fujita S. SFA-1, a novel cellular gene induced by human T-cell leukemia virus type 1, is a member of the transmembrane 4 superfamily. J Virol. 1996;70:3258–3263. doi: 10.1128/jvi.70.5.3258-3263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ho SH, Martin F, Higginbottom A, Partridge LJ, Parthasarathy V, Moseley GW, et al. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J Virol. 2006;80:6487–6496. doi: 10.1128/JVI.02539-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Protty MB, Watkins NA, Colombo D, Thomas SG, Heath VL, Herbert J, et al. Identification of Tspan9 as a novel platelet tetraspanin and the collagen receptor GPVI as a component of tetraspanin microdomains. Biochem J. 2009;417:391–400. doi: 10.1042/BJ20081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miller JL, Kupinski JM, Hustad KO. Characterization of a platelet membrane protein of low molecular weight associated with platelet activation following binding by monoclonal antibody AG-1. Blood. 1986;68:743–751. [PubMed] [Google Scholar]
- 98.Miao WM, Vasile E, Lane WS, Lawler J. CD36 associates with CD9 and integrins on human blood platelets. Blood. 2001;97:1689–1696. doi: 10.1182/blood.v97.6.1689. [DOI] [PubMed] [Google Scholar]
- 99.Israels SJ, McMillan-Ward EM. CD63 modulates spreading and tyrosine phosphorylation of platelets on immobilized fibrinogen. Thromb Haemost. 2005;93:311–318. doi: 10.1160/TH04-08-0503. [DOI] [PubMed] [Google Scholar]
- 100.Jennings LK, Fox CF, Kouns WC, McKay CP, Ballou LR, Schultz HE. The activation of human platelets mediated by anti-human platelet p24/CD9 monoclonal antibodies. J Biol Chem. 1990;265:3815–3822. [PubMed] [Google Scholar]
- 101.Rubinstein E, Boucheix C, Worthington RE, Carroll RC. Anti-platelet antibody interactions with Fc gamma receptor. Semin Thromb Hemost. 1995;21:10–22. doi: 10.1055/s-2007-1000375. [DOI] [PubMed] [Google Scholar]
- 102.Worthington RE, Carroll RC, Boucheix C. Platelet activation by CD9 monoclonal antibodies is mediated by the Fc gamma II receptor. Br J Haematol. 1990;74:216–222. doi: 10.1111/j.1365-2141.1990.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 103.Carroll RC, Worthington RE, Boucheix C. Stimulus-response coupling in human platelets activated by monoclonal antibodies to the CD9 antigen, a 24 kDa surface-membrane glycoprotein. Biochem J. 1990;266:527–535. doi: 10.1042/bj2660527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Griffith L, Slupsky J, Seehafer J, Boshkov L, Shaw AR. Platelet activation by immobilized monoclonal antibody: evidence for a CD9 proximal signal. Blood. 1991;78:1753–1759. [PubMed] [Google Scholar]
- 105.Hill SK. Memphis: University of Tennessee Health Science Center; The tetraspanin CD9 localizes to platelet-platelet contacts and regulates thrombus stability. PhD Thesis. [Google Scholar]
- 106.Brisson C, Azorsa DO, Jennings LK, Moog S, Cazenave JP, Lanza F. Co-localization of CD9 and GPIIb-IIIa (alpha IIb beta 3 integrin) on activated platelet pseudopods and alpha-granule membranes. Histochem J. 1997;29:153–165. doi: 10.1023/a:1026437522882. [DOI] [PubMed] [Google Scholar]
- 107.Hamamoto K, Ohga S, Nomura S, Yasunaga K. Cellular distribution of CD63 antigen in platelets and in three megakaryocytic cell lines. Histochem J. 1994;26:367–375. doi: 10.1007/BF00157770. [DOI] [PubMed] [Google Scholar]
- 108.Nishibori M, Cham B, McNicol A, Shalev A, Jain N, Gerrard JM. The protein CD63 is in platelet dense granules, is deficient in a patient with Hermansky-Pudlak syndrome, and appears identical to granulophysin. J Clin Invest. 1993;91:1775–1782. doi: 10.1172/JCI116388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Israels SJ, McMillan-Ward EM, Easton J, Robertson C, McNicol A. CD63 associates with the alphaIIb beta3 integrin-CD9 complex on the surface of activated platelets. Thromb Haemost. 2001;85:134–141. [PubMed] [Google Scholar]
- 110.Israels SJ, McMillan-Ward EM. Platelet tetraspanin complexes and their association with lipid rafts. Thromb Haemost. 2007;98:1081–1087. [PubMed] [Google Scholar]
- 111.Schroder JL-RR, Himmerkus N, Pleines I, Nieswandt B, Orinska Z, Koch-Nolte F, et al. Deficiency of the tetraspanin CD63 associated with kidney pathology but normal lysosomal function. Mol Cell Biol. 2009;29:1083–1094. doi: 10.1128/MCB.01163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ashman LK, Aylett GW, Mehrabani PA, Bendall LJ, Niutta S, Cambareri AC, et al. The murine monoclonal antibody, 14A2.H1, identifies a novel platelet surface antigen. Br J Haematol. 1991;79:263–270. doi: 10.1111/j.1365-2141.1991.tb04531.x. [DOI] [PubMed] [Google Scholar]
- 113.Goschnick MW, Lau LM, Wee JL, Liu YS, Hogarth PM, Robb LM, et al. Impaired ‘outside-in’ integrin alphaIIbbeta3 signaling and thrombus stability in TSSC6-deficient mice. Blood. 2006;108:1911–1918. doi: 10.1182/blood-2006-02-004267. [DOI] [PubMed] [Google Scholar]
- 114.Garcia-Espana A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 115.Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, et al. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics. 2005;86:674–684. doi: 10.1016/j.ygeno.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 116.Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, et al. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc Natl Acad Sci USA. 2001;98:6963–6968. doi: 10.1073/pnas.111132998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaji K, Oda S, Miyazaki S, Kudo A. Infertility of CD9-deficient mouse eggs is reversed by mouse CD9, human CD9, or mouse CD81; polyadenylated mRNA injection developed for molecular analysis of sperm-egg fusion. Dev Biol. 2002;247:327–334. doi: 10.1006/dbio.2002.0694. [DOI] [PubMed] [Google Scholar]