Abstract
The cytoplasmic dynein motor complex is known to exist in multiple forms, but few specific functions have been assigned to individual subunits. A key limitation in the analysis of dynein in intact mammalian cells has been the reliance on gross perturbation of dynein function, e.g., inhibitory antibodies, depolymerization of the entire microtubule network, or the use of expression of dominant negative proteins that inhibit dynein indirectly. Here, we have used RNAi and automated image analysis to define roles for dynein subunits in distinct membrane-trafficking processes. Depletion of a specific subset of dynein subunits, notably LIC1 (DYNC1LI1) but not LIC2 (DYNC1LI2), recapitulates a direct block of ER export, revealing that dynein is required to maintain the steady-state composition of the Golgi, through ongoing ER-to-Golgi transport. Suppression of LIC2 but not of LIC1 results in a defect in recycling endosome distribution and cytokinesis. Biochemical analyses also define the role of each subunit in stabilization of the dynein complex; notably, suppression of DHC1 or IC2 results in concomitant loss of Tctex1. Our data demonstrate that LIC1 and LIC2 define distinct dynein complexes that function at the Golgi versus recycling endosomes, respectively, suggesting that functional populations of dynein mediate discrete intracellular trafficking pathways.
INTRODUCTION
Cytoplasmic dynein (Schroer et al., 1989) plays diverse roles in mammalian cells that include mitosis, membrane trafficking, organelle positioning, intraflagellar transport, and cell migration (Ross et al., 2008; Scholey, 2008). The two mammalian cytoplasmic dynein complexes (cytoplasmic dynein 1 and 2, referred to here as dynein-1 and dynein-2) are built around the two distinct heavy-chain subunits: dynein-1 heavy chain (DYNC1H1) and dynein-2 heavy chain (DYNC2H1). Note that in the first instance we use the common subunits names followed by the standardized nomenclature (Pfister et al., 2005). Considerable work has gone in to defining the subunit architecture of dynein complexes, notably dynein-1, and the data suggest the existence of complexes comprising of a homodimer of heavy chains with associated homodimeric or heterodimeric intermediate chains, homodimeric light intermediate chains, and multiple light chains (King et al., 2002). Light chains can form homo- and heterodimers (Nikulina et al., 2004) and are known to participate in many dynein-independent processes through direct interactions with other proteins (Barbar, 2008). Recent structural data show that at least one of these interactions, with the protein Swallow, is mutually exclusive with interaction of LC8 light chain 1 [hereafter referred to as LC8 (DYNLL1)] with dynein (Benison et al., 2007), calling in to question a potential role for LC8 as a dynein-cargo adaptor. Thus, it is difficult to directly assign function to light chains in the specific context of the dynein complex, especially when using reagents that disrupt dynein function in a general manner.
Roles have been defined for some individual dynein subunits based on their interactions with other proteins. For example, light IC 1 (LIC1, DYNC1LI1), but not LIC2 (DYNC1LI2), interacts with pericentrin (Tynan et al., 2000), and LIC1 directs removal of key proteins from the spindle assembly checkpoint to allow mitotic progression (Sivaram et al., 2009). LIC1 (DYNC1LI1) also interacts with Rab4A (Bielli et al., 2001), but it has not been determined whether Rab4A also interacts with LIC2 (DYNC1LI2). Tctex-1 (DYNLT1) but not rp3 (DYNLT3), interacts with rhodopsin and mediates its trafficking in rod photoreceptor cells (Tai et al., 1999). Such roles are indicative of the existence of subpopulations of dynein that function in discrete pathways. Little is known about the differential function of the two roadblock (DYNLRB) subunits except that they bind to a distinct site on dynein from other light chains (Susalka et al., 2002), have the capacity to both homo- and heterodimerize (Nikulina et al., 2004), and show overlapping but different expression patterns in tissues. Similarly discrete roles for different LC8 (DYNLL1) subunits remain undetermined (see Pfister et al., 2006), but again, differences in tissue distribution are likely to be important. Distinct subunit composition is likely to be particularly important in neurons where different dynein subpopulations mediate fast-versus-slow components of axonal transport (see Susalka and Pfister, 2000). More recently, the neuron-specific splice form of IC 1, IC1B, has been shown to specifically associate and move TrkB-containing endosomes (Ha et al., 2008). These findings hint at the existence of individual dynein complexes that direct specific membrane-trafficking events in cells.
Cytoplasmic dynein-1 is the major motor for minus-end–directed membrane trafficking in mammalian cells and is known to play a key role in the organization of the Golgi complex (Corthesy-Theulaz et al., 1992; Roghi and Allan, 1999) and in endosomal sorting (Lakadamyali et al., 2006; Driskell et al., 2007). The importance of dynein in these processes is often understated; evidence clearly implicates dynein in driving these processes (such as in endosomal sorting; Driskell et al., 2007) and not simply in enhancing their efficiency. Work in this area has relied heavily on the use of nocodazole (which results in depolymerization of the entire microtubule network and not a specific inhibition of dynein function) or on use of reagents to perturb dynein function globally (such as use of inhibitory antibodies such as monoclonal 70.1 directed against intermediate chains), or reagents that interfere with the function of the dynein accessory dynactin (Gill et al., 1991). Examples of the latter include expression of regions of the critical p150glued subunit (Quintyne et al., 1999) or overexpression of the structurally important p50dynamitin subunit (Presley et al., 1997). It is important to note that experiments using these reagents in fact disrupt dynactin and not necessarily dynein. The exact mechanism by which dynein and dynactin operate together remains equivocal, notably in terms of recruitment and maintaining association of dynein with membranes where the microtubule-binding capability of dynactin appears unnecessary (Haghnia et al., 2007; Kim et al., 2007). Results in such experiments are further complicated by the fact that dynactin can also bind to kinesin family motors, at least kinesin-2 (Deacon et al., 2003) and so can operate in plus-end–directed transport events. Thus, reagents that disrupt dynactin do not necessarily specifically perturb dynein-mediated processes.
Cytoplasmic dynein-2 has been reported to localize to the Golgi apparatus in cultured cells and primarily functions in the assembly and function of cilia and flagella (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999; Mikami et al., 2002). The dynein-2 heavy chain (DYNC2H1) binds to a unique dynein-2 LIC [also known as D2LIC or LIC3, here termed D2LIC/LIC3 (DYNC2LI1); Grissom et al., 2002], which also localizes to the Golgi apparatus. Together, these data suggest that dynein-2 could function in the organization of, or transport through, the Golgi apparatus in addition to its function in cilia.
The precise function of the multiple possible forms of dynein that could exist within cells, especially at the level of individual components, remains undefined. We sought to examine the roles of each dynein subunit in well-characterized membrane-trafficking events using siRNA suppression of subunit expression. These data assign specific intracellular functions to defined dynein-1 complexes acting at the Golgi, endosomes, and provide insight into the key dynein components that are involved in mitosis and cytokinesis.
MATERIALS AND METHODS
All reagents were purchased from Sigma-Aldrich (Poole, United Kingdom) unless stated otherwise.
Cell Culture
HeLa (ATTC CRL-2) and HeLa cells stably expressing GRASP65-green fluorescent protein (GFP; Lane et al., 2002) were maintained in DMEM supplemented with 10% fetal calf serum. For imaging, cells were grown on 3.5-cm glass-bottom dishes (MatTek, Ashland, MA), glass-bottom 96-well plates (MatTek), or eight-well chamber slides (LabTek, Fisher Scientific, Loughborough, United Kingdom) all with no. 1.5 coverslips (tolerance: 0.16–0.19 mm thick; all measured as typically 0.17 mm).
Antibodies
Primary antibodies against the following antigens were used in this study: horseradish peroxidase (HRP)-conjugated anti-GAPDH (Abcam, Cambridge, United Kingdom); lamin A/C (Cell Signaling Technology, Danvers, MA); dynein heavy chain 1 (R-325, Santa Cruz Biotechnology, Santa Cruz, CA); dynein heavy chain 2 (Mikami et al., 2002), kindly provided by Richard Vallee (Columbia University, NY); Tctex1 (DYNLT1) (a kind gift from Viki Allan, University of Manchester); IC2 (DYNC1I2) (clone 70.1, Millipore, Watford, United Kingdom); LC8 (DYNLL1) (Pin1, BD Biosciences, Oxford, United Kingdom); LIC1 (DYNC1LI1) and LIC2 (DYNC1LI2) were generously provided by Richard Vallee (Tynan et al., 2000; GalT (CellMab, Göteberg, Sweden), β′-COP (Stephens laboratory, raised in rabbit against the following peptide sequence, KTDINLDEDILDD); ERGIC-53 (ER-Golgi intermediate compartment [ERGIC]; clone G1/93, Alexis Biochemicals, Nottingham, United Kingdom); Sec24C (Stephens laboratory; Townley et al., 2008), α-tubulin (clone DM1A, Sigma-Aldrich); GRASP65 (a generous gift from Jon Lane, University of Bristol; Chiu et al., 2008); phospho-histone H3 (Ser-10; Cell Signaling Technologies); and giantin (Covance, Berkeley, CA).
Expression Profiling
Cells were lysed and RNA extracted using Trizol (Invitrogen, Carlsbad, CA), cDNA was synthesized using Omniscript reverse transcription kit (Qiagen, Crawley, United Kingdom), and PCR was performed using Expand High Fidelity PCR kit (Roche, Burgess Hill, United Kingdom). Primer sequences used for RT-PCR were designed to amplify ∼500 base pairs toward the 3′ end of each transcript. Sequences can be made available on request. Quantitative PCR (qPCR) was performed as follows. RNA was isolated from cells using the TRIzol extraction method (Invitrogen). 50 μg RNA was used for reverse transcription using Omniscript reverse transcriptase (Qiagen) for 60 min at 37°C. Newly synthesized cDNA was then used for real-time qPCR using the DyNAmo SYBR green qPCR kit (New England Biolabs, Ipswich, MA). The following primers were used (designed using Primer3 software; Rozen and Skaletsky, 2000; available at http://primer3.wiki.sourceforge.net): DYNLRB1: fwd, 5′-AGGGAATCATCGTCGTGAAC-3′ and rev, 5′-GAGTGGCTTATTCGGTTGGA-3′; DYNC2H1a: fwd, 5′-CGACTTCCAAGTGCTGTCAA-3′ and rev, 5′-TGCATCACCAATTTCTTCCA-3′; DYNC2H1b: fwd, 5′-TTGGACTTCCTAGGGGGACT-3′ and rev, 5′-CTCCAACTCCCCAAAGATCA-3′; and DYNC2LI1: fwd, 5′-ACGATGCCCAGTGAAACTCT-3′ and rev, 5′-TGTGCCCTTTTGCTCTTCTT-3′. Each sample was run in triplicate using the target primers, together with primers against RNA polymerase II (fwd, 5′-GCACCACGTCCAATGACAT-3′ and rev, 5′-GTGCGGCTGCTTCCATAA-3′), which acted as the control. Amplification was performed and detected using an Opticon2 cycler (Bio-Rad, Hercules, CA), and data were analyzed using the comparative Ct method, which utilizes the formula 2(−ΔΔCT), where Ct is regarded as the threshold cycle). The amount of target relative to lamin A/C–suppressed samples and normalized to RNA polymerase II was calculated.
siRNA Transfection and Rescue
Duplexes were designed to target all known splice variants of each subunit by searching the NCBI database (October 2005). Duplexes were then designed using the online tool of Eurofins MWG Operon (Ebersberg, Germany) and synthesized with dTdT overhangs using the sequences shown in Supplementary Table S1. Duplexes were transfected using calcium phosphate as described previously (Watson and Stephens, 2006). In all cases, cells were used for experiments 72 h after transfection. For rescue experiments, cells were again transfected 48 h after small interfering RNA (siRNA) transfection with plasmids encoding either mouse LIC1 or mouse LIC2. cDNAs encoding the mLIC1 (IMAGE clone 5040044) and mLIC2 (IMAGE clone 6405596) were obtained from Geneservice (Cambridge, United Kingdom). mLIC1 was expression ready in vector pCMV-SPORT6, and mLIC2 was subcloned directly from the IMAGE clone in to pcDNA3.1+ using EcoRI-NotI restriction sites. Transfected cells were identified by immunolabeling of fixed cells 18–24 h after transfection.
Sucrose Density Gradient Centrifugation and Immunoblotting
HeLa cells in which dynein subunits had been suppressed were separated on 5–40% sucrose gradients as previously described (Watson et al., 2005) from which 1-ml fractions were taken. For detection of dynein heavy chain, 0.5 ml of each gradient fraction was concentrated using TCA precipitation (16% trichloroacetic acid, 60 min at 4°C followed by centrifugation at 20,000 × g for 15 min), resuspended in 20 μl of SDS-PAGE sample buffer before separating on 4% SDS-PAGE gels, and transfer to nitrocellulose. For other samples from the gradient, 15 ml of each fraction was analyzed directly by SDS-PAGE using precast polyacrylamide gels (Novex Bis-Tris or Tris-acetate, Invitrogen, Paisley, United Kingdom). For all other immunoblots, cells were lysed for immunoblotting, and samples were separated by SDS-PAGE followed by transfer to nitrocellulose membranes; primary antibodies were detected using HRP-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) and enhanced chemiluminescence (ECL, GE Healthcare, Cardiff, United Kingdom).
Imaging
In all cases, cells were imaged using an Olympus IX-series microscope (Olympus, Watford, United Kingdom) with either a UPLSAPO 40× NA 0.9 lens (all images in except those in Figures 2C and 6), a UIS2 PLAPON 60× 1.42 NA oil immersion lens (see Figure 2C), or a water immersion UIS2 UPLSAPO 60× NA 1.2 lens (Figure 6). An ASI PZ-2000 X-Y scanning stage was used for imaging multiple random points within wells of each 96-well plate; the integral Piezo z-plate of this stage was used for all acquisition in z. Illumination was provided by a Sutter DG-4 (Sutter, Novato, CA) using a BrightLine DA/FI/TR/Cy5-4X-A Quad-band “Pinkel” filter set from Semrock (Rochester, NY). Images were captured to a Hamamatsu Orca-ER camera (Bridgewater, NJ) at 12-bit depth. The system was controlled using Volocity 4 (Improvision, Coventry, United Kingdom). Image planes in z-series were separated by 1 μm. Images of fixed cells were acquired at room temperature (∼20°C), and live cells were imaged at 37°C using a heated Perspex chamber enclosing the microscope (Solent Scientific, Portsmouth, United Kingdom). Living cells were loaded with AlexaFluor-568-transferrin (Invitrogen) for 60 min to provide steady-state labeling of early endocytic compartments, minimizing effects of the time taken for image acquisition. Live cells were imaged in DMEM without Phenol Red supplemented with 0.1 g.l−1 sodium carbonate and 30 mM HEPES, pH 7.4. Fixed cells on 96-well plates and 35-mm glass-bottom dishes were imaged in phosphate buffered saline, pH 7.4. Representative images are shown, all experiments were repeated independently at least three times each, and all findings replicated with cells grown on 22-mm coverslips (Fisher Scientific) or 35-mm live cell dishes (MatTek). Cells for experiments shown in Figures 10 and 11 were processed on 0.17-mm-thick coverslips and imaged on a wide-field imaging system (Olympus IX-71 microscope with Exfo X-cite 120Q lamp, excitation and emission filter wheel; Ludl, Hawthorne, NY) containing a DA/FI/TR/Cy5-4 × 4M-B “Sedat” filer set (Semrock) with image capture on a CoolSnap HQ2 CCD (Photometrics, Tucson, AZ).
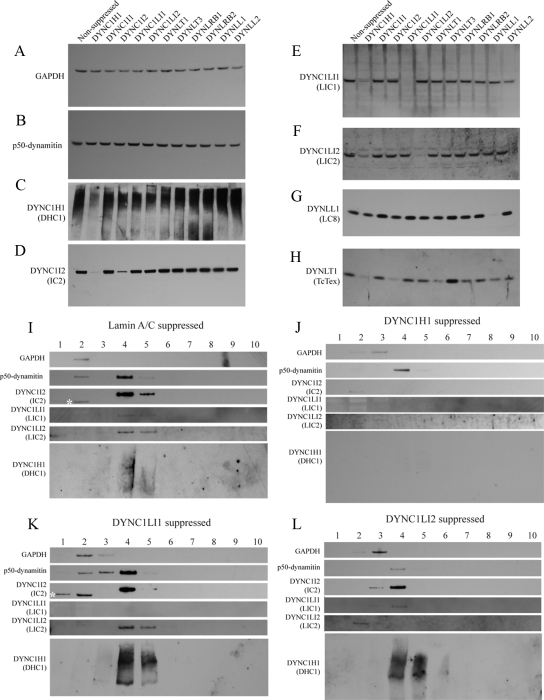
Figure 2.
Effects of dynein subunit suppression on dynein integrity. (A–H) Immunoblots show the expression of (A) GAPDH, (B) p50dynamitin, (C) DHC1, (D) IC2, (E) LIC1, (F) LIC2, (G) LC8, and (H) Tctex in lysates from HeLa cells transfected with siRNA duplexes directed against target proteins as indicated. (I–L) Fractions from 5 to 40% sucrose density gradients from cells depleted of (I) lamin A/C, (J) DHC1, (K) LIC1, or (L) LIC2 were immunoblotted for GAPDH, p50dynamitin, DHC1, IC2, LIC1, or LIC2 as indicated. Fraction 4 corresponds to 19S as judged by thyroglobulin sedimentation.
Figure 6.
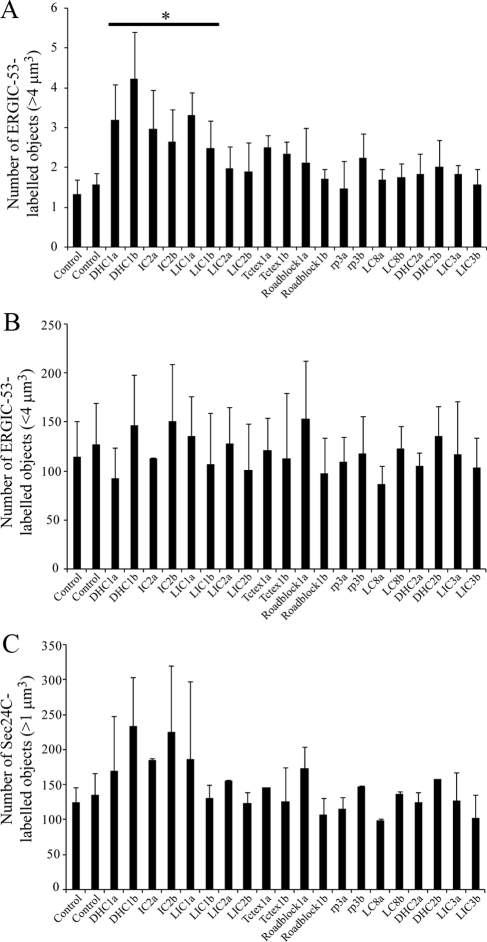
Quantification of disruption of ERGIC structures and ERES. Cells depleted of individual dynein subunits using one of two different siRNA duplexes (denoted a and b) were analyzed by automated object analysis as described in Materials and Methods. (A) Histogram showing total number of ERGIC-53–positive structures <4 μm3. (B) Histogram showing the total number of ERGIC-53–positive structures >4 μm3. (C) Histogram showing the total number of Sec24C-labeled objects >1 μm3. Error bars, SD; statistical significance, *p < 0.05.
Figure 10.
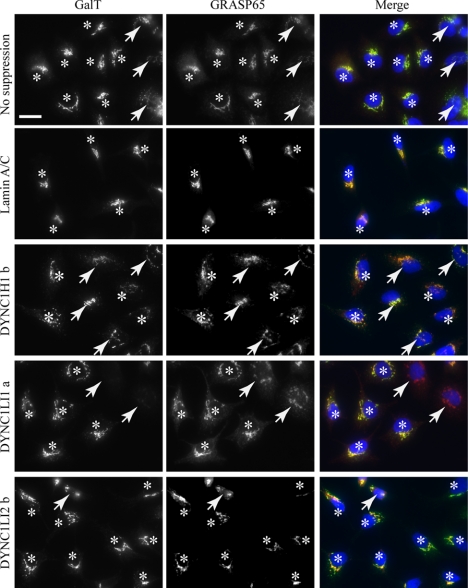
Rescue of LIC1 suppression by mLIC1 expression. Cells were suppressed for LIC1 followed by transfection with a plasmid encoding mLIC1. Asterisks indicate expressing cells (as adjudged by LIC1 immunofluorescence), and arrows indicate cells that were not detectably expressing mLIC1 above background. Note that suppression of DHC1 causes Golgi fragmentation that is not rescued by mLIC1 suppression. Golgi fragmentation in LIC1 suppressed cells is effectively rescued by expression of mLIC1. Bar (all panels), 20 μm.
Figure 11.
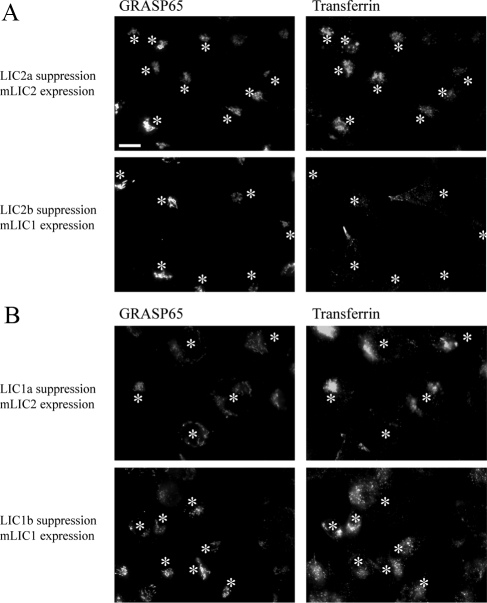
Rescue of LIC2 suppression by mLIC2 expression. Cells were suppressed by siRNA transfection and subsequently with plasmids encoding either mLIC1 or mLIC2 as indicated. After incubation with AlexaFluor-568-transferrin for 60 min, cells were extensively washed, fixed, and immunolabeled to detect GRASP65 and mLICs. Asterisks indicate mLIC-expressing cells. (A) The peripheral distribution of transferrin endosomes in LIC2 suppressed cells is rescued by mLIC2 expression but not by mLIC1 expression. (B) Golgi fragmentation caused by LIC1 suppression is rescued by mLIC1 but not mLIC2 expression. Bar (all panels), 20 μm.
Image Deconvolution and Analysis
Image stacks were deconvolved using the iterative deconvolution algorithm in Volocity 4.2 with 25 iterations and 95% confidence limits. The only other processing applied was adjustment of brightness and contrast, which was applied to whole extended focus images only. Deconvolved images were exported as extended focus TIFF files, and montages were produced in Adobe Illustrator CS3 (Adobe Systems, San Jose, CA). RGB and magenta green merges are shown; the latter was produced by copying all information from the red channel and pasting into the blue channel.
For image quantification, >200 cells were scored in total from three independent experiments (typically four fields of view from each well of a 96-well plate). Images were acquired as z-series covering the entire depth of each cell to ensure inclusion of all objects regardless of image plane. Automated analysis of object number was achieved using Volocity 4.2 (Improvision) by setting thresholds to detect objects with intensities >3 SDs above the mean, excluding objects <1 μm3, and counting the object number (for the Golgi, ERGIC, and endoplasmic reticulum exit sites [ERES]). Counts were normalized to the number of cells measured by counting the number of DAPI-labeled nuclei using the same thresholding method. ERGIC-53–positive objects >4 μm3 were counted without intensity thresholding. The thresholding protocol was established on three selected images from each data set, saved, and then applied to the entire data set automatically. Visual inspection of random images with thresholding applied was used to confirm the appropriateness of the method. Intensities of Sec24C-positive structures were measured following thresholding to include only those objects with >50% total intensity. Transferrin-positive structures in the periphery of cells were identified by manually drawing “donut” shapes around cells to define the periphery of cells only and to exclude the perinuclear region of cells and to count the area of fluorescence in 2D projections. The requirement to manually draw regions of interest for Sec24C intensity measurements and transferrin area intensity preclude fully automated image analysis. Data were exported to Microsoft Excel (Redmond, WA) and the SD was determined. Statistical significance was tested using the unpaired t test function of Excel (Microsoft) assuming nonequal variances. Differences were defined as discernable where p < 0.05. Error bars on all histograms show SDs.
RESULTS
Expression Profiling of Cytoplasmic Dynein Subunits and Efficacy of Gene Depletion
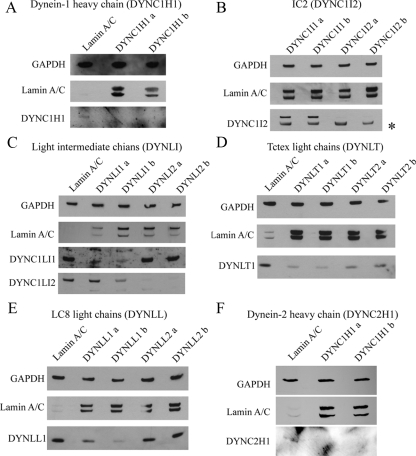
For this study, we chose to use HeLa cells owing to their widespread use and the efficacy of siRNA transfection and subsequent gene suppression that we have been able to obtain with these cells (Watson and Stephens, 2006). Importantly, use of siRNA depletion (i.e., a knockdown not a knockout) allows us to examine roles of dynein subunits even where they might be essential. We first established which subunits of dynein are expressed in HeLa and other cell lines (human telomerase immortalized retinal pigment epithelial cells [hTERT-RPE1], human dermal fibroblasts, and HepG2) using immunoblotting (Figure 1), RT-PCR (Supplementary Figure S1), and/or qPCR. IC 1 (DYNC1I1) is not widely expressed, and indeed we could not detect its expression in HeLa or other cells examined. In addition, we could not detect expression of Roadblock 2 (DYNLRB2) or LC8 light chain 2 (DYNLL2) in any cell type analyzed (Supplementary Figure S1). In addition to this, we validated the efficacy of our siRNA duplexes using immunoblotting. For each target, two independent siRNAs were used that were designed to target all splice variants of each gene; lamin A/C suppression was used as an RNA interference (RNAi) control, with GAPDH used as a loading control. Figure 1 shows that suppression of each target was achieved. There are several points to note from these data. Figure 1A shows effective suppression of dynein-1 heavy chain (DYNC1H1) expression by immunoblotting. Our batch of antibody IC74.1 directed against IC 2 [IC2 (DYNC1I2)] detects a nonspecific band (Figure 1B, marked with an asterisk; see Traer et al., 2007 for further validation of IC2 suppression). Here, we show that IC2 (DYNC1I2) is efficiently suppressed and not targeted by duplexes designed to be specific for IC1 (DYNC1I1), confirming duplex specificity. Both light intermediate chains can be efficiently and selectively suppressed (Figure 1C). The antibody to Tctex1 (DYNLT1) used here appears to detect both Tctex1 (DYNLT1) and rp3 (DYNLT3) because we see a partial suppression of expression of both isoforms (Figure 1D). Only DYNLL1 is expressed in our HeLa cells, but this can also be efficiently suppressed (Figure 1E). Dynein-2 heavy chain (DYNC2H1) is also effectively suppressed in HeLa cells as shown by Figure 1F. A lack of available antibodies with specificity for dynein-2 hampered further analysis here, but clear suppression in hTERT-RPE1 cells was also evident (not shown). Where suitable antibodies were not available, we used qPCR to monitor target suppression. This showed that in HeLa cells, Roadblock 1 (DYNLRB1) and D2LIC/LIC3 (DYNC2LI1) were suppressed to a maximum of 68% (DYNLRB1a) and 81% (DYNC2H1b), respectively. In hTERT-RPE1 cells Roadblock 1 (DYNLRB1) suppression was highly effective (92 and 96% average depletion with DYNLRB1a and DYNLRB1b, respectively); D2LIC/LIC3 (DYNC2LI1) depletion was also effective, showing 61% suppression with DYNC2LI1a and 52% suppression with DYNC2LI1b. All results obtained using HeLa cells were validated using hTERT-RPE1 cells.
Figure 1.
Suppression of dynein subunit expression using siRNA. Immunoblots show control suppression of lamin A/C from the same experiment, GAPDH as a loading control and the results of transfection with two independent duplexes targeting (A) dynein-1 heavy chain (DYNC1H1), (B) IC2 (DYNC1I2), (C) LIC1 (DYNC1LI1) and LIC2 (DYNC1LI2), (D) Tctex1 (DYNLT1) and rp3 (DYNLT3), (E) LC8 (DYNLL1), and (F) dynein-2 heavy chain (DYNC2H1). Inclusion of duplexes targeting isoforms that are not expressed in these cells provides an additional control and reinforces the specificity of our knockdowns.
Subsequent experiments used indirect immunofluorescence and live cell imaging to define the effects of depleting dynein subunits on cellular function. Cells were grown and transfected in either eight-well chamber slides or 96-well plates, and images were acquired using an XYZ scanning stage set up to image randomly selected areas within each well with a 40× objective lens. This approach provides an unbiased analysis of phenotype and includes multiple cells in each field of view. In all cases, high-resolution images were obtained using glass-bottomed dishes or coverslips with high-magnification oil immersion lenses to validate the data (not shown).
Biochemical Analysis of the Dynein Complex after Subunit Depletion
It is possible and, based on other studies likely (Caviston et al., 2007; Grigoriev et al., 2007; Levy and Holzbaur, 2008), that suppression of some dynein subunits results in concomitant loss of other subunits and therefore gross defects in dynein stability. We therefore blotted lysates of HeLa cells in which dynein subunits had been suppressed for components of dynein to assess expression of GAPDH and p50dynamitin as controls (Figures 2, A and B, respectively) and DHC1 (DYNC1H1, Figure 2C), IC2 (DYNC1I2, Figure 2D), LIC1 (DYNC1LI1, Figure 2E), LIC2 (DYNC1LI2, Figure 2F), LC8 (DYNLL1 Figure 2G), or Tctex1 (DYNLT1, Figure 2H). Dynein subunit suppression has no effect on GAPDH or p50dynamitin; in accordance with previous work, dynein heavy-chain suppression results in loss of DHC1 as expected (Figure 2C) as well as concomitant loss of IC2 (Figure 2D); notably we also see a partial (∼50% loss of each of LIC1, LIC2, and Tctex1 (Figure 2, E, F, and H, respectively) after DHC1 suppression. Interestingly, suppression of IC2 also causes a loss of Tctex1 (Figure 2H) but not of either LIC. LC8 is not lost from cells other than when directly targeted by RNAi (Figure 2G). Suppression of other dynein subunits (both light intermediate chains and all light chains) only results in loss of the specific subunits that are suppressed. We then sought to extend this work using sucrose density centrifugation to define the size of dynein after subunit suppression (Figure 2, I–L). As expected, suppression of lamin A/C did not shift dynein on gradient from its expected position of ∼20S (Figure 2I). Suppression of DHC1 (DYNC1H1, Figure 2J) resulted in a loss of DHC1 immunoreactivity (with a very small amount visible on 30-min exposures; not shown), and a shift of IC2 (DYNC1I2), LIC1 (DYNC1LI1), and LIC2 (DYNC1LI2) to lower fractions consistent with disassembly of the complex. Notably, dynactin integrity (p50dynamitin) was unaffected. Suppression of LIC1 (Figure 2K) resulted in a loss of reactivity for the target as expected, but importantly, the majority of DHC1, IC2, and all LIC2 remained associated with 20S fractions. Consistent with this, loss of LIC2 did not displace DHC1 or LIC1 from the 20S fraction (Figure 2L), indicating that there are indeed two distinct populations of dynein containing either LIC1 or LIC2. This is entirely consistent with previously published data (Tynan et al., 2000).
Analysis of Transport to the Golgi
To examine the effects of dynein subunit suppression on membrane trafficking, we first chose to examine transport of cargo from the ER to the Golgi, a transport step for which unequivocal evidence exists of a role for dynein. To define the requirement for dynein subunit function in ER-to-Golgi transport, we deliberately chose to assay transport of an endogenous cargo rather than an exogenous, overexpressed marker. We determined the effects of dynein subunit suppression on the localization of β1,4-galactosyltransferase I (GalT), an enzyme that localizes at steady state to the trans-Golgi cisternae (Nilsson et al., 1991). GalT cycles slowly between the ER and Golgi (Storrie et al., 1998), thereby acting as an endogenous indicator of transport between these compartments. A block of ER export or ER-to-Golgi transport results in a relocalization of GalT to the ER or other pre-Golgi compartments such as the ERGIC. Concomitantly we examined the distribution of the COPI coatomer complex, which is recruited to pre-Golgi membranes and to the Golgi apparatus itself (reviewed in Lippincott-Schwartz and Liu, 2006). COPI therefore provides an indicator of both Golgi integrity and of transport between the ER and Golgi.
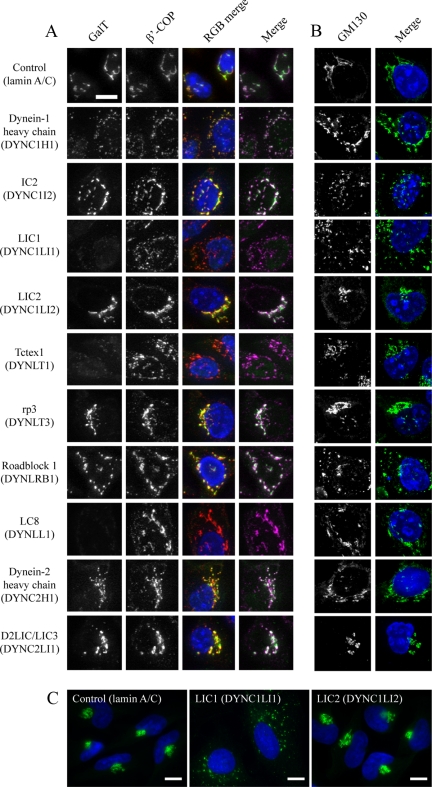
Depletion of LIC1, Tctex1, or LC8 caused a near complete loss of punctate labeling for GalT in cells (Figures 3A and Supplementary Figure S2). Higher exposures (not shown) revealed a diffuse labeling throughout cells consistent with a redistribution of GalT to the ER. Consistent with this, immunoblotting of cell lysates confirmed that GalT was not degraded in these cells (Supplementary Figure S3), rather redistributed. Similarly, no effect on expression levels of lamin A/C, p150glued, Sec24C, GAPDH, or EEA1 were observed (Supplementary Figure S3, these also serve to act as further validation of our siRNA specificity). We interpret these data as revealing a key role for each of these subunits in the transport of cargo from the ER to the Golgi (i.e., Golgi structure is perturbed but Golgi remnants are devoid of detectable GalT). An alternative explanation would be that dynein is required to retain GalT in the Golgi, but the weight of evidence presented here and in the literature (Corthesy-Theulaz et al., 1992; Burkhardt et al., 1997; Presley et al., 1997) is more consistent with a defect in ER-to-Golgi transport in these experiments. These experiments showed severe disruption of Golgi structure (shown by COPI labeling) after depletion of dynein-1 heavy chain, IC2, LIC1, Tctex1, Roadblock 1, or LC8 (Figure 3A, showing enlargements of cells; full images of multiple cells with each of the two siRNA duplexes used are shown in Figure S2). Surprisingly, suppression of several subunits that caused obvious disruption to Golgi architecture (dynein-1 heavy chain, IC2, and Roadblock 1) did not show the same defect in GalT localization (Figure 3A, Supplementary Figure S2). In these cases, GalT remained associated with the scattered Golgi fragments. We confirmed that GalT is redistributed under these conditions as opposed to being degraded by immunoblotting of whole cell lysates (Supplementary Figure S3).
Figure 3.
Effect of dynein subunit suppression on Golgi organization and function. (A) Cells depleted of targets as indicated were immunolabeled with antibodies to detect (A) GalT and β′-COP or (B) GM130. RGB and magenta/green merges show, respectively, GalT in green, with β′-COP in red or magenta. Symbols indicated where GalT localization (Golgi transport) or the integrity of the Golgi itself (Golgi structure) is (plus) or is not (minus) perturbed. Nuclei were labeled with DAPI (blue). Panels show enlargements from entire fields of view shown in Supplementary Figures S2 and S4. (C) Immunofluorescence labeling of cells depleted of lamin A/C, LIC1, or LIC2 with anti-giantin antibodies. Images in C were acquired with a 60× objective. Bar (all panels), 10 μm.
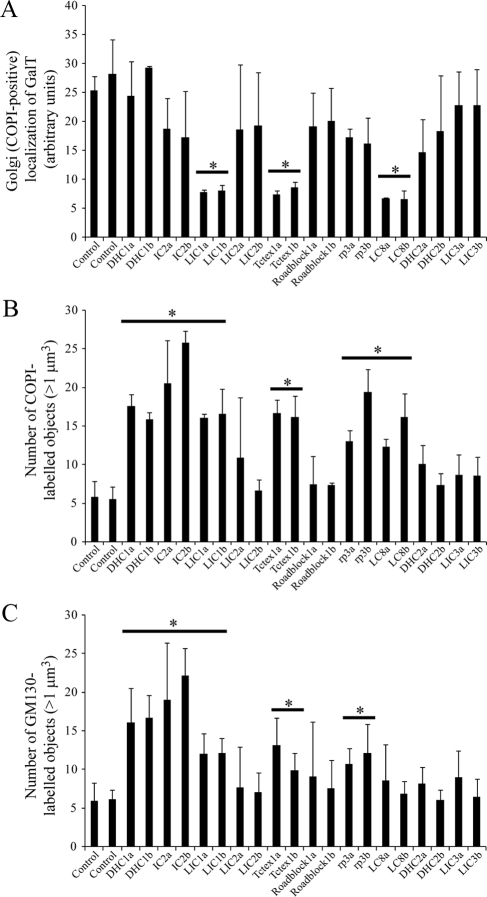
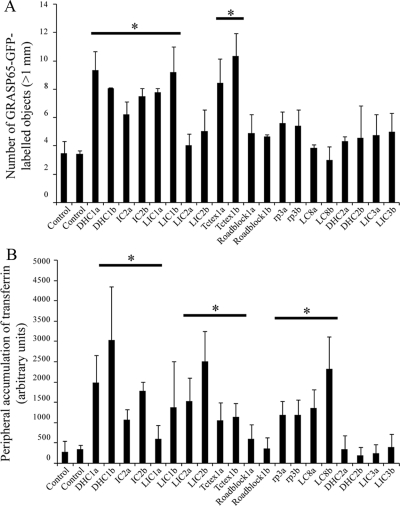
To provide a completely objective analysis of the data, we undertook automated object analysis as described in Materials and Methods. This approach, the results of which are shown in Figure 4, enabled an unbiased determination of the effects of dynein subunit suppression. We measured the number of COPI-positive structures (by thresholding the red channel) that were positive for GalT (by measuring objects above threshold intensity in the green channel). This provides a measure of the maintenance of GalT within intact or scattered Golgi elements and provides an endogenous marker for ongoing ER-to-Golgi transport. Here, suppression of LIC1, Tctex1, or LC8 resulted in a statistically discernable reduction in the localization of GalT to the Golgi (Figure 4A, horizontal bars and asterisks indicate statistical significance, where p < 0.05 in an unpaired t test). Quantification if the number of pre-Golgi and Golgi elements (COPI-positive structures, Figure 4B) also showed a statistically discernable (p < 0.05) increase after suppression of dynein-1 heavy chain, IC2, LIC1, Tctex1, Roadblock 1, or LC8. We obtained indistinguishable results using giantin (a transmembrane domain–containing marker of the Golgi; Figure 3C and data not shown).
Figure 4.
Quantification of disruption of Golgi architecture. Cells depleted of individual dynein subunits using one of two different siRNA duplexes (denoted a and b) were analyzed by automated object analysis as described in Materials and Methods. (A) The localization of GalT was quantified by detecting COPI-labeled structures based on intensity and measuring the intensity of GalT labeling within these areas. (B and C) Histograms showing the number of Golgi particles (>1 μm3) labeled with antibodies to detect (B) COPI or (C) GM130. Error bars, SD; statistical significance, *p < 0.05.
Data showing the redistribution of COPI labeling are consistent with a perturbation of Golgi organization; thus we determined the localization of other Golgi markers in cells suppressed of individual dynein subunits. GM130 is a peripheral membrane protein that forms part of the Golgi matrix (Nakamura et al., 1995). Figure 3B and Supplementary Figure S4 show that, as seen above, suppression of dynein-1 heavy chain, IC2, LIC1, Tctex1, or Roadblock 1 all resulted in disruption of Golgi structure, with a pronounced scattering of GM130-positive structures throughout the cytoplasm. These data were also quantified using automated analysis (Figure 4C); this confirmed suppression of each of these subunits, with either of the two independent siRNAs used, and showed a statistically discernable increase in the number of discrete Golgi (GM130-positive) elements per cell. These experiments define two populations of dynein subunits: those that are involved in transport to the Golgi and those that are involved in maintenance of Golgi structure itself.
Analysis of ER Exit Sites and the ERGIC
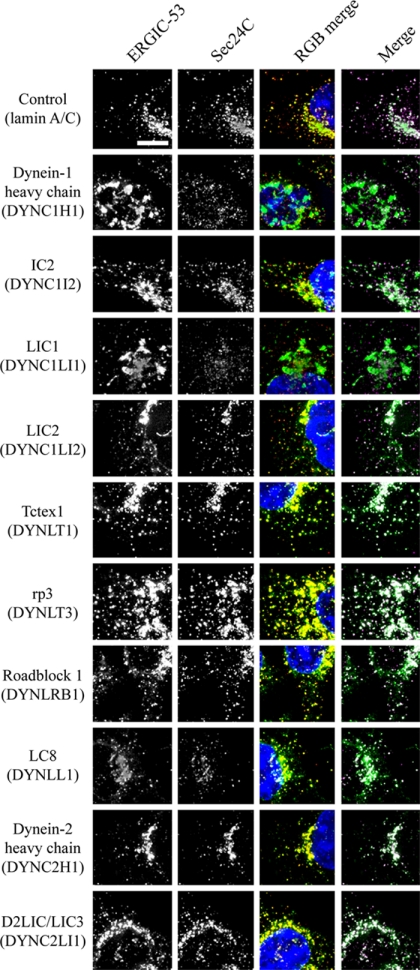
The first membrane-trafficking step in the secretory pathway is the formation of COPII-coated transport vesicles at discrete sites on the ER known as ER export sites (Hughes and Stephens, 2008). These vesicles bud and then coalesce to form the ERGIC, which is defined by the presence of a cargo-receptor, ERGIC-53. Antibody labeling of the COPII component Sec24C and of ERGIC-53 revealed key roles for dynein-1 heavy chain, LIC1, and to a lesser extent IC2 in maintaining the architecture of the ERGIC (Figure 5 showing enlargements of cells; full images of multiple cells with each of the two siRNA duplexes used are shown in Supplementary Figure S5, and these data are quantified in Figure 6). Notably, enlarged structures positive for ERGIC-53 were seen in cells, a phenotype most dramatically evident after suppression of LIC1 (Figures 5 and 6A and Supplementary Figure S5). Note the defects in ERGIC organization that are reflected by the gross changes in structure; the number of small ERGIC-53–positive objects was unchanged (Figure 6B). Despite dramatic effects caused by LIC1 suppression, LIC2 suppression did not cause detectable changes in ERGIC-53 or COPII localization. The apparent loss of juxtanuclear clustering of Sec24C (COPII) would correlate with our observations on ER-to-Golgi transport as defined by GalT localization and would be consistent with the reported cargo-dependent recruitment of COPII to ERES (Forster et al., 2006; Guo and Linstedt, 2006); however, using thresholding either based on Sec24C-positive object intensity or size, we were not able to define any difference in intensity, distribution, or number of Sec24C-positive puncta with any statistical confidence (Figure 6C). This might therefore be a result of epitope masking rather than a reduction of Sec24C at ERES. In addition, although an obvious change in the morphology of ERGIC-53-positive structures was observed after suppression of dynein heavy chain-1, IC2, or LIC1, no change in the number of these objects was evident.
Figure 5.
Effect of dynein subunit suppression on ERES (COPII) and the ERGIC. Cells depleted of targets as indicated were immunolabeled with antibodies to detect ERGIC-53 (green) and Sec24C (red, a core component of the COPII coat and marker of ERES). RGB and magenta/green merges show, respectively, ERGIC-53 in green, with Sec24C in red or magenta. Panels show enlargements from entire fields of view shown in Supplementary Figure S5. Bar (all panels), 10 μm.
Analysis of Golgi Structure and Endocytosis in Live Cells
Our data relating to Golgi architecture were further validated using a live cell imaging approach with HeLa cells stably expressing GRASP65-GFP (Lane et al., 2002). These cells were also labeled for 60 min with fluorescent transferrin to monitor endocytosis and early endosome distribution. These assays were performed on individual glass-bottom dishes rather than multiwell format plates because the timing of imaging is critical with regard to comparing transferrin localization between samples. Imaging of GRASP65-GFP–validated data obtained from COPI labeling in that suppression of dynein-1 heavy chain, IC2, LIC1, Tctex1, caused a disruption of Golgi structure (Figures 7 and Supplementary Figure S6, quantified in Figure 8A). Intriguingly, depletion of Roadblock 1 or LC8 did not cause such dramatic phenotypes with regard to Golgi structure as we observed in other experiments. This might reflect a difference in assay sensitivity when imaging live cells expressing relatively low levels of GFP-tagged GRASP65 versus strong labeling of markers using indirect immunofluorescence with fixed samples. Consistent with this, the automated detection that we have used counts about half the number of objects here compared with immunofluorescence labeling of either GM130 or COPI.
Figure 7.
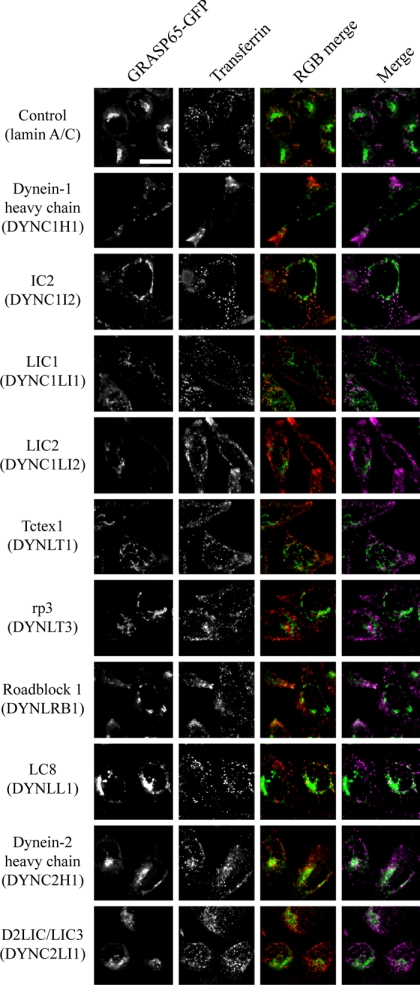
Effect of dynein subunit suppression on Golgi architecture and transferrin uptake in living cells. Cells stably expressing GRASP65-GFP (green) were depleted of targets as indicated and loaded with AlexaFluor-568-transferrin (red) for 1 h at 37°C followed by imaging also at 37°C. At this time AlexaFluor-568-transferrin labels a significant intracellular pool with little peripheral labeling in controls; peripheral accumulation indicates a defect in centripetal translocation of transferrin-positive endosomes. Symbols indicate where depletion of individual subunits does (plus) or does not (minus) result in peripheral accumulation of transferrin-positive endosomes. Panels show enlargements from entire fields of view shown in Supplementary Figure S6. Bar (all panels), 10 μm.
Figure 8.
Quantification of localization of transferrin-positive endosomes. Cells depleted of individual dynein subunits using one of two different siRNA duplexes (denoted a and b) were analyzed by automated object analysis as described in Materials and Methods. (A) The number of GRASP65-GFP–labeled Golgi structures (>1 μm3) was counted based on intensity thresholding. (B) The area covered by peripherally localized transferrin-positive endosomes was counted. Error bars, SD; statistical significance, *p < 0.05.
In these live cell imaging assays, discernable differences were observed in the localization of AlexaFluor-568-transferrin after a 60-min pulse of labeling (Figures 7 and Supplementary Figure S6, quantified in Figure 8B). We have previously shown that suppression of the LC8 subunit causes defects in endocytosis and endosomal sorting (Traer et al., 2007). Here, we include data on all other dynein subunits in addition to LC8. We found that suppression of dynein-1 heavy chain, IC2, LIC2, Tctex1, Roadblock 1, or LC8 all induced an accumulation of AlexaFluor-568-transferrin in the cell periphery. This defines a subset of dynein that is involved in endocytosis and/or endosomal function. Intriguingly, a key difference here to our experiments looking at Golgi structure is the role of LIC2 in endocytic function (Figures 7 and Supplementary Figure S6) as opposed to that of LIC1 in Golgi function (Figures 3 and Supplementary Figure S2).
Organization of the Microtubule Cytoskeleton
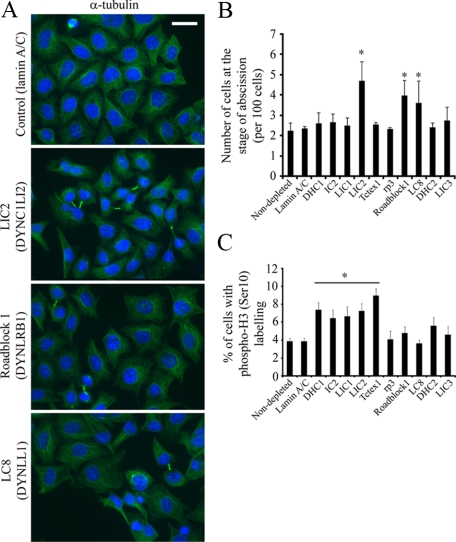
The organization of the Golgi apparatus correlates closely with the integrity of the microtubule cytoskeleton (Thyberg and Moskalewski, 1985). Dynein has also been shown to play a key role in the radial organization of the microtubule network (Quintyne et al., 1999; Malikov et al., 2004). Thus, we analyzed the radial array of microtubules in cells (Figures 9 and Supplementary Figure S7). No obvious differences were seen between cells depleted of any dynein subunit in terms of a centrally organized microtubule array, suggesting that the defects in Golgi organization are not down to a gross disorganization of the microtubule cytoskeleton. Interestingly, cells depleted of LIC2, Roadblock 1, or LC8 showed a higher proportion of cells that show a failure to complete cytokinesis (Figure 9) and suggested that these subunits play important roles in the completion of abscission. We quantified these data by visual inspection (Figure 9B) and found that suppression of LIC2, Roadblock1, LC8 all cause a statistically detectable increase in the number of cytokinetic failures, which was not seen after suppression of other subunits. Further to this, we sought to quantify any possible defect in mitosis after dynein subunit suppression using phospho-histone H3 labeling. Figure 9C shows that suppression of DHC1, IC2, LIC1, LIC2, and LT1, all caused a statistically detectable increase in the number of phospho-H3–labeled cells. No difference was discernable for suppression of other dynein light chains or dynein-2 components.
Figure 9.
LIC2 (DYNC1LI2) and LC8 (DYNLL1) are required for the completion of abscission during cytokinesis. (A) Cells depleted of dynein subunits were processed for immunofluorescence using antibodies to detect α-tubulin (green in right column); cells were counterstained with DAPI to visualize cell nuclei (blue). Bar (all panels), 20 μm. (B) Images were scored by visual inspection for numbers of cells at the stage of abscission (intercellular tubulin bridges) and are expressed as a percentage of cells analyzed (>200 cells from three independent experiments). Error bars, SD, asterisks indicate statistically discernable differences; only LIC2 (DYNC1LI2, p = 0.004), Roadblock1 (DYNLRB1, p = 0.007), and LC8 (DYNLL1, p = 0.04) show statistically detectable differences to lamin sA/ C–depleted controls. (C) Cells depleted of dynein subunits were fixed and immunolabeled to detect phospho-histone H3 (Serine-10). Error bars, SD; asterisk and horizontal bar indicate statistical significance: DHC1 (DYNC1H1) p = 0.002, IC2 (DYNC1I1) p = 0.01, LIC1 (DYNC1LI1) p = 0.01, LIC2 (DYNC1LI2) p = 0.004, Tctex1 (DYNLT1) p = 0.01, and Roadblock1 (DYNLRB1) p = 0.03.
Rescue of LIC1 and LIC2 Suppression
The major findings of this study were that two subpopulations of dynein-1 exist that are involved in trafficking at different locations within the cell. LIC1-containing dynein primarily operates at the Golgi and in ER-to-Golgi trafficking. In contrast LIC2-containing dynein operates in the endosomal system. To ensure the specificity of our siRNA experiments, we used two independent duplexes against each target and further validated our major findings (Golgi scattering, relocalization of GalT, and redistribution of recycling endosomes) using two additional siRNA duplexes against each of LIC1 and LIC2 (DYNC1LI1c, DYNC1LI1d, DYNC1LI2c, and DYNC1LI2d). In addition, we sought to “rescue” the effects of suppression of these subunits by recombinant expression of mouse orthologues of the two light intermediate chains, mLIC1 and mLIC2; these are not predicted to be targeted by our siRNAs, and immunofluorescence labeling confirms this (not shown). Cells in which LIC1 or 2 had been suppressed were transfected with cDNAs to express untagged versions of mLIC1 or mLIC2 and then either fixed and immunolabeled these cells for GalT and GRASP65 (Figure 10) or loaded with AlexaFluor-568-transferrin for 60 min followed by fixation and labeling with anti-GRASP65 antibodies (Figure 11).
Figure 10 shows siRNA-transfected cells that have subsequently been transfected with a plasmid encoding mLIC1; asterisks mark the transfected cells with arrows, indicating cells that were not detectably expressing mLIC1 by immunofluorescence. In nonsuppressed or lamin A/C cells, the Golgi is located in a classical juxtanuclear position and contains GalT. This is largely unaffected by mLIC1 expression; it is possible that mLIC1 expression enhances the juxtanuclear clustering of the Golgi, but we have not investigated this further. In DHC1 suppressed cells, expression of mLIC1 does not alter the scattered Golgi or intensity of GalT labeling. In cells suppressed for LIC1, expression of mLIC1 rescues the localization of GalT to the Golgi and substantially restores the juxtanuclear localization (indistinguishable results were obtained using the DYNC1LI1b duplex; see Figure 11). In contrast, cells depleted of LIC2 do not show significant Golgi scattering or loss of GalT labeling from the Golgi, and this is unchanged by expression of mLIC1.
Our data show that cells depleted of LIC2 show a scattering of juxtanuclear recycling endosomes to the cell periphery. We used this assay, in combination with Golgi labeling to determine whether mLIC2 could reverse the loss of LIC2 from HeLa cells. Figure 11A shows that cells that have been transfected with duplexes to suppress LIC2 and subsequently with a plasmid encoding mLIC2 show classical juxtanuclear clustering of both the Golgi and transferrin-positive recycling endosome. In contrast, mLIC1 expression cannot rescue the peripheral distribution of recycling endosomes. We then sought to test the specificity of these rescue experiments. Figure 11B shows that in cells depleted of LIC1 the Golgi is scattered, but the transferrin-labeled endosomes retain their juxtanuclear localization. Significantly, in most cells (∼70%) the dispersal of the Golgi is unaffected by mLIC2 expression, indicating that mLIC2 cannot substantially replace the function of endogenous LIC1 in HeLa cells. Some cells (∼30%) show a greater juxtanuclear clustering of Golgi elements, but this does not correlate with mLIC2 expression level. These cells could reflect those that were poorly suppressed for LIC1 initially or potentially indicate that under certain circumstances, mLIC2 can functionally replace LIC1. In contrast, as indicated by the results in Figure 10, expression of mLIC1 can restore the juxtanuclear localization of the Golgi in cells where LIC1 had previously been suppressed. Together these data confirm our findings that LIC1 acts primarily in Golgi function and ER-to-Golgi transport, whereas LIC2 plays a key role in recycling endosome localization.
DISCUSSION
We have used siRNA suppression to define the roles of all individual dynein subunits in mammalian cells. Other studies have used a similar strategy, and this experimental approach has the key advantage of leaving the microtubule cytoskeleton largely intact but reducing the amount of the dynein subunits in cells. This contrasts with other approaches previously used that either target the dynein accessory complex dynactin or globally block dynein function through the introduction of inhibitory antibodies. Notably microinjection of function blocking antibodies routinely targets IC2 (e.g., Driskell et al., 2007) and therefore would interfere with the function of all dynein-1 subcomplexes in most nonneuronal cells.
Immunoblotting and sucrose density gradient centrifugation reveal some requirements for the structural integrity of dynein. Suppression of DHC1 also results in a loss of IC2 from cells, as has been shown by others (Caviston et al., 2007; Grigoriev et al., 2007; Levy and Holzbaur, 2008). Further to this, we find that Tctex1 is lost from cells suppressed for DHC1, IC2, or Tctex1 itself. This clearly has important implications for our own data as well as that of others in which either DHC1 or IC2 is suppressed. Targeting these subunits should be seen perhaps as a more global suppression of dynein function. Together with our other data, DHC1, IC2, and Tctex1 can very much be seen as central to the function of dynein overall. One of the two dynein-1 light intermediate chains is required in addition to these. The requirements for associated light chains differ, and their analysis is significantly hampered by nondynein functions of these proteins. Given that suppression of DHC1 causes at least a partial loss of IC2 as well, one might expect all assays to show near identical results after either DHC1 and IC2 suppression. This does not reflect what we see in all assays, for example, the effects on ERGIC structure. A likely explanation for this is that direct suppression of IC2 is more effective in our hands than suppression of DHC1 and certainly results in a lower remaining amount of IC2 within cells. Thus, IC2 suppression provides a robust block in almost all trafficking assays. That IC2 suppression also results in loss of Tctex perhaps compounds the strength of these phenotypes because our work implicates Tctex1 in multiple steps of membrane trafficking and mitosis.
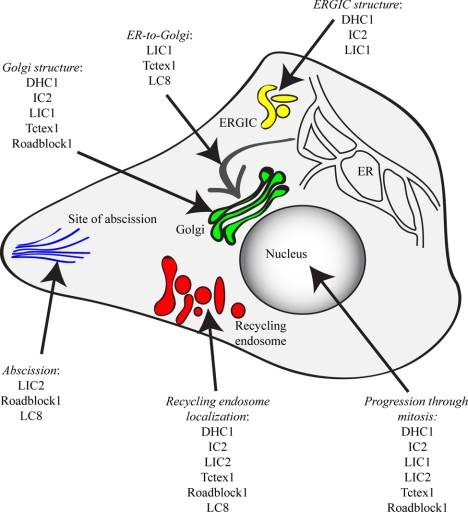
Our data clearly define the individual subunits of the dynein motor that are involved in particular membrane-trafficking steps (summarized in Table 1 and Figure 12). Our work shows that almost all known subunits of the cytoplasmic dynein-1 complex are required to maintain the structure of the Golgi with the exception of LIC2 and rp3; our use of multiple markers for the Golgi (GM130, GRASP65-GFP and β′-COP) validates these findings in both living and fixed cells. Obvious defects in Golgi organization are consistent with the role of this organelle, located adjacent to the centrosome in HeLa cells in the vicinity of microtubule minus ends, as a central hub for diverse dynein-driven membrane trafficking from many parts of the cell.
Table 1.
Summary of findings
| GalT localization phenotype (ER-to-Golgi transport) | Golgi scattering phenotype (GM130, GRASP65 labelling) | ERGIC structure phenotype (ERGIC-53 labeling) | Transferrin-labeled endosome phenotype (peripheral distribution) | Abscission phenotype (anaphase bridges) | Phospho-histone H3 labeling phenotype (mitotic progression) | |
|---|---|---|---|---|---|---|
| Dynein-1 heavy chain, DHC1 (DYNC1H1) | NO | YES | YES | YES | NO | YES |
| IC2 (DYNC1LI2) | NO | YES | YES | YES | NO | YES |
| LIC1 (DYNC1LI1) | YES | YES | YES | NO | NO | YES |
| LIC2 (DYNC1LI2) | NO | NO | NO | YES | YES | YES |
| Tctex1 (DYNLT1) | YES | YES | NO | YES | NO | YES |
| rp3 (DYNLT3) | NO | NO | NO | NO | NO | NO |
| Roadblock 1 (DYNLRB1) | NO | YES | NO | YES | YES | YES |
| LC8 (DYNLL1) | YES | NO | NO | YES | YES | NO |
| Dynein-2 heavy chain, DHC2 (DYNC2H1) | NO | NO | NO | NO | NO | NO |
| D2LIC1, LIC3 (DYNC2LI1) | NO | NO | NO | NO | NO | NO |
Defects are summarized here based on visual inspection of images and statistical relevance from automated object quantification. Consequently, as an example, LC8 (DYNLL1) scores negative for Golgi structure because we only see a strong phenotype when using COPI as a marker that also reports ongoing transport between the ER and Golgi. When measured using GM130 or GRASP65-GFP as markers, Golgi structure is not strongly perturbed after LC8 suppression.
Figure 12.
Summary model of the roles for dynein subunits identified in this study. The summary is compiled from the quantitative imaging data and is designed to highlight the most prominent of the phenotypes that we observe. All steps require the dynein heavy chain (DHC1 subunit) and IC (IC2 subunit) because these are required for complex assembly, but we do not always observe strong phenotypes in our assays after suppression of these subunits, which is most likely due to incomplete suppression of expression. These data are also summarized in Table 1.
We show that LIC1 is a major component of the machinery that maintains the steady-state architecture of the ERGIC and plays a key role in supporting stable recruitment of COPII to the ERES. We have previously shown that dynactin is important for the function of ER exit sites through a direct interaction between its p150glued subunit and the COPII component Sec23A (Watson et al., 2005). More recently, we have found that dynein itself plays a role in the anchoring of the Sec23-labeled ERES (Gupta et al., 2008). Our data reveal key roles for LIC1, Tctex1, and LC8 in ER-to-Golgi transport because their suppression results in a loss of endogenous GalT from the Golgi. This is consistent with the localization of Tctex1 to the Golgi (Tai et al., 1998) and with our other experiments on COPII localization and ERGIC structure. These data are also consistent with a role for dynein in ER export; if centripetal transport rather than ER export is perturbed, one might expect to have seen an accumulation in punctate ERGIC or vesicular-tubular clusters; instead, we observe the relocation of GalT to the ER, consistent with an inhibition of ER export. ER-to-Golgi transport has long been known to be mediated by dynein in concert with dynactin (Corthesy-Theulaz et al., 1992; Presley et al., 1997); perhaps most importantly, our data show the importance of dynein, rather than the global organization of microtubules, in centripetal transport from the ER to the Golgi, maintenance of the steady-state composition, and structural organization of the Golgi.
Significantly our data identify distinct functions of dynein in Golgi organization versus ongoing ER-to-Golgi transport. The presence of these two pools is exemplified by the role of LC8. Suppression of LC8 expression results in a loss of GalT from the Golgi but no statistically discernable perturbation of Golgi organization (at least at the level of our analyses). The loss of GalT correlates with an increase in number of COPI-positive structures, consistent with a block in transport to the Golgi, but the overall architecture of the Golgi matrix (GM130, GRASP65-GFP) is not significantly perturbed. Further finer detail from our data is also consistent with this: those dynein subunits that, when suppressed, lead to an increase in the number of COPI-positive structures overlaps with both the set of subunits, which show strong GalT phenotypes and also those that show strong GM130 and GRASP65 phenotypes (in terms of Golgi scattering). This is entirely consistent with roles for COPI in both ER-to-Golgi transport and in maintaining the structural integrity of the Golgi.
The role of dynein in the endocytic pathway is also well established (Aniento et al., 1993; Burkhardt et al., 1997; Habermann et al., 2001), and recent work has provided considerable molecular detail regarding the mechanism of recruitment of dynein to endosomal membranes (Caviston et al., 2007; Traer et al., 2007), the requirement for dynein in receptor sorting (Lakadamyali et al., 2006; Driskell et al., 2007), and specific roles for individual dynein subunits in endosomal sorting and trafficking (Traer et al., 2007; Ha et al., 2008). Notable was the severity of disruption of endosome distribution after LIC2 depletion, which was a much more obvious phenotype that we see with other subunits such as LC8 that we have previously implicated in this process (Traer et al., 2007). There are two immediate possibilities to explain this: either LIC2 plays a more direct role in endosome motility or loss of one light chain is more easily compensated for by the presence of the other dynein light chains. Our data reveal a core complex of “endosomal” dynein controlling the localization of these structures (Table 1 and Figure 12).
The differential role of the dynein light intermediate chains in Golgi (LIC1) and endosome (LIC2) function is perhaps the most striking finding of this study. This specificity is consistent with the presence of distinct dynein complexes that either contain homooligomers of LIC1 or LIC2 but not heterooligomers of both (Tynan et al., 2000). Other recent data support the idea of light intermediate chains as potential linkers to cargo molecules; LIC1 was shown to remove specific components from the kinetochore during spindle assembly checkpoint silencing (Sivaram et al., 2009). In addition this work also shows that LIC1 silencing alone does not significantly affect dynein complex integrity, consistent with our own data. Furthermore, the difference in requirement for core dynein subunits, notably DYNC1LIs, in movement of different organelles provides an obvious means to control their differential recruitment to membranes. An alternative explanation would be that these two populations of dynein in fact localize to multiple organelles yet the affinity of LIC1-dynein for the Golgi is lower than that for the endosome; thus, one observes Golgi-related defects more readily than endosomal defects. Likewise the converse could be true for LIC2. Current reagents do not allow us to test this possibility. Given other data suggesting similar specific roles for LIC1 (Sivaram et al., 2009), we favor a model in which LIC1 and LIC2 can direct recruitment of dynein to the Golgi and endosomes, respectively. Clearly there are multiple potential mechanisms by which these subunits direct dynein recruitment and binding to membranes, proteins, and other cargoes.
We also identified Roadblock 1 in our screen for subunits affecting endosome localization; this is consistent with its recent identification as a binding partner for Rab6 (Wanschers et al., 2008). The peripheral distribution of transferrin after Roadblock 1 suppression would be predicted from the recently proposed model implicating Rab6 in anchoring Rab11-positive recycling endosomes in the juxtanuclear area via interaction of both Rab6 and Rab11 with Rab6IP1 (Miserey-Lenkei et al., 2007). These and other data hint at complex mechanisms for the recruitment and control of activity of dynein on endosomes that clearly requires further investigation.
Roles for dynein in cell division are also clearly established (Pfarr et al., 1990; Yang et al., 2007). Previous work using siRNA depletion of dynein has previously found no defects in mitosis or cytokinesis (Kittler et al., 2004; Zhu et al., 2005). From our studies, suppression of LIC2, Roadblock1, and LC8 causes failure in the completion of cytokinesis. We note that these subunits are also implicated in endocytic function in our assays; notably, depletion of LIC2 shows the strongest defect in endosome distribution. Thus, it is possible that the observed defect in cytokinesis is in fact due indirectly to a defect in the endocytic pathway that is required for the completion of cytokinesis (Eggert et al., 2006 and references therein). Our analyses also identify the major dynein subunits, DHC1, IC2, both LICs, TcTex, and Roadblock1 as important for mitosis because suppression of any one of these results in an increase in cells labeling with phospho-histone H3 antibodies. This is entirely consistent with considerable published work, and we have not investigated these phenotypes further.
We did not identify any role for dynein-2 in any of the trafficking assays we undertook. Assays were performed with five independent siRNA duplexes targeting dynein-2 heavy chain (data not shown), and none revealed any role for dynein-2 in Golgi or early endosome organization. Dynein-2 and its associated light IC LIC3 have been shown to have key roles in the biogenesis and function of primary cilia (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999; Grissom et al., 2002). We were able to confirm the role of dynein-2 heavy chain and LIC3 in primary cilia function (K. J. Palmer and D. J. Stephens, unpublished observations), which validates the efficacy of these siRNA duplexes. Thus, our data are not consistent with a direct role for dynein-2 in Golgi or endosome function.
In summary, our data identify populations of dynein that are involved in ER-to-Golgi transport, in maintaining the structure of the Golgi, or in endosome distribution. This could reflect distinct pools of dynein within the cell that have specific subunit compositions or could reflect differential usage of subunits within common complexes (e.g., we cannot explicitly state that “Golgi” pools of dynein do not contain rp3 subunits, but our experiments can identify no role for them at this location). Our results provide a framework for further analysis of the molecular mechanisms by which individual dynein complexes are recruited to and function in these individual trafficking steps, as well as in other intracellular events including cell polarization and migration, autophagy, and the movement and positioning of organelles.
Supplementary Material
ACKNOWLEDGMENTS
We thank Viki Allan (University of Manchester), Phil Woodman (University of Manchester), Anna Akhmanova (Erasmus Medical Center), Pete Cullen (University of Bristol), Harry Mellor (University of Bristol), Richard Vallee (Columbia University), Stephen King (University of Connecticut Health Center), and particularly Jon Lane (University of Bristol) for reagents, suggestions, and helpful discussion relating to this work, and members of the Stephens laboratory for constructive discussion relating to this project. We also thank Caroline McKinnon and Virginie Betin for advice and assistance with qPCR and to the anonymous reviewers of this work for their comments that greatly benefited this study. This work is funded by a Medical Research Council Non-Clinical Senior Fellowship to D.J.S. (G117/553) and doctoral training studentship from the Biotechnology and Biological Sciences Research Council.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1160) on April 22, 2009.
REFERENCES
- Aniento F., Emans N., Griffiths G., Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J. Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 2008;47:503–508. doi: 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- Benison G., Karplus P. A., Barbar E. Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J. Mol. Biol. 2007;371:457–468. doi: 10.1016/j.jmb.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Bielli A., Thornqvist P. O., Hendrick A. G., Finn R., Fitzgerald K., McCaffrey M. W. The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem. Biophys. Res. Commun. 2001;281:1141–1153. doi: 10.1006/bbrc.2001.4468. [DOI] [PubMed] [Google Scholar]
- Burkhardt J. K., Echeverri C. J., Nilsson T., Vallee R. B. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J. P., Ross J. L., Antony S. M., Tokito M., Holzbaur E. L. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. F., Ghanekar Y., Frost L., Diao A., Morrison D., McKenzie E., Lowe M. ZFPL1, a novel ring finger protein required for cis-Golgi integrity and efficient ER-to-Golgi transport. EMBO J. 2008;27:934–947. doi: 10.1038/emboj.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy-Theulaz I., Pauloin A., Pfeffer S. R. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J. Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon S. W., Serpinskaya A. S., Vaughan P. S., Lopez Fanarraga M., Vernos I., Vaughan K. T., Gelfand V. I. Dynactin is required for bidirectional organelle transport. J. Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell O. J., Mironov A., Allan V. J., Woodman P. G. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat. Cell Biol. 2007;9:113–120. doi: 10.1038/ncb1525. [DOI] [PubMed] [Google Scholar]
- Eggert U. S., Mitchison T. J., Field C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Forster R., Weiss M., Zimmermann T., Reynaud E. G., Verissimo F., Stephens D. J., Pepperkok R. Secretory cargo regulates the turnover of COPII subunits at single ER exit sites. Curr. Biol. 2006;16:173–179. doi: 10.1016/j.cub.2005.11.076. [DOI] [PubMed] [Google Scholar]
- Gill S. R., Schroer T. A., Szilak I., Steuer E. R., Sheetz M. P., Cleveland D. W. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., et al. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Grissom P. M., Vaisberg E. A., McIntosh J. R. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol. Biol. Cell. 2002;13:817–829. doi: 10.1091/mbc.01-08-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Linstedt A. D. COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J. Cell Biol. 2006;174:53–63. doi: 10.1083/jcb.200604058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Palmer K. J., Spence P., Hudson A., Stephens D. J. Kinesin-1 (uKHC/Kif5b) is required for bidirectional motility of ER exit sites and efficient ER-to-Golgi transport. Traffic. 2008;9:1850–1866. doi: 10.1111/j.1600-0854.2008.00811.x. [DOI] [PubMed] [Google Scholar]
- Ha J., Lo K. W., Myers K. R., Carr T. M., Humsi M. K., Rasoul B. A., Segal R. A., Pfister K. K. A neuron-specific cytoplasmic dynein isoform preferentially transports TrkB signaling endosomes. J. Cell Biol. 2008;181:1027–1039. doi: 10.1083/jcb.200803150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A., Schroer T. A., Griffiths G., Burkhardt J. K. Immunolocalization of cytoplasmic dynein and dynactin subunits in cultured macrophages: enrichment on early endocytic organelles. J. Cell Sci. 2001;114:229–240. doi: 10.1242/jcs.114.1.229. [DOI] [PubMed] [Google Scholar]
- Haghnia M., Cavalli V., Shah S. B., Schimmelpfeng K., Brusch R., Yang G., Herrera C., Pilling A., Goldstein L. S. Dynactin is required for coordinated bidirectional motility, but not for Dynein membrane attachment. Mol. Biol. Cell. 2007;18:2081–2089. doi: 10.1091/mbc.E06-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Stephens D. J. Assembly, organization, and function of the COPII coat. Histochem. Cell Biol. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ling S. C., Rogers G. C., Kural C., Selvin P. R., Rogers S. L., Gelfand V. I. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J., Bonilla M., Rodgers M. E., Schroer T. A. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 2002;11:1239–1250. doi: 10.1110/ps.2520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R., et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Lakadamyali M., Rust M. J., Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J. D., Lucocq J., Pryde J., Barr F. A., Woodman P. G., Allan V. J., Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 2002;156:495–509. doi: 10.1083/jcb.200110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J. R., Holzbaur E. L. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Liu W. Insights into COPI coat assembly and function in living cells. Trends Cell Biol. 2006;16:e1–4. doi: 10.1016/j.tcb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Malikov V., Kashina A., Rodionov V. Cytoplasmic dynein nucleates microtubules to organize them into radial arrays in vivo. Mol. Biol. Cell. 2004;15:2742–2749. doi: 10.1091/mbc.E03-10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami A., Tynan S. H., Hama T., Luby-Phelps K., Saito T., Crandall J. E., Besharse J. C., Vallee R. B. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S., et al. Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic. 2007;8:1385–1403. doi: 10.1111/j.1600-0854.2007.00612.x. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulina K., Patel-King R. S., Takebe S., Pfister K. K., King S. M. The Roadblock light chains are ubiquitous components of cytoplasmic dynein that form homo- and heterodimers. Cell Motil. Cytoskelet. 2004;57:233–245. doi: 10.1002/cm.10172. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Lucocq J. M., Mackay D., Warren G. The membrane spanning domain of beta-1,4-galactosyltransferase specifies trans Golgi localization. EMBO J. 1991;10:3567–3575. doi: 10.1002/j.1460-2075.1991.tb04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Witman G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr C. M., Coue M., Grissom P. M., Hays T. S., Porter M. E., McIntosh J. R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Pfister K. K., et al. Cytoplasmic dynein nomenclature. J. Cell Biol. 2005;171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister K. K., Shah P. R., Hummerich H., Russ A., Cotton J., Annuar A. A., King S. M., Fisher E. M. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Quintyne N. J., Gill S. R., Eckley D. M., Crego C. L., Compton D. A., Schroer T. A. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi C., Allan V. J. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J. Cell Sci. 1999;112(Pt 24):4673–4685. doi: 10.1242/jcs.112.24.4673. [DOI] [PubMed] [Google Scholar]
- Ross J. L., Ali M. Y., Warshaw D. M. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics Methods and Protocols. Vol. 132. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Scholey J. M. Intraflagellar transport motors in cilia: moving along the cell's antenna. J. Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T. A., Steuer E. R., Sheetz M. P. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- Signor D., Wedaman K. P., Orozco J. T., Dwyer N. D., Bargmann C. I., Rose L. S., Scholey J. M. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaram M. V., Wadzinski T. L., Redick S. D., Manna T., Doxsey S. J. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 2009;28:902–914. doi: 10.1038/emboj.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., White J., Rottger S., Stelzer E. H., Suganuma T., Nilsson T. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susalka S. J., Nikulina K., Salata M. W., Vaughan P. S., King S. M., Vaughan K. T., Pfister K. K. The roadblock light chain binds a novel region of the cytoplasmic Dynein intermediate chain. J. Biol. Chem. 2002;277:32939–32946. doi: 10.1074/jbc.M205510200. [DOI] [PubMed] [Google Scholar]
- Susalka S. J., Pfister K. K. Cytoplasmic dynein subunit heterogeneity: implications for axonal transport. J. Neurocytol. 2000;29:819–829. doi: 10.1023/a:1010995408343. [DOI] [PubMed] [Google Scholar]
- Tai A. W., Chuang J. Z., Bode C., Wolfrum U., Sung C. H. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tai A. W., Chuang J. Z., Sung C. H. Localization of Tctex-1, a cytoplasmic dynein light chain, to the Golgi apparatus and evidence for dynein complex heterogeneity. J. Biol. Chem. 1998;273:19639–19649. doi: 10.1074/jbc.273.31.19639. [DOI] [PubMed] [Google Scholar]
- Thyberg J., Moskalewski S. Microtubules and the organization of the Golgi complex. Exp. Cell Res. 1985;159:1–16. doi: 10.1016/s0014-4827(85)80032-x. [DOI] [PubMed] [Google Scholar]
- Townley A. K., Feng Y., Schmidt K., Carter D. A., Porter R., Verkade P., Stephens D. J. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J. Cell Sci. 2008;121:3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- Traer C. J., Rutherford A. C., Palmer K. J., Wassmer T., Oakley J., Attar N., Carlton J. G., Kremerskothen J., Stephens D. J., Cullen P. J. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 2007;9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- Tynan S. H., Purohit A., Doxsey S. J., Vallee R. B. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 2000;275:32763–32768. doi: 10.1074/jbc.M001536200. [DOI] [PubMed] [Google Scholar]
- Wanschers B., van de Vorstenbosch R., Wijers M., Wieringa B., King S. M., Fransen J. Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil. Cytoskelet. 2008;65:183–196. doi: 10.1002/cm.20254. [DOI] [PubMed] [Google Scholar]
- Watson P., Forster R., Palmer K. J., Pepperkok R., Stephens D. J. Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat. Cell Biol. 2005;7:48–55. doi: 10.1038/ncb1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Stephens D. J. Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J. Cell Sci. 2006;119:2758–2767. doi: 10.1242/jcs.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U. S., Wadsworth P., Rieder C. L. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.