Abstract
In the filamentous fungus Aspergillus nidulans, cytokinesis/septation is triggered by the septation initiation network (SIN), which first appears at the spindle pole body (SPB) during mitosis. The coiled-coil protein SNAD is associated with the SPB and is required for timely septation and conidiation. We have determined that SNAD acted as a scaffold protein that is required for the localization of the SIN proteins of SIDB and MOBA to the SPB. Another scaffold protein SEPK, whose localization at the SPB was dependent on SNAD, was also required for SIDB and MOBA localization to the SPB. In the absence of either SEPK or SNAD, SIDB/MOBA successfully localized to the septation site, indicating that their earlier localization at SPB was not essential for their later appearance at the division site. Unlike their functional counterparts in fission yeast, SEPK and SNAD were not required for vegetative growth but only for timely septation. Furthermore, down-regulation of negative regulators of the SIN suppressed the septation and conidiation phenotypes due to the loss of SNAD. Therefore, we conclude that SPB localization of SIN components is not essential for septation per se, but critical for septation to take place in a timely manner in A. nidulans.
INTRODUCTION
Timely execution of cytokinesis is essential for growth and development of multicellular organisms. Temporal deregulation of cytokinesis often has the catastrophic consequences of forming aneuploid cells and causing fatality. In fungi, cytokinesis/septation involves septum assembly coupled with the contraction of the actomyosin ring at the cell division site. Studies in fission yeast have revealed that actomyosin ring contraction is triggered by a signaling cascade known as the septation initiation network (SIN; Krapp et al., 2004; Wolfe and Gould, 2005). In fission yeast, the SIN has a core of three cascaded kinases: Cdc7p, Sid1p, and Sid2p. On activation by the small G-protein Spg1p when in a GTP-bound form, the kinase cascade initiates the contraction of the actomyosin ring after the completion of mitosis (Wolfe and Gould, 2005).
In the filamentous fungus Aspergillus nidulans, genes homologous to most yeast SIN components are present in the genome (see Table 2), implying that a signaling network resembling the SIN in fission yeast likely regulates septation. In fact, orthologues of Cdc7p and Sid2p, known to us as SEPH and SIDB, play essential role in septation in A. nidulans (Bruno et al., 2001; Kim et al., 2006). Moreover, SIDB and MOBA exhibit a similar localization pattern to their fission yeast counterparts (Kim et al., 2006).
Table 2.
SIN components in S. pombe and A. nidulans
| S. pombe | A. nidulans | Protein feature |
|---|---|---|
| Spg1p | SPGA (AN7206) | Small G-protein |
| Cdc7p | SEPH | Protein kinase |
| Sid1p | SEPL (AN8033) | Protein kinase |
| Sid2p | SIDB | Protein kinase |
| Cdc14p | SEPM (AN0655) | Sid1p/SEPM regulator |
| Mob1p | MOBA | Sid2p/SIDB regulator |
| Cdc16p | BUBA (AN0281) | Two-component GAP |
| Byr4p | BYRA (AN9413) | |
| Cdc11p | SEPK (AN2459) | Leu-rich scaffold protein |
| Sid4p | No homologue | Coiled coil scaffold protein |
| No homologue | SNAD | Coiled coil scaffold protein |
Cell cycle–dependent localization patterns of SIN components reflect their regulatory roles in coordinating septation with mitosis. SIN molecules first appear at the spindle pole body (SPB), some starting at interphase and others appearing after mitotic entry (Simanis, 2003). Among them, the Sid2p/SIDB kinase and its associated cofactor Mob1p/MOBA later decorate the cell division site after appearing at the SPB at early stages of mitosis (Kim et al., 2006; Simanis, 2003). Unlike in fission yeast, the SIDB/MOBA signal contracts with the actomyosin ring in A. nidulans (Kim et al., 2006). Although it has been speculated that Sid2p/SIDB and Mob1p/MOBA might translocate from the SPB to the division site, it has not been experimentally proven. One possibility is that their localization at the SPB could be essential for their activation by their upstream components like Cdc7p/SEPH, which likely communicate with other essential cell cycle regulators located at the site. Indeed a recent study by laser ablation shows that in dividing cells segregated SPBs act as essential regulatory sites, and either one can trigger septation (Magidson et al., 2006).
In fission yeast, SIN kinases and their associated proteins are anchored to the SPB via the scaffold proteins Cdc11p and Sid4p (Chang and Gould, 2000; Krapp et al., 2001; Tomlin et al., 2002; Morrell et al., 2004). Although Cdc11p depends on Sid4p for its SPB localization, Sid4p localizes to the SPB via interaction with Ppc89p (Morrell et al., 2004; Rosenberg et al., 2006). The fact that both Cdc11p and Sid4p are indispensable for septation suggests that the localization of SIN molecules to the SPB might be required for septation in fission yeast.
Based on direct sequence comparisons, the complete A. nidulans genome (http://www.broad.mit.edu/annotation/genome/aspergillus_nidulans) does not contain genes encoding obvious homologues of Sid4p and Ppc89. Thus, SIN molecules are anchored to the SPB either via a different scaffold or by a different mechanism.
Our previous work showed that the coiled-coil protein SNAD is localized to the SPB and is required for timely septation and conidiation in A. nidulans (Liu and Morris, 2000). It was not known how SNAD participates in temporal regulation of septation. The results described in this report document that SNAD is required for the anchoring of currently identified SIN components to be anchored at the SPB. Our results also indicate that the spindle pole localization is important for timely execution of septation, but is not essential for this process.
Because SIN components and its anchoring scaffold are essential for septation and consequently for viability in fission yeast, it was not possible to assess how the scaffold would affect other developmental events in this organism. Filamentous fungi like A. nidulans form mycelia of multinucleate cells. Septation is essential for conidiation (asexual spore formation), but not for viability as demonstrated by mutations at the mobA and sidB loci (Kim et al., 2006). Moreover, snaD mutations also do not abolish septation, but completely abolish conidiation (Liu and Morris, 2000). Therefore, the SPB localization of SIDB/MOBA is not absolutely required for their appearance at the division site to trigger septation. This suggests that SIDB/MOBA can be activated in the cytosol before appearing at the division site to exercise their activation role for the assembly and contraction of the actomyosin ring.
MATERIALS AND METHODS
Strains and Growth Condition
A. nidulans strains used in this study are listed in Table 1, with their genotypes and sources indicated. The Neurospora crassa pyr4 and A. fumigatus AfpyrG genes were used for genetic suppression/complementation of the pyrG89 mutation, and the A. fumigatus AfpyroA gene for the pyroA4 mutation. Recipes for rich and minimal growth media were as reported (Kafer, 1977). Induction condition for alcA(p) strains was as described previously (Liu and Morris, 2000). Standard crosses were carried out to combine mutations in single strains.
Table 1.
A. nidulans strains described in the text
| Strain | Genotype | Source |
|---|---|---|
| CZA2 | alcA(p)::mCherry-byrA::AfpyrG; alcA(p)::GFP-bubA::AfpyroA; pyrG89; pyroA4; WA2 | This work |
| CZA14 | MRLC::GFP::AfpyrG; pyrG89; pyroA4; nkuA::argB; argB2; riboB2; pabaA1 yA2 | This work |
| CZA16 | snaD290; alcA(p)::mCherry-byrA::AfpyrG; alcA(p)::GFP-bubA::AfpyroA; pyrG89; pyroA4 | This work |
| CZA17 | alcA(p)::mCherry-byrA::AfpyrG; pyrG89; pyroA4; nkuAΔ::argB; riboB2 | This work |
| CZA18 | alcA(p)::GFP-bubA::AfpyroA; pyrG89; pyroA4; nkuAΔ::argB; riboB2 | This work |
| CZA28 | pyrG89; mobAΔ::pyrG; MRLC::GFP::AfpyrG; yA2 | This work |
| GR5 | pyrG89; pyroA4; wA2 | FGSC |
| JKA8 | alcA(p)::GFP-sidB::pyrG; pyrG89; pabaA1 yA2 | Kim et al. (2006) |
| JKA23-1 | alcA(p)::GFP-sidB::pyrG; snaDΔ::pyr4; pyrG89; wA2; pyroA4 | This work |
| JKA63-1 | apsBΔ::argB; alcA(p)::GFP-snaD::pyr4; pyrG89; pyroA4 | This work |
| JKA64-1 | apsBΔ::argB; snaDΔ::pyr4; pyroA4 | This work |
| JKA69-1 | alcA(p)::GFP-sidB::pyrG; apsB6; pabaA1; pyrG89 | This work |
| JKA72-4 | alcA(p)::GFP-snaD::pyr4; apsB6; wA2; pyroA4; biA1 | This work |
| JKA76-2 | alcA(p)::mCherry-sepK::AfpyrG; snaD-GFP::AfpyrG; pyrG89, pabaA1 yA2 | This work |
| LB04 | pyroA4; chaA1 | Kim et al. (2006) |
| LB15 | snaD290; pyroA4; pyrG89; chaA1 | Liu and Morris (2000) |
| LB30 | alcA(p)::GFP-snaD::pyr4; pyrG89; pyroA4; wA2 | Liu and Morris (2000) |
| LB31 | snaDΔ::pyr4; pyrG89; pyroA4; wA2 | Liu and Morris (2000) |
| LBA49 | alcA(p)::GFP-mobA::pyrG; pyrG89; pyroA4; wA2 | Kim et al. (2006) |
| LBA53 | alcA(p)::GFP-mobA::pyrG; pyrG89; chaA1 | Kim et al. (2006) |
| LBA54 | alcA(p)::GFP-mobA::pyrG; nudA1; pyrG89; pyroA4 | This work |
| LBA69 | alcA(p)::GFP-mobA::pyrG; snaD290; pyrG89; pyroA4; chaA1 | This work |
| LO1591 | pyrG89, pyroA4, riboB2, nkuAΔ::argB, sepK-GFP-AfpyrG | Nayak et al. (2008) |
| LO1906 | nkuAΔ::argB; pyrG89; pyroA4; riboB2; HH1-mRFP::AfriboB; sepKΔ::AfpyrG | Nayak et al. (2008) |
| R21 | pabaA1 yA2 | C. F. Roberts |
| RSA121 | snaD290; pyr4::alcA(p)::GFP-apsB; pyroA4; chaA1 | This work |
| RSA122 | pyr4::alcA(p)::GFP-apsB; pyrG89; snaD290; pyroA4 | This work |
| RSA123 | apsBΔ::argB; alcA(p)::GFP-mobA::pyrG; pyroA4 | This work |
| RSA126 | snaD290; sepK-GFP:: AfpyrG; pyroA4; chaA1; riboB2 | This work |
| RSA129 | alcA(p)::GFP-mobA::pyrG; sepKΔ::AfpyrG; pyroA4; pyrG89; HH1-mCherry::AfpyrG | This work |
| RSA131 | sepKΔ::AfpyrG; alcA(p)::GFP-snaD::pyr4; riboA1 | This work |
| RSA143 | MRLC::GFP::AfpyrG; snaD290 | This work |
| RSA147 | MRLC::GFP::AfpyrG; sepKΔ::AfpyrG; pyrG89 | This work |
| SEa3 | pyr4::alcA(p)::GFP-apsB; pyrG89; wA3; pyroA4 | Veith et al. (2005) |
| SRS1 | biA1; apsBΔ::argB | Suelmann et al. (1998) |
| TN02A7 | pyrG89; pyroA4; nkuAΔ::argB; riboB2 | Nayak et al. (2006) |
| XX3 | nudA1; pyrG89; chaA1 | Xiang et al. (1994) |
Recombinant DNA Techniques
DNA fragments were amplified by PCR from either plasmids or genomic DNA of A. nidulans as indicated in the text by the KOD DNA polymerase (EMD Chemicals, Gibbstown, NJ). The pBluescript KS+ plasmid (Stratagene, La Jolla, CA) was used as a general cloning vector. The host strain for plasmid propagation was Escherichia coli DH5α.
Creation of alcA(p)::GFP-bubA and alcA(p)::mCherry-byrA Strains
To make a plasmid for creating the alcA(p)::GFP-bubA strain, we first made the pJK20 plasmid by replacing the pyrG fragment in pLB04B (Kim et al., 2006) with a AfpyroA fragment using BamHI and HindIII. Next the bubA fragment was amplified using the primers, 5′-GC TCTAGA ATG TCG TCC CAA CCA CCA ACG-3′ and 5′-CG GGATCC GAC GTG CTA CTG TTG CTG CT-3′ (sites designated with underlines), and cloned into pJK20 at XbaI and BamHI sites. The plasmid was then transformed into the A. nidulans strain TN02A7. After confirmation by Southern blotting (Supplementary Figure S1), the strain 4 was used for further experiments.
The plasmid used for creating the alcA(p)::mCherry-byrA strain was made by the following steps. First, the mCherry fragment was amplified from the pRSET-B mCherry plasmid (Shaner et al., 2004) using the primer sets of 5′-CC ATCGAT ATG GTG AGC AAG GGC GAG G-3′ and 5′-T GATATC CTT GTA CAG CTC GTC CAT GCC G-3′. The amplified fragment was digested with ClaI and EcoRV (sites designated with underlines in the primers) before being cloned into the pBluescript KS+ plasmid at the corresponding sites to create the plasmid pBS-mCherry. Next the alcA(p) fragment was excised from the plasmid pMCB17 (Fernández-Abalos et al., 1998) with EcoRI (blunt-ended by overhang fill-in) followed by SalI, and cloned into pBS-mCherry at XhoI (blunt-ended by overhang fill-in) and SalI sites to create the plasmid pAlc-mCherry. The AfpyrG fragment from A. fumigatus, amplified from the pFN03 plasmid (Yang et al., 2004) using the primers 5′-TGT TCG CAA TGC CTC CTC AC-3′ and 5′-ACA GCC ATT GCG AAA CCT CA-3′, was cloned into pAlc-mCherry at the blunt end after fill in the overhang of the NotI site to give rise to pJK13. Finally, the 5′ end byrA genomic fragment was amplified from genomic DNA using the primers 5′-C GAATTC GCG CCA TCC ACC GTC GAT GTG-3′ and 5′-GC ACTAGT CCC AAA TCG TAC ACC AAT GC-3′ (sites designated with underlines) and cloned into pJK13 at EcoRI and SpeI before being transformed into the TN02A7 host strain. After confirmation by Southern blotting (Supplementary Figure S1), the strain 2 with a single integration at the desired site was chosen for our experiment.
Creation of the RLCA-Green Fluorescent Protein Strain
The Green Fluorescent Protein (GFP)-coding sequence was inserted in frame with the RLCA locus AN0267 in front of the stop codon using a published fusion PCR protocol (Nayak et al., 2006; Szewczyk et al., 2006). Briefly, the 5′ genomic fragment was amplified with the primers of 5′-ATG CCC GTA GTA TCG CCC AG-3′ and 5′-CTC CAG CGC CTG CAC CAG CTC CCT TCG GTT TCG CCG CTT GTC -3′, and the 3′ genomic fragment was amplified with 5′- CAG TGC CTC CTC TCA GAC AGA TAT CTA AGT CAT TGT TCT ACG GG -3′ and 5′-GCC ATC GAC GCT GCA GGC G-3′ using the AccuPrime Pfx DNA polymerase (Invitrogen, Carlsbad, CA). The fragment containing the GFP-coding sequence plus the AfpyrG gene was amplified from pFN03 using the primers, 5′-GGA GCT GGT GCA GGC GCT GGA G-3′ and 5′-GAT ATC TGT CTG AGA GGA GGC ACT G-3′ with the same enzyme. The fusion PCR cassette amplified by AccuPrime Taq High Fidelity DNA polymerase (Invitrogen) was used to transform the RSA135 strain.
Microscopy and Image Analysis
GFP and mCherry signals were observed in live cells by having growing mycelia under coverslips. To visualize the nucleus and the septum, cells were first fixed with 4% paraformaldehyde (Polyscience, Warrington, PA). Then DNA and chitin were stained using DAPI (4′,6-diamidino-2-phenylindole) and Calcofluor white (both from Sigma Aldrich, St. Louis, MO), respectively.
Cells were observed with an Eclipse E600 microscope equipped with DIC and epifluorescence optics using 60× Plan-Apo or 100× Plan-Fluor objectives (Nikon, Tokyo, Japan). GFP, mCherry, and DAPI/Calcofluor signals were observed using standard Chroma filter sets for GFP, Texas red, and DAPI, respectively (Chroma, Brattleboro, VT). Images were recorded using an Orca-100 CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan) driven by the Metamorph software package (Molecular Devices, Sunnyvale, CA). All images were assembled using Photoshop 7.0 software (Adobe, San Jose, CA).
RESULTS
SNAD Is Required for SPB Localization of MOBA and SIDB
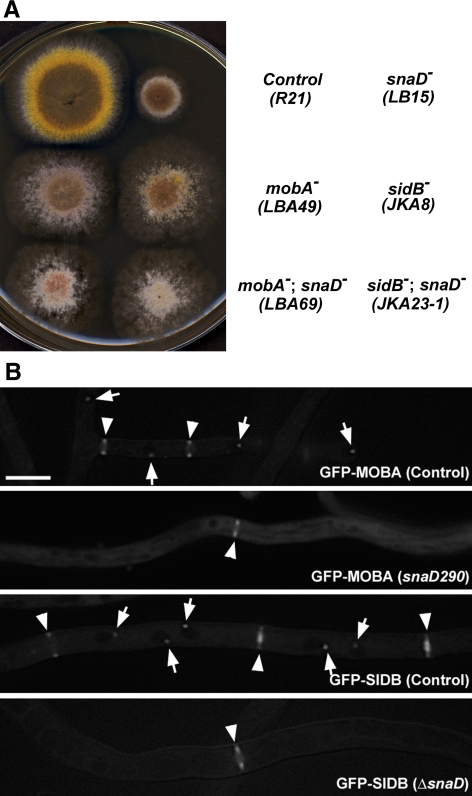
A simple BLAST search of the A. nidulans genome with the SIN components of the fission yeast S. pombe resulted in the identification of homologues for most of them, except for Sid4p (Table 2). Because the SNAD protein localizes to the SPB and is involved in temporal regulation of septation, we wondered whether it had any interaction with the SIN components at the site. Loss of function of either mobA or sidB, orthologues of the yeast mob1 and sid2, abolishes septation and conidiation (Kim et al., 2006). The snaD290 and deletion (snaDΔ) mutations exhibit identical phenotypes of a delay in septation and failed conidiation (Liu and Morris, 2000). Double mutants of snaD; mobA and snaD; sidB were created. Because null mutations of SIN genes like mobA resulted in complete abolishment of conidiation and formation of colonies with a fluffy appearance (Kim et al., 2006), subculture of the mutant was a problem. We consequently used conditional mutations in which the mobA or sidB gene was under the control of the inducible/repressible alcA promoter. The mutants conidiated when grown on nonrepressing medium (i.e., with glycerol as the sole carbon source) and produced fluffy colonies resembling the null mutants when grown on repressing medium (i.e., containing glucose; Kim et al., 2006). The snaD single mutant produced nonconidiating, but otherwise robust colonies (Figure 1A). On the medium containing glucose, the double mutants snaD290; alcA(p)::GFP-mobA and snaDΔ; alcA(p)::GFP-sidB resembled single mutants when either mobA or sidB was not expressed (Figure 1A). Therefore, the loss of SNAD did not cause a phenotype in addition to that caused by the loss of SIN activity. This suggested to us that SNAD possibly functions in the SIN pathway.
Figure 1.
SNAD functions in the SIN pathway. (A) Colony growth phenotype of single and double mutants. Strains with names marked on the right were grown on rich medium containing glucose plus uridine and uracil. Under these growth conditions, the mobA and sidB genes in the respective strains were repressed, which caused nonconidiating colonies due to abolishment of septation. Note the mobA; snaD and sidB; snaD double mutant did not show more severe phenotypes than mobA and sidB single mutants, respectively. (B) Dependence of the SPB localization of SIDB/MOBA on SNAD. In control strains, both GFP-MOBA and GFP-SIDB localized to SPBs (arrows) and septation sites (arrowheads). In the mutants carrying either the snaD290 or ΔsnaD (null) mutation, GFP-MOBA and GFP-SIDB were no longer localized at the SPB, but still localized to the septation sites (arrowheads). Scale bar, 5 μm.
Next we determined whether SNAD was required for the localization of SIDB and MOBA. On the completion of mitosis, both SIDB and MOBA appeared at both the SPB and the division site as a ring (Figure 1B). In the snaD290 or snaDΔ mutation background, functional GFP-MOBA and GFP-SIDB were never detected at the SPB throughout the cell cycle. However, they exhibited localization at the division site as rings (Figure 1, B and C). As seen in the wild-type snaD background, the GFP-labeled rings contracted as septation progressed in the snaD290 or snaDΔ mutant. These results thus suggested to us that SNAD was required for the localization of SIDB and MOBA at the SPB, but not at the septation site.
The Cdc11p Homolog SEPK Is Required for SIN Localization at the SPB
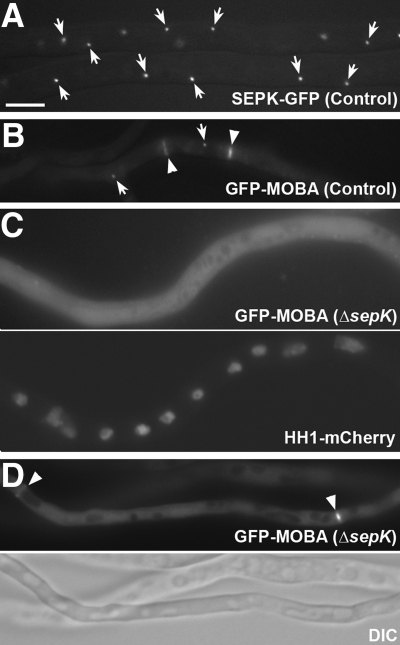
Cdc11p was initially identified as a homolog of the budding yeast protein Nud1p with very limited identity (Tomlin et al., 2002). The AN2459 locus was annotated to be a 1781-amino acid protein with 35% identity and 57% similarity to the fission yeast Cdc11p protein in the leucine-rich region toward the C-terminus and was named sepK (T. Nayak and B. Oakley, unpublished data). A null sepK mutation causes a significant delay in septation and almost abolishes conidiation (T. Nayak and B. Oakley, unpublished data). The SEPK protein decorated the SPB as shown by a functional SEPK-GFP fusion protein (Figure 2A).
Figure 2.
Dependence of SPB localization of MOBA on SEPK. (A) SEPK-GFP localization to the SPB (arrows). (B) GFP-MOBA appeared at SPBs (arrows) and septation sites (arrowheads) in control cells. (C) In mutant cells carrying the sepKΔ (null) mutation, the GFP-MOBA signal appears diffuse in the cytoplasm. The nuclei in the same hypha were detected using a histone H1 (HH1)-mCherry fusion protein. (D) GFP-MOBA still decorated the septation site (arrowhead) in the sepKΔ mutant. Scale bar, 5 μm.
The fission yeast Cdc11p contains leucine-rich repeats toward its C-terminus and is required for its interaction with Sid4p and its localization to the SPB (Krapp et al., 2001; Tomlin et al., 2002), but its N-terminal sequence shows very little, if any, homology to the budding yeast Nud1p, which is required for the interaction with the SIN molecules via direct protein–protein interaction (Tomlin et al., 2002; Morrell et al., 2004). Similarly, SEPK showed very limited sequence homology to Cdc11p in the SIN-interacting domain. Therefore, we asked whether it also played a role in the localization of SIN molecules to the SPB like SNAD. In the presence of SEPK, GFP-MOBA almost exclusively appeared at the SPB and the septation site during mitosis and cytokinesis, and very little if any signal was detected in the cytoplasm (Figure 2B). When a functional GFP-MOBA fusion was expressed in the sepK null mutation background, however, GFP-MOBA no longer appeared at the SPB (Figure 2C), but it still appeared at the septation site (Figure 2D). It was noticed that the diffuse cytoplasmic signal of GFP-MOBA became greatly enhanced compared with that in control cells expressing SEPK (Figure 2C). Therefore, we concluded that SEPK, like SNAD, is required for SPB localization of the SIN components SIDB and MOBA, but not for their localization to the septation site. This finding also suggests that septation continues to take place in the absence of SEPK because of the sustained activity of the SIN at the septation site.
SNAD Is Required for SEPK Localization at the SPB, But Not Vice Versa
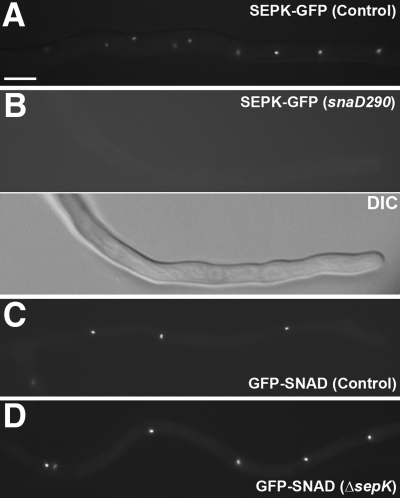
Because SNAD and SEPK were both required for the localization of SIN molecules at the SPB, we wondered whether there was a dependency of localization between them. This test was also inspired by the fact that Sid4p is required for Cdc11p localization in fission yeast (Krapp et al., 2001; Tomlin et al., 2002). When SEPK-GFP was expressed in the snaD290 background, SEPK-GFP was no longer detected at the SPB in contrast to control cells (Figure 3, A and B). When GFP-SNAD was expressed in the sepK null mutation background, however, it still decorated the SPB as in control cells (Figure 3, C and D). These results, thus, indicated that SNAD localization to the SPB was independent of SEPK, but SEPK depended on SNAD for its localization. Therefore, SNAD may act like Sid4p in fission yeast as a scaffold protein at the SPB, which is required for anchoring SIN molecules to the SPB.
Figure 3.
SEPK localization depends on SNAD. (A) SEPK-GFP decorated SPB in control cells. (B) In the snaD290 mutation background, the SEPK-GFP signal was no longer detected at the SPB and was barely detected in the cytoplasm. The same hypha was visualized by the DIC optics. (C) In control cells, GFP-SNAD localized to SPB. (D) GFP-SNAD localization was not affected in cells carrying sepKΔ. Scale bar, 5 μm.
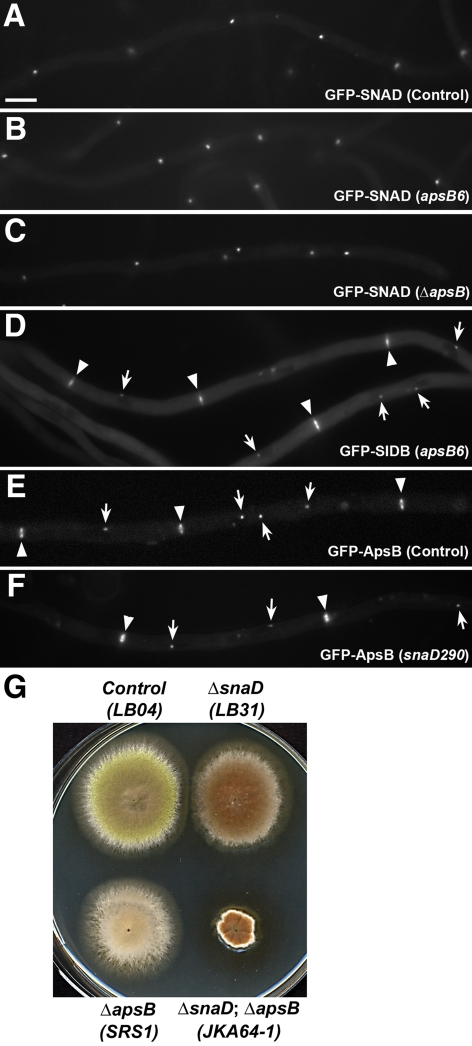
ApsB Acts in a Different Pathway from SNAD
Loss-of-function mutations of SNAD completely abolish conidiation and partially suppress mutations in genes of the cytoplasmic dynein pathway (Liu and Morris, 2000). Coincidently, another spindle pole body–associated coiled coil protein ApsB is also known to play a critical role in conidiation, and mutant alleles of the apsB gene partially suppress poor colony growth caused by the loss of the dynein heavy chain in A. nidulans as well (Suelmann et al., 1998; Veith et al., 2005). These similarities prompted us to test whether the two coiled coil proteins ApsB and SNAD interacted with each other at the SPB. We first tested whether ApsB was required for SNAD to localize to the SPB. A GFP-SNAD fusion protein (encoded by the only copy of snaD in the genome) decorated the SPB throughout the cell cycle, as shown previously (Figure 4A; Liu and Morris, 2000). The apsB deletion (apsBΔ) and apsB6 alleles exhibit an identical phenotype of anucleate primary sterigmata resulting in failure of conidiation (Clutterbuck, 1994; Suelmann et al., 1998), and both were tested for their effects on GFP-SNAD localization. In strains carrying either apsB6 or ΔapsB, the GFP-SNAD fusion protein was still detected at the SPB (Figure 4, B and C), indicating that SNAD localization did not require functional ApsB. We further tested whether ApsB was required for the SPB localization of the SIN molecules SIDB. Again, the SIDB localization at the SPB and the septation site was not dependent on ApsB (Figure 4D).
Figure 4.
ApsB acts in a different pathway from SNAD. (A) GFP-SNAD localized to the SPB. (B) GFP-SNAD localization was not affected by the apsB6 mutation. (C) In the apsBΔ (null) background, GFP-SNAD still localized to the SPB. (D) GFP-SIDB localization to SPB (arrows) and septation sites (arrowheads) were not affected by the apsB6 mutation. (E) GFP-ApsB decorated SPBs (arrows) and septation sites (arrowheads) in control cells. (F) GFP-ApsB localization was not altered in the snaD290 mutation background. (G) The apsBΔ; snaDΔ double mutant formed a much smaller colony than the single mutants. Scale bar, 5 μm.
Conversely, we tested whether SNAD was required for ApsB localization. The localization of GFP-ApsB was indistinguishable in the presence or absence of SNAD (Figure 4, E and F). This result suggested that ApsB and SNAD might act in different pathways in spite of the similarities of their localization patterns and mutant phenotypes. To test this possibility, double null mutant of apsB; snaD was created. The apsBΔ; snaDΔ double null mutant failed to conidiate as did the single null mutants. In addition, the double null mutant formed colonies of significantly smaller size than that of wild-type and null mutants, indicating an additive phenotype in colony growth (Figure 4G). Thus, despite overlapping ApsB and SNAD localizations and similar genetic interactions of snaD-nud and apsB-nud mutations, apsB and snaD, do not appear to share a function in regulating septation in A. nidulans.
Actomyosin Ring Dynamics in the sepK and snaD Mutant Background
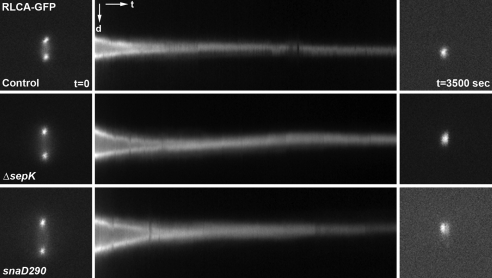
Because snaD mutations delay septation during vegetative growth, we wanted to test whether septation took place at a slower rate than wild-type cells. As revealed in published studies in fission yeast, the actomyosin contractile ring can be marked by tagging the regulatory light chain of type II myosins known as Rlc1p (Le Goff et al., 2000). The A. nidulans homolog of Rlc1p, RLCA (for regulatory light chain A) is encoded by the AN0267 locus based on sequence homology. Indeed an RLCA-GFP fusion protein expressed under the control of its native promoter tracked the contraction of the actomyosin ring during septation (control cell in Figure 5A, note that the Kymograph in Figure 5A depicts the contraction process). In the sepK null and snaD290 mutation backgrounds, the actomyosin rings labeled by RLCA-GFP contracted similarly to that in control cells (Figure 5A). Therefore, we concluded that sepK and snaD do not play a regulatory role in actomyosin ring contraction. They are probably only required for initiating the assembly of the actomyosin ring in a timely manner.
Figure 5.
Actomyosin ring contraction in sepK and snaD mutants. RLCA-GFP was expressed in a control strain and strains carrying the sepKΔ or snaD290 mutation. Time-lapse images were taken at 10-s intervals. For each strain, 350 images starting at the beginning of ring contraction were used to generate kymographs shown in the middle column. Kymograph images have the orientation with distance (d) and time (t) illustrated. Images at t = 0 and t = 3500 s are shown in the left and right columns, respectively.
A previous study indicated that the sepH1 allele abolished actomyosin ring assembly when the mutant was grown at a restrictive temperatures (Bruno et al., 2001). However, in the fission yeast the SIN pathway is only required for ring contraction but not for ring assembly (Krapp et al., 2004). We wanted to further test whether the SIN kinase cascade was required for the assembly of the actomyosin ring using a null mutation. Using a strain (CZA28) in which RLCA-GFP was expressed in a mobA null mutation background, we attempted to observe an actomyosin ring-like signal. We did not detect a defined localization site of RLCA-GFP, suggesting that an actomyosin ring failed to form.
The snaD Mutation Can Be Suppressed by Down-Regulation of a Two-Component GAP
In fission yeast, Cdc16p and Byr4p function as a two-component GAP (GTPase-activating protein) that negatively regulates the initiation of septation, and deletion of either component hyperactivates septation, similar to the overexpression of SIN components (Song et al., 1996; Furge et al., 1998; Jwa and Song, 1998). The AN0281 and AN9413 loci encode proteins homologous to Cdc16p and Byr4p, respectively (Table 2). Thus, we named them bubA and byrA, respectively. Strains were created in which only functional copy of bubA or byrA gene was under the control of the inducible/repressible alcA promoter (alcA(p)::GFP-bubA or alcA(p)::mCherry-byrA; Supplementary Figure S1), as described previously (Kim et al., 2006). When the expression of bubA, byrA, or both genes was turned off by growth on medium containing glucose, surprisingly, no noticeable growth phenotype was observed (Figure 6A). The lack of growth phenotypes was further confirmed by null mutations (T. Nayak and B. Oakley, unpublished data). This result was rather surprising because in fission yeast cdc16 and byr4 are essential genes. Thus, we were prompted to test whether the two homologous proteins negatively regulate the SIN pathway. When the expression of either GFP-bubA or mCherry-byrA was elevated by growth on medium containing ethanol, colony growth and conidiation were similar to those of the control strain (Figure 6A). When both GFP-bubA and mCherry-byrA were expressed at elevated levels, however, conidiation was almost completely abolished (Figure 6A). We wondered whether the conidiation phenotype was linked to possible septation defects. Under repressive conditions using glucose as the carbon source, hyphae of the double mutant formed ample septa as revealed by Calcofluor white labeling (Figure 6B). On induction of overexpression of both GFP-bubA and mCherry-byrA induced by ethanol, the double mutant did not form a single septum among thousands of 16-h germlings examined (Figure 6B). Typically, germlings form the first septum 7–8 h after conidial germination (Liu and Morris, 2000). Therefore, we concluded that BUBA and BYRA most likely function as negative regulators of septation in A. nidulans, as is the case in fission yeast. In the double mutant, the failure of conidiation was a direct consequence of the abolition of septation due to overexpression of both BUBA and BYRA. The only surprise was that the overexpression of both proteins was required to abolish septation, which was different from findings in fission yeast.
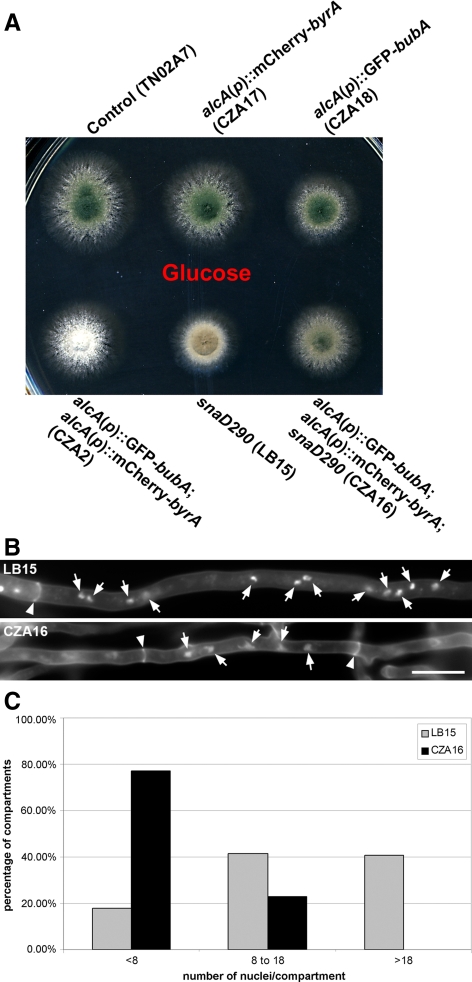
Figure 6.
Regulation of septation by the BUBA and BYRA proteins. (A) Colony phenotypes of the control strain, alcA(p):: GFP-bubA and alcA(p)::mCherry-byrA single mutants, and the double mutant under repressing conditions (with glucose) and inducible conditions (with ethanol). All strains conidiated identically on the glucose medium. On the ethanol medium, only the alcA(p)::GFP-bubA; alcA(p)::mCherry-byrA double mutant exhibited a phenotype of nonconidiation. (B) The alcA(p)::GFP-bubA; alcA(p)::mCherry-byrA double mutant formed septa separating either one or two nuclei when both genes were turned off. No septum was detected when the strain was grown under inducing conditions. Septa were detected in the control (TN02A7) hyphae bordering one or a few nuclei under both growth conditions. Nuclei were highlighted by arrows, and septa by arrowheads. Scale bar, 5 μm. (C) Quantitative assessment of septum formation after conidial germination. Germlings were scored for having 0, 1, 2, or 3 septa in the control (R21) and the alcA(p)::GFP-bubA; alcA(p)::mCherry-byrA double mutant (CZA2) grown on media containing glucose.
We further assessed whether down-regulation of bubA and byrA expression would affect the timing of septation after conidial germination. Germlings typically form the first septum after three rounds of mitosis to render a five-nucleus apical cell and a three-nucleus basal cell (Liu and Morris, 2000; Harris, 2001; Kim et al., 2006). After grown for 6 h, 76.7% of the control germlings had not septated and in average had nine nuclei/germling (n = 236; Figure 6C). When both bubA and byrA were down-regulated, however, 74.7% of the germlings have already septated once and had in average eight nuclei/germling (n = 392; Figure 6C). Surprisingly, 4.3% of the mutant germlings had two septa already, which were not observed in any control germlings. This result supported the notion that down-regulation of bubA and byrA expression did not affect mitotic progression, but accelerate the execution of septation in growing germlings.
Because snaD and sepK mutations abolished conidiation by delaying septation, we tested whether down-regulation of bubA and byrA expression would suppress the conidiation phenotype. In the snaD290 mutation background, conidiation was restored to a significant level when the expressions of both bubA and byrA were turned off (Figure 7A). The suppression of a sepK mutation was also observed when byrA and bubA expression was repressed although not to the same level observed with the snaD290 mutation. The suppression of the nonconidiation phenotype caused by the snaD290 mutation was only brought about by the down-regulation of both bubA and byrA expressions, not by down-regulation of each gene. Because down-regulation of bubA and byrA expression accelerated septation timing, we further tested whether the down-regulation suppressed the septation delay brought about by the snaD mutation. To do so, we examined the number of nuclei in hyphal compartments/cells between two septa in the snaD/bubA/byrA triple mutant compared with snaD single mutant. Septa were detected in hyphae grown at 42°C for 13 h. The numbers of nuclei in hyphal compartments were categorized into three groups: <8, 8–18, and >18. In the snaD290 mutant, only 17.7% of the compartments contained <8 nuclei, and 41.6% and 40.7% of them had 8–18 and >18 nuclei, respectively (n = 113; Figure 7, B and C). In the triple mutant, however, 77.1% of the compartments contained <8 and the rest 22.9% had 8–18 nuclei, respectively (n = 179; Figure 7, B and C). Thus, we concluded that activation of septation by down-regulation of both BUBA and BYRA could suppress the negative impacts of septation and conidiation due to the loss of either SNAD or SEPK. This result further strengthened the notion that SNAD and SEPK functioned in anchoring the SIN components to the SPB, to trigger the onset of septation in a timely manner. Nevertheless, both SNAD and SEPK are not essential for septation in A. nidulans.
Figure 7.
Suppression of a snaD mutation by down-regulation of both bubA and byrA expressions. (A) When both bubA and byrA genes were down-regulated, the conidiation phenotype caused by the snaD290 mutation was suppressed. Although colonies carrying the snaD290 mutation did not form conidia so that they were brown, the triple mutant produced conidia causing the colony color to be green. (B) Staining of nuclei and septa in the snaD290 single mutant (LB15) and the snaD290; alcA(p)::GFP-bubA; alcA(p)::mCherry-byrA triple mutant (CZA16) grown on media containing glucose. Nuclei were highlighted by arrows, and septa by arrowheads. Scale bar, 10 μm. (C) Quantitative assessment of the number of nuclei in hyphal compartments in the snaD290 single mutant (LB15) and the snaD290; alcA(p)::GFP-bubA; alcA(p)::mCherry-byrA triple mutant (CZA16) grown on media containing glucose. The compartments were grouped together for having <8 nuclei, 8–18 nuclei, or >18 nuclei.
DISCUSSION
In this study, we report that the coiled-coil protein SNAD acts as a scaffold protein that plays a role in anchoring components of the SIN pathway to the SPB. SNAD is also required for the localization of another scaffold protein SEPK. Our data indicate that the localization of SIN components to the SPB is critical for timely execution of septation, but not essential for the event to take place during vegetative growth. Our results also support the notion that temporal regulation of septation is essential for conidiation.
Localization of SIN Proteins to the SPB
It has been well established that proteins of the SIN kinase cascade require the scaffold protein Cdc11p for their localization to the SPB in fission yeast (Krapp et al., 2001; Tomlin et al., 2002). The budding yeast homolog Nud1p also plays a critical role in directing MEN (mitotic exit network, analogous to the SIN) proteins to the SPB, and such a role is mediated by Spc72p, which interacts with the γ-tubulin complex (Gruneberg et al., 2000). Cdc11p and the A. nidulans homolog, SEPK, both have leucine-rich C-termini, and they share 35% sequence identity. Not surprisingly, SEPK is also required for spindle pole localization of SIN proteins. Unlike cdc11 or nud1, however, the sepK gene is not essential.
It has been shown that the leucine-rich repeats are required for Cdc11p to localize to the SPB through its interaction with the other scaffold protein Sid4p (Tomlin et al., 2002). Based on sequence homology, no obvious homolog of Sid4p is encoded by the A. nidulans genome. Because of our results on the requirement of SNAD for the localization of SEPK and SIN proteins at the SPB, we wonder if SNAD might be an analogue of Sid4p. This notion is supported by the structural features of Sid4p and SNAD. Both proteins are predicted to form extensive coiled-coil domains and reside at the SPB in a cell cycle–independent manner. No Sid4p homolog has been identified in budding yeast either, so functional conservation may not be reflected in amino acid sequence conservation. It would be interesting to find out whether SEPK directly interacts with SNAD as Cdc11p and Sid4p. Although it plays a critical role in temporal regulation of septation, snaD, like sepK, is not an essential gene.
Recently, in fission yeast Ppc89p was shown to be the linker between Sid4 and the core of the SPB (Rosenberg et al., 2006). Again, no Ppc89p homolog could be identified based on sequence homology when the A. nidulans genome was searched. Taken together, we would like to suggest that the difference beyond SEPK/Cdc11p between A. nidulans and fission yeast could arise from the structural difference between the spindle pole bodies in the two organisms.
Septation in the Absence of SNAD or SEPK
In budding and fission yeasts, SPB localization of MEN and SIN proteins is essential for septation and ultimately for mitotic exit and cell viability (Wolfe and Gould, 2005). Despite their critical role in temporal regulation of septation in A. nidulans, SEPK and SNAD are not essential for septation. Using laser ablation tools, the SPB has been shown to be essential for cytokinesis in fission yeast (Magidson et al., 2006). This result supports the notion that Sid2-Mob1p might need to be activated at the SPB before being translocated to the cell division site. Apparently, such a requirement is not absolutely needed in A. nidulans and probably in other filamentous ascomycetes as well. In the absence of either SEPK or SNAD, the efficiency of activation is discounted, which leads to a delay in septation. Nevertheless, it is possible that SIDB-MOBA can be activated in the cytosol before decorating the contractile ring. The fission yeast polo kinase Plo1p can induce septum formation in G1 and G2 phases upon overexpression (Ohkura et al., 1995). It would be tempting to suggest that the A. nidulans polo kinase PLKA might activate the SIN in the cytosol. However, the fact that overexpression of PLKA inhibits septation tends to rule out such a possibility in A. nidulans (Bachewich et al., 2005). It remains to be characterized how SIDB-MOBA can be activated in the cytoplasm and translocated to the septation site.
Earlier studies have indicated that septation is absolutely required for conidiation in A. nidulans (Harris et al., 1994). Our studies show that simply delaying septation significantly inhibits if not abolishes conidiation although it does not seriously affect vegetative hyphal growth as shown in the sepK and snaD mutants. Conidia contain single nuclei in A. nidulans (Adams et al., 1998). Temporal deregulation of septation would have much more impact on conidiation than on hyphal growth. Surprisingly, delay of septation does not result in multinucleate conidia. This fact suggests to us that temporal deregulation of septation might trigger an inhibitory signal to turn off conidiation. Alternatively, the deregulation might prevent proper activation of conidiation due to, for example, abnormal spatial distribution and/or accumulation of developmental regulators caused by the delay.
Although septation is not always coupled with mitosis during vegetative growth in A. nidulans, they are most likely coupled during conidiation as the spores are uninucleate. We suggest that tight coupling of the two events is dependent on SPB localization of the SIN components. Because of failed SIN localization to the SPB in the snaD and sepK mutants, inability to execute septation on time after mitosis may eventually cause the abortion of conidiation.
Nonoverlapping Roles of BUBA and BYRA
Our results showed that overexpression of either BUBA or BYRA did not show an obvious phenotype, but simultaneous overexpression of both proteins abolished septation and conidiation. The results were distinct from those in fission yeast in which Byr4p acts as a scaffold protein for mediating the interaction between Spg1p and its GAP catalytic subunit Cdc16p (Song et al., 1996; Furge et al., 1998). The results in fission yeast indicate that compromising the function of either protein would hyperactivate septation. Our results suggest that BUBA and BYRA negatively regulate their G-protein target SPGA by two different mechanisms. Although there is little if any doubt that BUBA acts as a GAP, BYRA might not be part of this GAP. This hypothesis is supported by recent findings on the budding yeast homologues of Cdc16p and Byr4p, Bub2, and Bfa1 (Ro et al., 2002; Fraschini et al., 2008). Although Bub2 and Bfa1 form a complex, their functions are not completely overlapping in terms of regulating mitotic exit (Ro et al., 2002; Fraschini et al., 2006). As a matter of fact, a recent report suggests that Bfa1 negatively regulates its target G-protein Tem1 by acting as a guanine-nucleotide dissociation inhibitor (Fraschini et al., 2008). In our experiments, we did not observe an mCherry-BYRA signal (even upon overexpression) at the SPB where GFP-BUBA was clearly detected (data not shown). Therefore, our results suggest that being negative regulators of SPGA, BUBA, and BYRA act independently at different sites.
Actomyosin Ring Contraction without an Active SIN
Earlier conclusions suggest that the primary function of the SIN pathway is also essential to trigger the contraction of the actomyosin ring during cytokinesis in yeasts (Krapp et al., 2004). In our experiments, when the mobA gene was deleted, we no longer observed the ring using the RLCA-GFP fusion as a marker. This finding echoed an earlier report showing that sepH1 mutant cells failed to form the ring at restrictive temperature (Bruno et al., 2001). More recently, it has been found that the SIN is also involved in actomyosin ring assembly in fission yeast (Hachet and Simanis, 2008; Huang et al., 2008). In yeast the SIN and the aniline-like protein Mid1p play partially redundant roles for ring assembly. Using the yeast Mid1p sequence, we did not detect an obvious homolog in A. nidulans. Hence, the absence of a Mid1p homolog would explain why the SIN becomes indispensible for regulating the assembly of the actomyosin ring in this filamentous fungus.
To date, substrate(s) of the SIN kinase cascade at the actomyosin ring have not been firmly identified or confirmed. It has been widely speculated that proteins like Etd1p might link the SIN to actomyosin ring contraction in fission yeast (Daga et al., 2005). Unfortunately, so far no Etd1p homologues have been identified in other fungi. Because the conclusion that the SIN is not required for ring assembly was made using temperature-sensitive mutations, it would be intriguing to know if an actomyosin ring could still be formed in a null SIN mutant in fission yeast. Our results suggest that putative SIN substrate(s) might lie more upstream than previously thought. They could play essential roles in assembling the actomyosin ring. Clearly, identification of SIN substrates would be a critical task in order to understand how the SIN pathway regulates the onset of septation in A. nidulans and other fungi.
Suppression of Cytoplasmic Dynein Mutations by snaD
The nud mutations in the cytoplasmic dynein pathway lead to the formation of anucleate apical cells after failure of nuclear migration in developing germlings (Liu and Morris, 2000). This results in significant inhibitions of hyphal extension and colony growth. snaD alleles partially suppress nud mutations (Goldman and Morris, 1995; Liu and Morris, 2000). After the phenotype of the snaD mutation was determined, it was suggested that the suppression of nudA and mutations in other genes of the cytoplasmic dynein pathway was due to the delay in septation (Liu and Morris, 2000). Because, based on current work, SNAD acts in the SIN pathway, suppression of the nudA mutation would be expected if septation were inhibited by down-regulation of SIN components. Indeed we tested this prediction by constructing a nudA1; alcA(p)::GFP-mobA strain, which would allow us to turn off the expression of the only mobA gene on rich medium containing glucose. At the restrictive temperature of 42°C, a strain carrying the nudA1 temperature-sensitive allele formed a tiny colony as previously reported (Xiang et al., 1994). The nudA1; alcA(p)::GFP-mobA double mutant formed a colony larger than the nudA1 single mutant (Supplementary Figure S2). Therefore, this result confirms our earlier hypothesis that down-regulating septation leads to suppression of nud.
SNAD and ApsB Are Involved in Different Pathways at the SPB
Because of the phenotypic similarities of snaD and apsB mutations, and of the fact that SNAD and ApsB are SPB proteins, we first suspected that snaD and apsB might act in a common pathway. Our results indicate, however, that this is not the case. Thus, these two SPB proteins are likely involved in two different pathways. Results presented here point to a critical role for SNAD as a scaffold protein to allow SIN components to localize to the SPB. ApsB seem to be directly involved in microtubule organization as apsB mutants cause a dramatic reduction of astral microtubules has been observed (Veith et al., 2005). In fact, the fission yeast ApsB homologue Mto1p associates with the γ-tubulin complex and is required for recruiting this complex to interphase microtubule-organizing centers and microtubules (Zimmerman and Chang, 2005). Thus, ApsB is involved in microtubule organization while SNAD plays a role in temporal regulation of septation. The suppression of nud mutations by snaD could be attributed to the effect of delay in septation so that anucleate apical cells are formed less frequently. The suppression of nud mutations by apsB could be directly linked to microtubule-dependent nuclear migration. It was reported that modification of microtubule dynamics by apsB mutations could improve nuclear migration when the primary motor dynein was absent (Veith et al., 2005). It is noteworthy that destabilization of microtubules by an α-tubulin mutation or by treatment with low doses of the antimicrotubule agent benomyl could also suppress nud mutations (Willins et al., 1995). Therefore, snaD and apsB mutations cause suppression of nud mutations, but through very different mechanisms.
In summary, our results suggest that the coiled-coil protein SNAD acts at the SPB and plays a role in anchoring the SIN components. When the SIN fails to localize to the SPB in the absence of SNAD, septation is delayed although not eliminated, and conidiation is completely abolished. Therefore, timely execution of septation is a prerequisite for conidiation in the filamentous ascomycete A. nidulans.
Supplementary Material
ACKNOWLEDGMENTS
We want to thank Ms. Elizabeth Oakley for generating the LO strains used in this study. We thank Dr. Roger Tsien (University of California, San Diego) for generously providing the mCherry plasmid and Dr. Reinhard Fischer (University of Karlsruhe, Germany) for providing apsB strains. This report is based on work supported by the National Science Foundation (NSF) Grant MCB-0615892 to B. L., and the National Institutes of Health (NIH) Grant GM31837 to R.B.O. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of NSF or NIH.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1177) on April 22, 2009.
REFERENCES
- Adams T. H., Wieser J. K., Yu J. H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C., Masker K., Osmani S. The polo-like kinase PLKA is required for initiation and progression through mitosis in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2005;55:572–587. doi: 10.1111/j.1365-2958.2004.04404.x. [DOI] [PubMed] [Google Scholar]
- Bruno K. S., Morrell J. L., Hamer J. E., Staiger C. J. SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol. Microbiol. 2001;42:3–12. doi: 10.1046/j.1365-2958.2001.02605.x. [DOI] [PubMed] [Google Scholar]
- Chang L., Gould K. L. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck A. J. Mutants of Aspergillus nidulans deficient in nuclear migration during hyphal growth and conidiation. Microbiology. 1994;140:1169–1174. doi: 10.1099/13500872-140-5-1169. [DOI] [PubMed] [Google Scholar]
- Daga R. R., Lahoz A., Munoz M. J., Moreno S., Jimenez J. Etd1p is a novel protein that links the SIN cascade with cytokinesis. EMBO J. 2005;24:2436–2446. doi: 10.1038/sj.emboj.7600705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Abalos J. M., Fox H., Pitt C., Wells B., Doonan J. H. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- Fraschini R., D'Ambrosio C., Venturetti M., Lucchini G., Piatti S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 2006;172:335–346. doi: 10.1083/jcb.200507162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Venturetti M., Chiroli E., Piatti S. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem. Soc. Trans. 2008;36:416–420. doi: 10.1042/BST0360416. [DOI] [PubMed] [Google Scholar]
- Furge K. A., Wong K., Armstrong J., Balasubramanian M., Albright C. F. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Goldman G. H., Morris N. R. Extragenic suppressors of a dynein mutation that blocks nuclear migration in Aspergillus nidulans. Genetics. 1995;139:1223–1232. doi: 10.1093/genetics/139.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Campbell K., Simpson C., Grindlay J., Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. D. Septum formation in Aspergillus nidulans. Current Opin. Microbiol. 2001;4:736–739. doi: 10.1016/s1369-5274(01)00276-4. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Morrell J. L., Hamer J. E. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics. 1994;136:517–532. doi: 10.1093/genetics/136.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yan H., Balasubramanian M. K. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J. Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jwa M., Song K. Byr4, a dosage-dependent regulator of cytokinesis in S-pombe, interacts with a possible small GTPase pathway including Spg1 and Cdc16. Mol. Cell. 1998;8:240–245. [PubMed] [Google Scholar]
- Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- Kim J. -M., Lu L., Shao R., Chin J., Liu B. Isolation of mutations that bypass the requirement of the septation initiation network for septum formation and conidiation in Aspergillus nidulans. Genetics. 2006;173:685–696. doi: 10.1534/genetics.105.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Gulli M. P., Simanis V. SIN and the art of splitting the fission yeast cell. Curr. Biol. 2004;14:R722–R730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Krapp A., Schmidt S., Cano E., Simanis V. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 2001;11:1559–1568. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- Le Goff X., Motegi F., Salimova E., Mabuchi I., Simanis V. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J. Cell Sci. 2000;113:4157–4163. doi: 10.1242/jcs.113.23.4157. [DOI] [PubMed] [Google Scholar]
- Liu B., Morris N. R. A spindle pole body-associated protein, SNAD, affects septation and conidiation in Aspergillus nidulans. Mol. Gen. Genet. 2000;263:375–387. doi: 10.1007/s004380051181. [DOI] [PubMed] [Google Scholar]
- Magidson V., Chang F., Khodjakov A. Regulation of cytokinesis by spindle-pole bodies. Nat. Cell Biol. 2006;8:891–893. doi: 10.1038/ncb1449. [DOI] [PubMed] [Google Scholar]
- Morrell J. L., Tomlin G. C., Rajagopalan S., Venkatram S., Feoktistova A. S., Tasto J. J., Mehta S., Jennings J. L., Link A., Balasubramanian M. K., et al. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 2004;14:579–584. doi: 10.1016/j.cub.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., Murray S. L., Hynes M. J., Osmani S. A., Oakley B. R. A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Hagan I. M., Glover D. M. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Ro H. S., Song S., Lee K. S. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:5436–5441. doi: 10.1073/pnas.062059999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J. A., Tomlin G. C., McDonald W. H., Snydsman B. E., Muller E. G., Yates J.R.R., Gould K. L. Ppc89 links multiple proteins, including the septation initiation network, to the core of the fission yeast spindle-pole body. Mol. Biol. Cell. 2006;17:3793–3805. doi: 10.1091/mbc.E06-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Simanis V. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- Song K. W., Mach K. E., Chen C. Y., Reynolds T., Albright C. F. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J. Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suelmann R., Sievers N., Galetzka D., Robertson L., Timberlake W. E., Fischer R. Increased nuclear traffic chaos in hyphae of Aspergillus nidulans: molecular characterization of apsB and in vivo observation of nuclear behaviour. Mol. Microbiol. 1998;30:831–842. doi: 10.1046/j.1365-2958.1998.01115.x. [DOI] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S. A., Oakley B. R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tomlin G. C., Morrell J. L., Gould K. L. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol. Biol. Cell. 2002;13:1203–1214. doi: 10.1091/mbc.01-09-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith D., Scherr N., Efimov V. P., Fischer R. Role of the spindle-pole-body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans. J. Cell Sci. 2005;118:3705–3716. doi: 10.1242/jcs.02501. [DOI] [PubMed] [Google Scholar]
- Willins D. A., Xiang X., Morris N. R. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics. 1995;141:1287–1298. doi: 10.1093/genetics/141.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Xiang X., Beckwith S. M., Morris N. R. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C.P.C., Dou X., Perez-Balaguer A., Osmani S. A. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S., Chang F. Effects of γ-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell. 2005;16:2719–2733. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.