Abstract
When haploid cells of Saccharomyces cerevisiae are crossed, parental nuclei congress and fuse with each other. To investigate underlying mechanisms, we have developed assays that evaluate the impact of drugs and mutations. Nuclear congression is inhibited by drugs that perturb the actin and tubulin cytoskeletons. Nuclear envelope (NE) fusion consists of at least five steps in which preliminary modifications are followed by controlled flux of first outer and then inner membrane proteins, all before visible dilation of the waist of the nucleus or coalescence of the parental spindle pole bodies. Flux of nuclear pore complexes occurs after dilation. Karyogamy requires both the Sec18p/NSF ATPase and ER/NE luminal homeostasis. After fusion, chromosome tethering keeps tagged parental genomes separate from each other. The process of NE fusion and evidence of genome independence in yeast provide a prototype for understanding related events in higher eukaryotes.
INTRODUCTION
In yeast zygote formation, cell fusion is followed by congression of parental nuclei, which results from depolymerization of the microtubule bundles that connect their spindle pole bodies (SPBs), which are embedded in the nuclear envelope (NE; Marsh, 1997; Jaspersen and Winey, 2004; Molk and Bloom, 2006). Nuclear fusion (karyogamy) then leads to encounter of the two parental genomes.
Among proteins that are known to be required for karyogamy are the endoplasmic reticulum (ER) luminal proteins, Kar2p, and Kar8p/Jem1p; the ER transmembrane protein, Kar5p, which localizes near the SPB; Kar7p/Sec71p and Sec63p of the ER translocon; and Prm3p, a protein that, surprisingly, has been localized to the nucleoplasmic face of the inner nuclear membrane (Marsh, 1997; Beilharz et al., 2003). Because karyogamy follows establishment of SPB contact, the mechanism of fusion could be distinct from nuclear fusion in organisms that lack SPBs. An alternate view is that the SPBs ensure spatial proximity and that fusion of the outer membranes, which initiates with contact of their cytoplasmic aspect, involves Snares and the AAA ATPase, Sec18p/NSF, as in topologically equivalent fusion events along the secretory and endocytic paths (Jahn and Scheller, 2006).
For organelles that are enclosed by double membranes, one can envisage distinct models of fusion (see Figure 1A). In the “trans-first” model, the outer membranes (cis, trans) fuse with each other, and the inner membranes fuse with each other. These events could be simultaneous or sequential. If the outer membrane fuses before the inner membrane, fusion of the inner membranes would initiate with contact of their luminal aspect. It is therefore plausible that proteins of the lumen between the outer and inner membranes would be required. Alternatively, the outer and inner membranes of a single organelle could first fuse with each other, and this unit(s) could subsequently fuse with equivalent intermediates generated from the target organelle. This second model is followed during the mitotic cell cycle of higher eukaryotes as part of the events of NE breakdown and reformation (Sheehan et al., 1988; Holaska et al., 2002; Hetzer et al., 2005; Baur et al., 2007; Stewart et al., 2007).
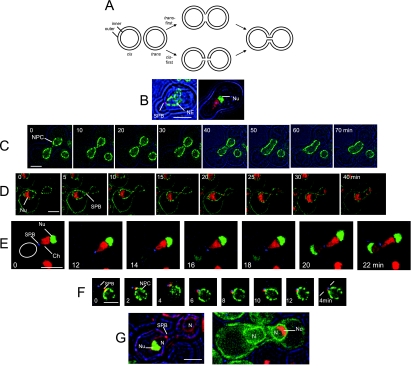
Figure 1.
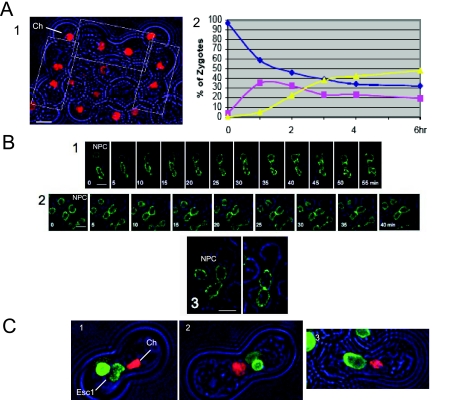
Overview of NE Fusion. (A) Models of fusion of organelles with double membranes. According to both models, the two nuclei diagrammed at the left accomplish fusion of both outer and inner membranes, generating a single nucleus with a continuous envelope. In the trans-first model, the two outer membranes fuse with each other before the two inner membranes. In the cis-first model, the inner and outer membranes of each nucleus first fuse with each other. The cis-first model is considered in the Discussion. (B) Polarization of nuclei of cells after 3-h exposure to α-factor. Left, a cell expressing Spc42p-mRFP and Nup49p-GFP (ATY1816). Right, a cell expressing Htb2p-mRFP and nucleolar Gar1p-GFP (ATY2594). (C) Cross between strains that express Nup49p-GFP (ATY3405 × ATY3358). The nexus is established just after t = 0 min and persists until just before the 40-min time point. (D) Cross between a strain that expresses Mid2p-GFP, Nup49p-GFP, Sik1p-mRFP, and Spc42p-mRFP (ATY1897) with a strain that expresses Mid2p-GFP, Nup49p-GFP, and Spc42p-mRFP (ATY1916). The nexus persists from +5 until +30 min. Note that the flux of Sik1p-mRFP is detected before dilation of the nexus. Disengagement of the SPB from one face of the NE is evident at the last time point. (E) A cross between a strain expressing Gar1p-GFP and Htb2p-mRFP (ATY2416), with a strain expressing Spc42p-CFP (ATY1455). The corresponding nucleus is circled. Note that contact of the SPB (blue) with the trans nucleus (red) is followed by a pause before the initial transfer of Gar1p-GFP (and Htb2p-mRFP), and a further delay before dilation of the nexus and disengagement of the SPB is seen at 22 min. (F) Delay of SPB coalescence. Cross between a strain that expresses Nup49p-GFP and Spc42p-mRFP (ATY1817) and a strain that expresses Spc42p-CFP (ATY1455). The two SPBs, although adjacent, remain separate throughout. The first images precede establishment of contact. (G) Left, cross between kar1-1 expressing mRFP-HDEL (ATY3004) and a strain that expresses Spc42p-mRFP and Gar1p-GFP (ATY3198). Images were collected after 3 h. Note the absence of Gar1p-GFP from the kar1-1 nucleus, implying that this protein does not shuttle to the cytoplasm and that its continued synthesis during the cross does not generate a de novo signal in the trans nucleus. Right, similar observations were made following Sik1p-mRFP in crosses with kar1-1 that expresses GFP-HDEL (ATY3373 × ATY1513).
Consistent with the trans-first model for NE fusion, electron micrographs of fertilization in marine algae and sea urchins capture putative intermediates that show selective continuity of outer membranes (Longo and Anderson, 1968; Urban, 1969). A recent report concludes that the outer membrane also fuses seconds before the inner membrane in yeast (Melloy et al., 2007); however, as is explained below, the underlying experimental strategy used in those studies is flawed.
Apart from the importance of microtubules for nuclear congression, there is little information on the cytoskeleton in zygote formation (Hasek et al., 1987). Septins have however been detected at the cell cortex near the waist of zygotes (Ford and Pringle, 1991; Kim et al., 1991).
Little is known regarding the possible independence of parental nuclear genomes during zygote genesis, although studies of fertilization in plants and mice indicate lack of intermixing for at least several generations (Odartchenko and Keneklis, 1973; Rechsteiner and Parsons, 1976; Gleba et al., 1987; Brandriff et al., 1991; Callimassia et al., 1994; Mayer et al., 2005). The relevant genetic and cytological data for diploid yeast are not obviously all in agreement with each other (Kadyk and Hartwell, 1992; Haber and Leung, 1996; Jin et al., 2000; Lorenz et al., 2002).
This study inquires whether fusion of outer and inner nuclear membranes is simultaneous, identifies sequential intermediates in karyogamy, novel conditions that inhibit karyogamy, and examines the extent of genome intermixing which occurs upon karyogamy.
MATERIALS AND METHODS
Cells, Plasmids, and Drugs
Strains (Supplemental Table SI) were grown in complete synthetic medium (CSM) at room temperature, with 200 μg/ml adenine sulfate. Plasmids are in Supplemental Table SII.
Protein Tagging
Tagging of histone Htb2p and Sik1p was achieved by generating PCR fragments based on pFA6-mRFP-KanMX6 or pFA6-GFP(S65T)-KanMX6 and transforming by standard methods. The viability of the resulting haploids demonstrates the functionality of these integrants.
Two-Step Assay for Zygote Formation
Cells grown to OD600 2–4 were diluted 10 times in growth medium and allowed to grow for 2 more hours before mixing equal numbers of appropriate pairs at OD = 8, adding an equal volume of fresh medium, and reincubating with shaking for 1–2 h. One hundred microliter samples of the cell mixture were then applied to the surface of CSM plates and spread to cover twice the original surface area. After excess liquid had been absorbed (10–15 min), the plates were covered and incubated 3 h at room temperature. The plates included 2 μg/ml nocodazole (Sigma, St. Louis, MO) and 0.5% DMSO, as well as 0.1 M hydroxyurea, to block DNA synthesis. Cells were then rinsed off the plates with complete medium lacking sugar at room temperature, washed two times with medium lacking sugar (including 0.5% DMSO), and reincubated at OD = 1 in appropriate medium with 0.5% DMSO and 0.1 M hydroxyurea at 23 or 37°C, as appropriate. For quantitation of congression and karyogamy, samples were fixed by addition of an equal volume of 4% formaldehyde in PBS, washed, and examined. For examination by Deltavision (Applied Precision Instruments\, Issaquah, WA) they were processed as described in the next paragraph.
Real-Time Deltavision Microscopy
Samples of rapidly growing cells (or cells recovered from nocodazole plates) were mixed and sedimented. One-microliter aliquots of the pellet were applied to 1.5% agarose pads including CSM and additives of interest. After overlaying a coverslip and sealing with petroleum jelly, they were examined at 23°C (unless specified otherwise) using a 100× oil immersion objective without binning (Olympus, Melville, NY; UPlanApo 100×/1.40; ∞/0.17/FN26.5). Images were deconvolved using Softworks (Olympus) and processed minimally. A minimum of 20 cells was observed for each condition, and the selected illustrations are representative of the large majority. Brightfield images are in blue.
Photobleaching
Samples on agarose pads were studied with a Zeiss 510 confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY). Typically, squares [0.2–0.5μ]2 were bleached ∼80% at 50% laser intensity and then were imaged at 5–15-s intervals with a Plan-Apochromat 100× oil-immersion objective (NA 1.4), 1% laser intensity, using the acquisition software LSM510 (Carl Zeiss MicroImaging).
RESULTS
Overview of Karyogamy
When cells treated with α-factor develop a shmoo, the nucleus elongates and becomes highly polarized, with the SPB at the apex toward the shmoo tip (Figure 1, B and C). Chromatin fills the apical volume, and the nucleolus is at the distal extremity (Stone et al., 2000).
After cell fusion and nuclear congression, the point of contact of the nuclei (“nexus”), at or immediately adjacent to the SPB (Figure 1, D and E), persists for 10–30 min. In crosses between strains that express tagged SPB proteins (Spc42p-mRFP and Spc42p-CFP) one can see that the two colored foci remain distinct (Figure 1F). Although the nexus is present—and the inner membrane and outer membrane fuse (see below)—the nucleoli remain at the opposed ends of the nucleus (Figure 1, D and E). After dilation of the nexus, the SPB disengages from one face of the NE (Figure 1D).
In crosses in which a tagged nucleolar protein (Gar1p, Sik1p; or histone Htb2p) is contributed by one parent, these proteins gradually relocate to the trans nucleus, with the first trans signal becoming detectable after nuclear contact, but before the nexus is visibly dilated (Figure 1, D and E). Because no such signal is detected in the trans nucleus in crosses of kar1-1 (Figure 1G), which inhibits nuclear congression (Vallen et al., 1992), the trans signal results from intranuclear shuttling of these proteins. Moreover, these observations show that the production of new fluorescent copies of these proteins is negligible over the time period studied. We therefore use the arrival of the tagged nucleolar proteins in the trans nucleus as a temporal landmark.
Mechanism of NE Fusion: HMG1-Green Fluorescent Protein
Investigation of the mechanism and path by which NE membrane proteins such as HMG CoA Reductase 1 (HMG1) access the trans-NE is complicated because known proteins of the outer membrane are also found throughout the ER. On cell–cell fusion, transfer therefore could involve a combination of ER-to-ER fusion and NE-to-NE fusion.
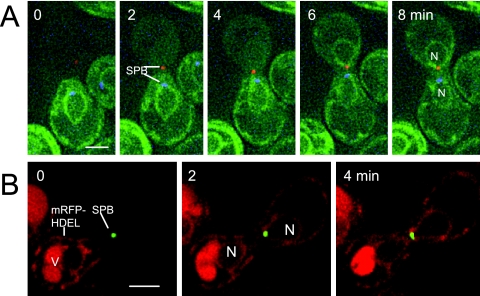
In crosses in which HMG1-green fluorescent protein (GFP; Profant et al., 2000; Wiederkehr et al., 2003) is contributed by one parent, we observe that the labeled nucleus first becomes somewhat pear-shaped (reminiscent of the impact of α-factor; Figure 1B). An intriguing focal discontinuity in the HMG1-GFP signal then appears at the apex of the NE that is oriented toward the midline (Figure 2A). Two to 4 min later, visible transfer of HMG1-GFP to the trans-NE is detected. When transfer begins, the signal appears to emerge from the apical discontinuity (Figures 2, A, B, and B′ and Supplemental Figure S1). It is striking that little or no signal is seen in the trans cortical ER and that the labeled elements of the cis cortical ER end abruptly just before the midline. Thus, transfer of HMG1-GFP is not primarily via the cortical ER.
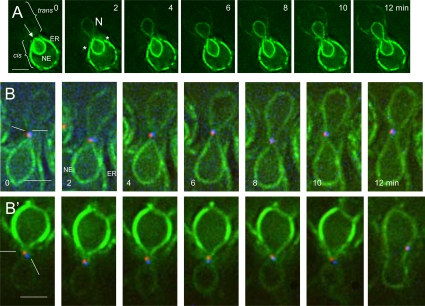
Figure 2.
Transit of HMG1-GFP. (A) Time course of transfer of HMG1-GFP from (ATY1529) to a nonfluorescent recipient (ATY2112). Note the parting of the apex of the cis-NE (arrow), followed by the appearance of signal in the trans-NE before it acquires equal intensity in the trans-cortical ER. The increase in intensity of HMG1-GFP in the trans-NE is gradual. See Supplemental Figure S2 for a further example. The asterisks indicate the abrupt interruption of the cortical ER signal. (B and B′) Crosses between a strain expressing HMG1-GFP and Spc42p-mRFP (ATY3525) with a strain expressing Spc42p-CFP (ATY1455). Note the position of the SPB sentinels of distinct parentage during HMG1-GFP transfer. In many images they flank the axis of transfer.
A further surprise is that the transfer of HMG1-GFP is gradual, i.e., although the entire perimeter of the trans-NE becomes labeled simultaneously, its intensity increases progressively over several minutes before equaling that of the cis-NE. A simple way to rationalize this metered flux of HMG1-GFP (as well as the inner membrane protein, GFP-Prm3p, and the protein of the nuclear periphery, GFP-Esc1p;Hattier et al., 2007; see below) is to recognize that it must pass through the focal nexus, whose surface area is certainly very small. For HMG1-GFP, as well as GFP-Prm3p and GFP-Esc1p, photobleaching shows that bleaching of one fourth to one third of the perimeter of the NE is followed by recovery with a t1/2 of ∼3 s (Supplemental Figure S2).
When nuclei establish contact, the parental SPBs do not fuse (Figure 1F). To position the path of transfer of HMG1-GFP relative to the SPBs, we have studied crosses in which each parent expresses a distinct form of Spc42p and one parent expresses HMG1-GFP. Figure 2, B and B′, show that the two “sentinel” SPBs remain adjacent to each other during HMG1-GFP transfer and consistently continue to be distinguishable well after karyogamy. Many images suggest that they bracket the site of cis-trans flux of HMG1-GFP. Judging from EM studies, the SPB encounters occur at their “half-bridges” (Byers and Goetsch, 1975; Melloy et al., 2007).
Mechanism of NE Fusion: Transfer of Outer Membrane Proteins, Inner Membrane Proteins, and NPCs Occur Sequentially
To learn whether outer and inner membrane fusion are simultaneous, we have performed a set of crosses in which membrane markers are followed in parallel with a nucleolar marker (Sik1p-mRFP), which indicates the establishment of nucleoplasmic continuity. Crosses between strains that express both HMG1-GFP and Sik1p-mRFP show that HMG1-GFP begins to transfer when the nexus is established, 13.4 ± 5.4 min before transfer of the nucleolar marker is detected (Figure 3A).
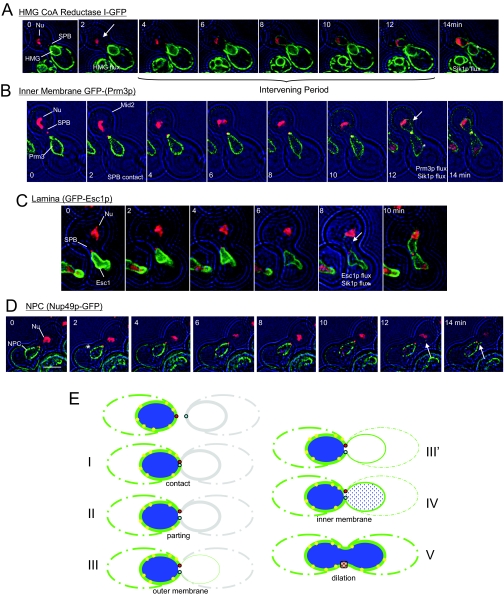
Figure 3.
Outer membrane fusion precedes fusion of the inner membrane and transfer of NPCs. (A) Transfer of HMG1-GFP compared with Sik1p-mRFP (and Spc42p-mRFP). Note that the HMG1-GFP is detected in the trans-nucleus long before Sik1p-mRFP (asterisk, arrow; ATY1917 × ATY1528). (B) Transfer of the tagged inner membrane protein, GFP-Prm3p, compared with Sik1p-mRFP (and Spc42p-mRFP). GFP-Prm3p and Sik1p-mRFP are detected in the trans-nucleus at essentially the same time (asterisk, arrow). The cell that expresses the mRFP-tagged proteins also expresses Mid2p-GFP (ATY1919 × ATY3149). Note: GFP-Prm3p was not preinduced. (C) Transfer of the tagged protein of the periphery of the nucleoplasm, GFP-Esc1p, compared with Sik1p-mRFP (and Spc42p-mRFP). GFP-Esc1p and Sik1p-mRFP are detected in the trans-nucleus at approximately the same time (asterisk, arrow; ATY1917 × ATY1550). (D) A cell expressing Sik1p-mRFP and Spc42p-mRFP (ATY1917) was crossed with a cell expressing Nup49p-GFP (ATY2226). Sik1p-mRFP arrives in the trans-nucleus well before the NPCs (asterisk), and the tagged NPCs invade the trans-NE (arrow) when dilation of the nexus is seen. Dilation of the nexus is accompanied by separation of the tagged SPBs from one face of the NE. Note: Experiments that follow a mRFP-tagged inner membrane protein along with HMG1-GFP also indicate initial fusion of the outer membrane, but the mRFP signal is not sufficiently strong to allow definitive imaging. (E) Summary diagram of the sequential events of NE fusion. The steps are (I) SPB contact, (II) appearance of the apical discontinuity of the NE, (III) initial transfer of outer membrane proteins, (III′) continued transfer of outer membrane and spreading of the trans-NE signal to the cortical ER, (IV) transfer of inner membrane proteins and the lamina, as well as nucleoplasmic continuity, and (V) visible dilation of the nexus, transfer of NPCs and disengagement of the SPB from one face of the NE. There are obvious delays between steps III and IV and between IV and V, suggesting that, at the molecular level, these intervening periods allow appropriate preparations for the succeeding events. HMG1-GFP, green; NPCs, yellow.
By contrast to HMG1-GFP, transfer of the inner membrane protein, GFP-Prm3p, occurs well after the nexus has been established, essentially coincident with the first detection of Sik1p-mRFP in the trans nuclear volume (Figure 3B). Parallel studies demonstrate that transfer of tagged Esc1p slightly precedes first detection of the nucleolar signal in the trans compartment (Figure 3C). Transfer of the nucleolar marker is detected 5.0 ± 5.6 min before visible dilation of the nexus. Thus, although there is variability in exact timing, outer membrane fusion precedes fusion of the inner membrane and dilation of the nexus occurs several minutes after inner membrane fusion.
NPCs span the inner membrane and outer membrane of the NE, and previous studies indicate that nucleoporins can transfer upon karyogamy as part of intact NPCs (Belgareh and Doye, 1997; Bucci and Wente, 1997). To establish the timing of their transfer relative to dilation of the nexus, we have crossed strains that express Nup49p-GFP with strains that do not express this tagged protein. Obvious transfer occurs after dilation of the nexus, when the SPB has disengaged from one face of the NE. Interestingly, although several NPCs can remain adjacent to each other, one often finds what appear to be single tagged NPCs deep in trans-NE territory.
Figure 3E summarizes the steps that accomplish karyogamy.
Mechanism of NE Fusion: Interruption in Known Mutants
We have followed pairs of strains what express either Nup49p-GFP or the tagged histone, Htb2p-mRFP, in order to evaluate congression and fusion (Table 1). As an indicator of the extent to which karyogamy is inhibited, we calculate an arrest index (% of cells with nuclei in contact divided by the % of cells with fused nuclei).
Table 1.
Evaluation of congression and fusion
| Separate (%) | Contact (%) | Fused (%) | Arrest indexa | Strain names | |
|---|---|---|---|---|---|
| One-step crosses, 3 hr 23°C | |||||
| wt × wt | 7 ± 4 | 22 ± 4 | 78 ± 8 | ∼0.3 | ATY3405 × ATY3358ATY2835 × ATY2289 |
| prm3Δ × prm3Δ | 4 ± 3 | 95 ± 2 | 2 ± 2 | ∼47 | ATY3130 × ATY3131 |
| wt [pHAC1i] × wt [pHAC1i] | 10 ± 6 | 9 ± 6 | 82 ± 12 | ∼0.1 | ATY3528 × ATY3529 |
| Two-step crosses, 3 h nocodazole + 2 h 23°C | |||||
| wt × wt | 37 ± 12 | 22 ± 4 | 41 ± 10 | ∼ 0.5 | ATY3405 × ATY3358 |
| wt × wt, 2 mM DTT | 51 ± 9 | 47 ± 10 | 0.5 ± 1 | ∼50 | ATY3405 × ATY3358 |
| ire1Δ × ire1Δ | 42 ± 7 | 24 ± 6 | 34 ± 8 | ∼0.7 | ATY3474 × ATY3476 |
| ire1Δ × ire1Δ, 2 mM DTT | 39 ± 6 | 58 ± 7 | 2 ± 0.1 | ∼30 | ATY3474 × ATY3476 |
| wt × wt, cycloheximide (0.1 mM) | 39 ± 8 | 56 ± 7 | 5 ± 3 | ∼10 | ATY2835 × ATY2289 |
| wt × wt, nocodazole (15 μg/ml) | 95 ± 5 | 3 ± 4 | 1 ± 0.7 | NA | ATY2835 × ATY2289 |
| wt × wt, latrunculin A (1.25 μM) | 79 ± 3 | 9 ± 2 | 7 ± 5 | NA | ATY2835 × ATY2289 |
| Two-step crosses: 3 h nocodazole + 2 h 37°C | |||||
| wt × wt | 27 ± 9 | 29 ± 3 | 44 ± 6 | ∼0.6 | ATY2835 × ATY2289 |
| sec1-1 × sec1-1 | 43 ± 3 | 15 ± 3 | 42 ± 5 | ∼0.3 | ATY3871 × ATY3872 |
| sec18-1 × sec18-1 | 12 ± 5 | 80 ± 11 | 12 ± 3 | ∼8 | ATY2538 × ATY2138 |
| cdc12-6 × cdc12-6 | 22 ± 11 | 43 ± 15 | 35 ± 6 | ∼1.2 | ATY3456 × ATY3458 |
| cdc48-3 × cdc48-3 | 32 ± 6 | 16 ± 8 | 52 ± 7 | ∼0.3 | ATY2229 × ATY2632 |
a The arrest index is calculated as the percent of zygotes in which nuclei contact each other divided by the percent in which they have fused. NA, not applicable.
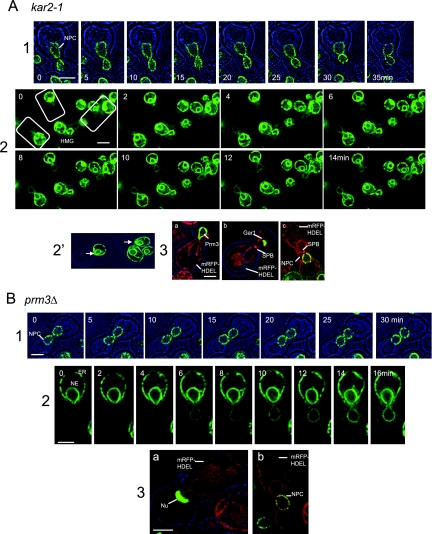
To investigate the mechanisms of NE fusion, we have followed the transfer of NE and nucleoplasmic markers in crosses between mutants in which karyogamy is strongly interrupted. In Figure 4, A (kar2-1) and B (prm3Δ), the first panels illustrate the distribution of Nup49p-GFP and document the establishment and persistence of nuclear contact. The second panels show the transfer of HMG1-GFP. In the prm3Δ cross, as for wild type, the trans-NE becomes labeled well before the cortical ER. Interestingly, this is somewhat less obvious for kar2-1, perhaps signifying that this mutation does not allow HMG1-GFP transfer via the nexus. The third panels show that the inner membrane does not fuse even well after nuclear contact has been established, as judged by monitoring the distribution of markers of the inner membrane (GFP-Prm3p), nucleolus (Gar1p-GFP), or Nup49p-GFP. For this purpose, these labels are introduced by a single parent.
Figure 4.
NE Fusion in kar2-1 and prm3Δ crosses. (A) kar2-1 × wt. (1) Nuclear encounter of kar2-1 and wt, which express Nup49p-GFP (ATY1713 × ATY3359). (2) Transfer of HMG1-GFP from wt to kar2-1. Note the more uniform timing of arrival of signal in the trans-NE and trans-cortical ER than in wt. (2′) The parting of the apex of the cis-NE in the same preparation (ATY3197 × ATY1528). Unlike wt crosses, when HMG1-GFP arrives in the trans-NE of kar2-1, its intensity in the trans-ER is nearly equal to that of the trans-NE. Thus, arrival of HMG1-GFP in the trans-NE could be via the cortical ER in kar2-1 crosses. (3) To learn whether the inner membranes fuse in kar2-1× wt crosses, we have performed experiments in which one of the parental strains expresses GFP-Prm3p, Gar1p-GFP, or Nup49p-GFP (which would be expected to transfer only if both membranes fused). In each case, the kar2-1 cell expresses mRFP-HDEL. Crosses were examined after 3 h to follow GFP-Prm3p: panel a, ATY3197 × ATY3149; a nucleolar protein, Gar1p-GFP: panel b, ATY3197 × ATY3004; or tagged NPCs, Nup49p-GFP: panel c, ATY3197 × ATY1916. As shown, none are transferred. The partners that express Gar1p-GFP or Nup49p-GFP also express Spc42p-mRFP. ATY1916 additionally expresses Mid2p-GFP. GFP-Prm3p itself concentrates at the nexus (panel a). (B) prm3Δ× prm3Δ. (1) Encounter of two prm3Δ strains that express Nup49p-GFP (ATY3130 x ATY3131). (2) Transfer of HMG1-GFP (ATY3277 × ATY2782). Note that the intensity of the trans-NE signal generally exceeds that of the trans-cortical ER and note the focal gap. As in wt × wt crosses, during arrival of HMG1-GFP in the trans-cell, the intensity in the NE exceeds that of the cortical ER. (3) prm3Δ x prm3Δ crosses were examined after 3 h to learn whether nucleolar Gar1p-GFP (a: ATY3216 × ATY3298) or tagged NPCs (b: ATY3216 × ATY3131) are transferred. The MATa partner expresses mRFP-HDEL in both cases. Note the lack of transfer in each case.
Mechanism of NE Fusion: New Mutants/Inhibitors
We developed an assay in which cells first fuse and then arrest reversibly before nuclear congression, due to the presence of low concentrations of nocodazole (Figure 5A, panel 1). At this point, soluble proteins (GFP, dsRed) and tagged ribosomes (Rps3p-GFP) have fully equilibrated (not shown). On removal of nocodazole and reincubation, congression and karyogamy occur (Figure 5A, panel 2). This is the only assay for karyogamy that makes it possible to assess the impact of conditional mutations or agents that also affect earlier steps in zygote formation.
Figure 5.
NE fusion assay. (A) Two-step assays to identify factors required for karyogamy. (1) Two strains that express Htb2p-mRFP (ATY2835 × ATY2289) were crossed for 3 h on a nocodazole plate and examined. Note the designated prezyogotes in which the nuclei are separate. Ch, chromatin. (2) Congression and karyogamy of the wt strains illustrated in panel 1 after recovery from a nocodazole plate and reincubation at 23°C. Individual zygotes were classified as showing separate nuclei (blue), nuclei in contact with each other (pink), or fused nuclei (yellow). Some wt crosses yield a higher efficiency of fusion and more rapid kinetics than this example. (B) Karyogamy: novel conditions cause arrest. (1) Time lapse of two sec18-1 strains that express Nup49-GFP (ATY2538 × ATY2138) which were crossed on a nocodazole plate at 23°C, recovered, and reincubated at 37°C. (2) Time lapse of two wt strains that express Nup49p (ATY3405 × ATY3359). The strains were crossed on a nocodazole plate and reincubated with 2 mM DTT. (3) Two wt strains that express Nup49p (ATY3405 x ATY3359) were crossed on a nocodazole plate and reincubated with 100 μg/ml cycloheximide for 2 h. (C) Evaluation of inner membrane fusion in novel conditions. To learn whether inner membrane fusion occurs in sec18-1× sec18-1 crosses, in the presence of DTT or cycloheximide, we have followed a copy of GFP-Esc1p that is preinduced from a galactose-inducible promoter and silenced by the glucose in the nocodazole plates. We observe that GFP-Esc1p is restricted to the cis-nucleus in each case. Thus, inner membrane fusion does not occur. (1) sec18-1 strains that express either GFP-Esc1p or Htb2p-mRFP (ATY3522 × ATY3384) were crossed, recovered from a nocodazole plate, and reincubated at 37°C for 2 h. ATY3522 was preinduced. Note that both the green signal and the red histone are confined to one nucleus, indicating lack of inner membrane fusion. V, vacuole; Ch, chromatin. (2) Wild-type strains that express GFP-Esc1p or Htb2p-mRFP (ATY2102 × ATY2289) were crossed, recovered from a nocodazole plate, and reincubated for 2 h with 2 mM DTT. Note that each signal is confined to one nucleus. ATY2102 was pregrown overnight in galactose medium, and the cross was conducted in glucose medium. (3) As in panel 2, with the 2-h chase in the presence of 100 μg/ml cycloheximide. Note 1: To determine whether the outer membrane can fuse in sec18-1 crosses or in the presence of DTT or cycloheximide, one might express HMG1-GFP in one parent and follow its distribution through time using two-step protocols. We observe that HMG1-GFP already surrounds both nuclei upon removal of nocodazole. It therefore is not possible to judge whether flux through the nexus occurs. Note 2: Expression of GFP-Esc1p often distorts the contour of the NE. Cell growth is only slightly slowed. Note 3: In experiments equivalent to those shown, we have followed Htb2p-GFP and observe that it also does not pass between nuclei after arrest in sec18-1 crosses or after treatment with DTT or cycloheximide.
The experiments summarized in Table 1 show that congression is inhibited by nocodazole (as expected) and by latrunculin A, implying a role for the actin cytoskeleton. Congression and karyogamy are not inhibited in crosses between a pair of temperature-sensitive (ts) septin mutants (cdc12-6) or in crosses between strains that carry ts mutations in the AAA ATPase, Cdc48p, which is required for homotypic ER fusion (Latterich et al., 1995; Cao et al., 2003). By contrast, karyogamy is inhibited in sec18-1× sec18-1 crosses, but not in crosses between sec1-1 strains that are temperature sensitive for exocytosis (Carr et al., 1999), indicating that one or more Snares is required. Interestingly, after establishment of contact between the nuclei in sec18-1 crosses, the shape of the nuclei becomes progressively more complex, perhaps because vesicle delivery to the NE/ER remains blocked. Karyogamy is also inhibited upon inclusion of cycloheximide or the reducing agent, dithiothreitol (DTT; Figure 5B and Supplemental Figure S4), which interrupts ER disulfide bond formation and therefore is expected to sequester Kar2p (Bernales et al., 2006).
The impact of DTT is not a consequence of the unfolded protein response (UPR; Bernales et al., 2006), because 1) induction of such a response in the absence of unfolded proteins in the cisternal space of the ER allows karyogamy to occur, e.g., upon expression of an active (spliced) form of the mRNA encoding the HAC1 transcription factor (HAC1i; Cox and Walter, 1996) and 2) DTT is inhibitory in crosses between ire1Δ strains, which cannot generate a UPR (Sidrauski and Walter, 1997; Table 1). We therefore propose that sequestration of folding equipment within the ER accounts for the impact of DTT. As attempts at overexpression of Kar2p alone do not protect against the effect of DTT (Supplemental Figure S4), it is likely that multiple factors are sequestered, more than one of which is required.
As for kar2-1 and prm3Δ crosses (Figure 4, A and B), Figure 5C illustrates the arrest of karyogamy in sec18-1 crosses and after treatment with DTT or cycloheximide and shows that the inner membrane does not fuse.
Transfer of ER/NE Content Before Nuclear Contact
In the establishment of ER/NE continuity, different markers are transferred at different times. Thus, content markers (GFP-HDEL, mRFP-HDEL, Figure 6) are actually transferred before nuclear contact (n = 20), whereas HMG1-GFP transfer begins only when the nexus is established. This asynchrony leads us to suspect, as at the bud neck (Luedeke et al., 2005), that the cortical ER does establish cis-trans continuity (allowing flux of tagged HDEL), but allows only a subset of proteins to pass. This is consistent with photobleaching studies that show little cis-trans continuity of HMG1-GFP during zygote formation (Supplemental Figure S4).
Figure 6.
Transfer of content before membrane proteins. (A) Cross between a strain that expresses Spc42p-mRFP and a strain that expresses GFP-HDEL and Spc42p-CFP (ATY1774 × ATY3365). mRFP-HDEL transfers before SPB contact. (B) Cross between a strain that expresses Spc42p-GFP (ATY1454) and a strain that expresses mRFP-HDEL (ATY3196). Note redistribution of the HDEL signal before nuclear contact. V, Vacuole.
Do Parental Genomes Intermix after Karyogamy?
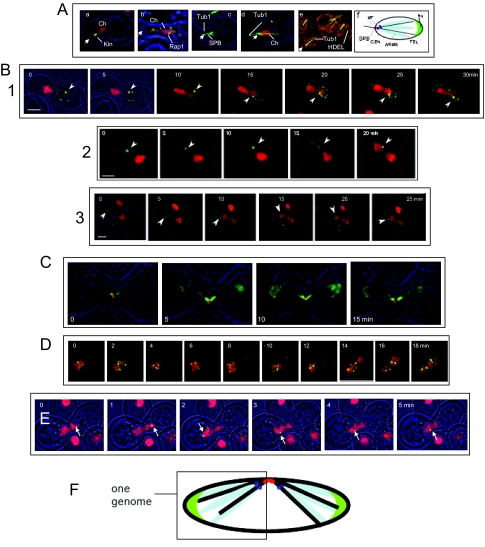
On treatment with mating factor, the genome becomes spatially polarized, with the SPB and centromeres near the shmoo tip and the nucleolus at the distal end of the nucleus. Telomere-associated foci are widely distributed and conspicuous microtubule cables run from the SPB across the nucleoplasm and through the nucleolus to terminate at the inner surface of the NE (Figures 1B and 7A, a–f). The SPB-nucleolus axis is maintained through karyogamy and the nucleoli remain separate (Figure 1, D and E).
Figure 7.
Spatial separation of parental genomes in prezygotes. (A) Polarization of the haploid genome. MATa cells after 3-h exposure to α-factor. Cell outlines (brightfield) are in blue. In panels a–e the approximate position of the SPB is designated by an arrowhead. (a) Htb2p-mRFP-tagged chromatin compared with a pan-kinetochore marker, Cse4p-GFP (Kin). Note that kinetochores are at the apex (ATY3083); (b) the telomere-associated protein, Rap1p-GFP and Htb2p-mRFP-tagged chromatin (Ch). Note the broad distribution of Rap1p (ATY3138); (c) Tub1p-GFP and Spc42p-mRFP (SPB). Note the microtubule cables extending both toward the shmoo tip and into the interior of the nucleus (ATY3034); (d) Tub1p-GFP and Htb2p-mRFP. Note that the microtubule cables in the nucleus extend as far as the red dots that mark rDNA (*) (ATY2524); (e) Tub1p-GFP and mRFP-HDEL. Note that the microtubule cables in the nucleus contact the NE (ATY3342). (f) Model of genome polarization. Because rDNA is organized around the right arm of chromosome XII, chromosome XII may fold back from the nucleolus toward the interior of the nucleus (Loidl, 2003). (B) Crosses between strains that express lacO-tagged loci and lacI-GFP with strains that express Htb2p-mRFP. In each case, the presence of Htb2p-mRFP in the trans volume shows that nuclear fusion has occurred. In (1) the recipient strain also expresses Nup49p-GFP. (1) The lacO-repeat is 11 kb from TELXIVL (ATY1460 × ATY2937). (2) The lacO-repeat is 10 kb from CENIII (ATY1456 × ATY2289). Continuity between the two domains of the nucleus appears interrupted because the focal plane was adjusted to capture the lacO signal. (3) The lacO repeat is adjacent to rDNA (ATY2597 × ATY2289). Note in each case that the tagged locus can move but remains in the parental domain from which it originated. (C) Two bundles of microtubules extend toward the two parental nuclei after karyogamy. Cross between cells that express Tub1p-GFP and Spc42p-mRFP (ATY3004) or Gar1p-GFP (ATY3220). Note the persistence of intranuclear microtubule bundles long after the flux of Gar1p-GFP is detected. (D) An ARS plasmid can pass cis-to-trans after karyogamy. A cell expressing Htb2p-mRFP, a lacO-tagged ARS plasmid, and lacI-GFP (ATY2549) was crossed with an unlabeled wt cell (W303). Note the entry of Htb2p-mRFP, which is followed by the plasmid. (E) Loss of kinetochore function allows chromosome motion. Two ndc10-1 mutants were crossed, one of which carries a tetO insertion adjacent to CENV (Romao et al., 2008; ATY3768) and one of which expresses Htb2p-mRFP (ATY2784). After 2 h at room temperature to initiate zygote formation, they were shifted to 37°C and examined over 1 h, focusing on the ∼50% cells in which the labeled locus was no longer at the apex of the nucleus, indicating that Ndc1p function had been lost. Transit from one to the other domain was seen in ∼20% of these zygotes examined over 30 min. (F) Model indicating genome territoriality after karyogamy.
To inquire whether the two parental genomes intermix, we crossed cells that express individual lacO-tagged loci (and lacI-GFP) with strains that express Htb2p-mRFP. The lacO inserts are near CENIII, near TEL XIVL, or adjacent to rDNA. In each case, upon karyogamy, the tagged loci can move but remain within the nuclear volume from which they originated for as long as the nuclei retain their elongate shape (Figure 7B, 1–3). Moreover, two separate intranuclear bundles of microtubules are present, each directed toward one of the contributing nuclei (Figure 7C).
Restriction of genome intermixing could reflect their tethering to SPBs (Loidl, 2003; Kitamura et al., 2007). To learn whether such association is required for restriction, we have performed crosses in which one of the mating partners carries a lacO-tagged ARS plasmid (which lacks a centromere; Velmurugan et al., 2000). In this case tagged loci invade the trans nucleoplasm (Figure 7D). Further evidence that genome tethering is required comes from observations of strains that carry ts mutations in the kinetochore protein, Ndc10p (Goh and Kilmartin, 1993). As shown in Figure 7E, in crosses between an ndc10-1 strain that carries a tetO repeat near CENV (and TetR-GFP) and a ndc10-1 partner that expresses Htb2p-mRFP, transit of the tagged locus can occur. In the protocol used, the cross is initiated at room temperature and the prezygotes are shifted to 37°C once cell fusion has occurred.
Figure 7F provides a diagram of genome organization after karyogamy.
DISCUSSION
Cell fusion is a normal occurrence for selected cell types and has become an issue of more general interest in the context of experimental cell hybrid formation and stem cell biology (e.g., Harris, 1988; Ogle et al., 2005; Jaenisch and Young, 2008). During fertilization in certain organisms (e.g., marine algae, sea urchins) nuclei fuse with each other, as opposed to first disassembling their NEs, followed by assembly of a single NE around the composite genome. Unlike yeast, many cell types do not have SPBs; however, the present observations support the idea that the importance of SPBs for nuclear fusion is primarily to bring nuclei together, rather than being functionally central for membrane fusion per se.
In Saccharomyces cerevisiae, after nuclear congression, the sequence of events that accomplishes nuclear fusion begins with the focal opening of a gap in the HMG1-GFP signal at the site of imminent fusion. When the HMG1-GFP signal is first detected in the trans-NE, the cortical ER signal is still confined to the cis cell, making it highly likely that transfer occurs only via the nexus. The narrow dimensions of the nexus along with the SPBs that appear to bracket it could account for the slow tempo of transfer. Inner membrane continuity is not established until later, essentially at the same time as for the lamina (Esc1p) and entry of nucleolar proteins. Still later dilation of the waist of the nucleus could result from removal of yet-unidentified structural elements that also define its initial pear-like shape.
A recent study of karyogamy has concluded that the outer membrane of the yeast NE fuses ∼30 s before the inner membrane (Melloy et al., 2007). These conclusions are based on an EM tomogram and the timing of GFP-HDEL transfer versus nucleoplasmic markers, after correction. Because tagged HDEL transfers before nuclear contact, we consider it not to be a suitable tracer. In our experience, the delay between fusion of the outer membrane and inner membrane is at least 10 min.
The present observations divide karyogamy into five steps: 1) SPB contact, 2) appearance of the apical discontinuity of the NE, 3) transfer of outer membrane proteins, 4) transfer of inner membrane proteins and the lamina, as well as nucleoplasmic continuity, and 5) visible dilation of the nexus, transfer of NPCs and disengagement of the SPB from one face of the NE. There are obvious delays between steps 3 and 4 and between 4 and 5, suggesting that these intervening periods allow appropriate molecular preparations for subsequent events.
As mentioned in the Introduction, the outer membranes and inner membranes of the cis-NE might first fuse with each other (and the outer and inner membranes of the trans-NE do the same). Subsequently, the two “cis-fused” NEs could fuse with each other (Figure 1A). This model has the unattractive property of possibly allowing momentary leakage of nuclear content to the cytoplasm. It nevertheless appears topologically equivalent to mitotic NE breakdown and reformation in higher eukaryotes and in half-open mitosis (e.g., Straube et al., 2005). Moreover, chromosomes can escape from the yeast nucleus during congression in kar1-1 (Dutcher, 1981). Supplemental Figure S5 presents observations inconsistent with this model.
Our investigation of crosses of kar2-1, prm3Δ and the impact of novel inhibitors of karyogamy (DTT, cycloheximide, inhibition of Sec18p) all show that inner membrane fusion is blocked. The requirement for Sec18p immediately situates karyogamy in the context of cytoplasmic membrane fusion events. Because Sec18p is a Snare disassembly factor, its involvement suggests that the Snare(s) involved in karyogamy (which occurs only once) also participate in other (preceding) ER membrane fusion events. Judging from the lack of involvement of the UPR, we attribute the impact of DTT to sequestration of folding factors of the ER/NE lumen.
The outer and inner membranes fuse sequentially. Two topologically distinct mechanisms are therefore likely to be involved. Fusion initiated by interaction of the cytoplasmic surfaces of the outer membranes could account for the Sec18p requirement, as for vesicle fusion. Subsequent contact between the luminal surfaces of the inner membrane could account for the involvement of proteins that are present in the ER/NE lumen such as Kar2p, Kar5p, and Kar8p. Because the critical feature of DTT treatment appears to be its disruption of ER homeostasis, translocon mutants could have a similar effect, either by compromising the folding of newly synthesized ER proteins or by blocking the removal of those that are not well folded. The role of Prm3p remains obscure, although its accumulation at the SPB (Figure 4A) is consistent with its participation in either step. Perhaps, like other proteins of the inner nuclear membrane, it also gains access to the outer membrane, where it would face the cytoplasm and therefore could be critical for outer membrane fusion.
The kinetics of transfer of tagged HDEL are reminiscent of studies of mitotic cells, in which arrival of this tracer in the cortical ER of the bud occurs before the arrival of membrane proteins (Luedeke et al., 2005). This transfer could be mediated by specialized ER elements or might result from ER–ER fusion being only transient, as in “kiss-and-run” exocytosis (e.g., Sokac and Bement, 2006). In any event, this transfer is exempt from the dramatic restriction of transfer of the membrane protein, HMG1-GFP.
The actin cytoskeleton participates in nuclear orientation during the mitotic cell cycle (Pearson and Bloom, 2004), but its importance for congression is not understood. As in the mitotic cell cycle, it is likely to be required for proper orientation of the nucleus. Consistent with this hypothesis, when latrunculin A is present after cell fusion, nuclei remain for extended periods with their SPBs and microtubule bundles pointing in seemingly arbitrary directions (Supplemental Figure S6).
Several soluble nucleoplasmic proteins and even an ARS plasmid redistribute upon karyogamy, by contrast to chromosomal loci in wild-type strains. Moreover, inactivation of kinetochore function also allows redistribution of chromosomal loci. Mere tethering of the haploid genome to the SPB does not predict persistence of cis-trans genome segregation after karyogamy, a situation which should inhibit recombination between the two contributing genomes during this period. We propose that the lack of coalescence of parental SPBs causes each SPB to continue to coordinate a single genome and thereby imposes spatial restrictions on the distribution of the corresponding chromosomes. So long as the restriction does not preclude random assortment of chromosomes during meiosis, it is not obvious that it would have genetic consequences.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. D. MacDonald for use of microscopic facilities; D. Donato, A. Feczko, M. Khan, M. Lam, L. Mattera, M. Mattera, F. Najm, S. Roy and Y. Zhang for help; and the following for materials: C. Antony (European Molecular Biology Laboratory, Heidelberg, Germany), L. Aragon (Imperial College London, London, United Kingdom), J. Brodsky (University of Pittsburgh, Pittsburgh, PA), J. Cooper (Washington University, St. Louis, MO), N. Dean (Stony Brook University, Stony Brook, NY), V. Doye (Institute Curie, Paris, France), S. Emr (Cornell University, Ithaca, NY), A. Franzusoff (University of Colorado, Denver, CO), S. Ferro-Novick (University of California, Berkeley, CA), S. Gasser (Miescher Institute, Basel, Switzerland), P. E. Gleizes (Université Paul Sabatier, Toulouse, France), E. Grote (Johns Hopkins University, Baltimore, MD), W. Huh (Seoul National University, Seoul, Korea), J. Kilmartin (University of Cambridge, Cambridge, United Kingdom), T. Lithgow (University of Melbourne, Melbourne, Australia), S. Michaelis (Johns Hopkins University), M. Rose (Princeton University, Princeton, NJ), E. O'Shea (Harvard University, Cambridge, MA), L. Pemberton (University of Virginia, Charlottesville, VA), W. Printz (National Insitutes of Health), T. Rapoport (Harvard University), K. Runge (Cleveland Clinic Foundation, Cleveland, OH), R. Schekman (University of California, Berkeley, CA), D. Shore (University of Geneva, Geneva, Switzerland), R. Sternglanz (Stony Brook University), S. Velmurugan (University of Texas, Austin, TX), P. Walter (University of California, San Francisco, CA), and A. Weil (Vanderbilt University, Nashville, TN). This work was supported by National Institutes of Health Grant P30 CA43703-12.
Abbreviations used:
- DTT
dithiothreitol
- HMG1
HMG-CoA reductase I
- NE
nuclear envelope
- NPC
nuclear pore complex
- SPB
spindle pole body
- TBP1
TATA-binding protein
- UPR
unfolded protein response
- Vac8p
vacuolar membrane protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1193) on April 15, 2009.
REFERENCES
- Baur T., Ramadan K., Schlundt A., Kartenbeck J., Meyer H. H. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J. Cell Sci. 2007;120:2895–2903. doi: 10.1242/jcs.010181. [DOI] [PubMed] [Google Scholar]
- Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Belgareh N., Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S., Papa F. R., Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Brandriff B. F., Gordon L. A., Segraves R., Pinkel D. The male-derived genome after sperm-egg fusion: spatial distribution of chromosomal DNA and paternal-maternal genomic association. Chromosoma. 1991;100:262–266. doi: 10.1007/BF00344160. [DOI] [PubMed] [Google Scholar]
- Bucci M., Wente S. R. In vivo dynamics of nuclear pore complexes in yeast. J. Cell Biol. 1997;136:1185–1199. doi: 10.1083/jcb.136.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callimassia M. A., Murray B. G., Hammett K. R., Bennett M. D. Parental genome separation and asynchronous centromere division in interspecific F1 hybrids in Lathyrus. Chromosome Res. 1994;2:383–397. doi: 10.1007/BF01552798. [DOI] [PubMed] [Google Scholar]
- Cao K., Nakajima R., Meyer H. H., Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- Carr C. M., Grote E., Munson M., Hughson F. M., Novick P. J. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K. Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1981;1:245–253. doi: 10.1128/mcb.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Gleba Y. Y., Parokonny A., Kotov V., Negrutiu I., Momot V. Spatial separation of parental genomes in hybrids of somatic plant cells. Proc. Natl. Acad. Sci. USA. 1987;84:3709–3713. doi: 10.1073/pnas.84.11.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P. Y., Kilmartin J. V. NDC10, a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., Leung W. Y. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc. Natl. Acad. Sci. USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. The analysis of malignancy by cell fusion: the position in 1988. Cancer Res. 1988;48:3302–3306. [PubMed] [Google Scholar]
- Hasek J., Rupes I., Svobodova J., Streiblova E. Tubulin and actin topology during zygote formation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1987;133:3355–3363. doi: 10.1099/00221287-133-12-3355. [DOI] [PubMed] [Google Scholar]
- Hattier T., Andrulis E. D., Tartakoff A. M. Immobility, inheritance and plasticity of shape of the yeast nucleus. BMC Cell Biol. 2007;8:47–64. doi: 10.1186/1471-2121-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. W., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Holaska J. M., Wilson K. L., Mansharamani M. The nuclear envelope, lamins and nuclear assembly. Curr. Opin. Cell Biol. 2002;14:357–364. doi: 10.1016/s0955-0674(02)00329-0. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jin Q. W., Fuchs J., Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 2000;113(Pt 11):1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Haarer B. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Kitamura Y., Tanaka T. U. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M., Frohlich K. U., Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Loidl J. Chromosomes of the budding yeast Saccharomyces cerevisiae. Int. Rev. Cytol. 2003;222:141–196. doi: 10.1016/s0074-7696(02)22014-8. [DOI] [PubMed] [Google Scholar]
- Longo F. J., Anderson E. The fine structure of pronuclear development and fusion in the sea urchin, Arbacia punctulata. J. Cell Biol. 1968;39:339–368. doi: 10.1083/jcb.39.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., Fuchs J., Trelles-Sticken E., Scherthan H., Loidl J. Spatial organisation and behaviour of the parental chromosome sets in the nuclei of Saccharomyces cerevisiae× S. paradoxus hybrids. J. Cell Sci. 2002;115:3829–3835. doi: 10.1242/jcs.00066. [DOI] [PubMed] [Google Scholar]
- Luedeke C., Frei S. B., Sbalzarini I., Schwarz H., Spang A., Barral Y. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 2005;169:897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L., Rose M. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In: Pringle J. R., Broach J. R., Jones E. W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Mayer R., Brero A., von Hase J., Schroeder T., Cremer T., Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005;6:44–66. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy P., Shen S., White E., McIntosh J. R., Rose M. D. Nuclear fusion during yeast mating occurs by a three-step pathway. J. Cell Biol. 2007;179:659–670. doi: 10.1083/jcb.200706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk J. N., Bloom K. Microtubule dynamics in the budding yeast mating pathway. J. Cell Sci. 2006;119:3485–3490. doi: 10.1242/jcs.03193. [DOI] [PubMed] [Google Scholar]
- Odartchenko N., Keneklis T. Localization of paternal DNA in interphase nuclei of mouse eggs during early cleavage. Nature. 1973;241:528–529. doi: 10.1038/241528a0. [DOI] [PubMed] [Google Scholar]
- Ogle B. M., Cascalho M., Platt J. L. Biological implications of cell fusion. Nat. Rev. Mol. Cell Biol. 2005;6:567–575. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- Pearson C. G., Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Profant D. A., Roberts C. J., Wright R. L. Mutational analysis of the karmellae-inducing signal in Hmg1p, a yeast HMG-CoA reductase isozyme. Yeast. 2000;16:811–827. doi: 10.1002/1097-0061(20000630)16:9<811::AID-YEA579>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Parsons B. Studies on the intranuclear distribution of human and mouse genomes and formation of human-mouse hybrid cells. J. Cell. Physiol. 1976;88:167–179. doi: 10.1002/jcp.1040880206. [DOI] [PubMed] [Google Scholar]
- Romao M., Tanaka K., Sibarita J. B., Ly-Hartig N. T., Tanaka T. U., Antony C. Three-dimensional electron microscopy analysis of ndc10-1 mutant reveals an aberrant organization of the mitotic spindle and spindle pole body defects in Saccharomyces cerevisiae. J Struct. Biol. 2008;163:18–28. doi: 10.1016/j.jsb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Sheehan M. A., Mills A. D., Sleeman A. M., Laskey R. A., Blow J. J. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J. Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sokac A. M., Bement W. M. Kiss-and-coat and compartment mixing: coupling exocytosis to signal generation and local actin assembly. Mol. Biol. Cell. 2006;17:1495–1502. doi: 10.1091/mbc.E05-10-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. L., Roux K. J., Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- Stone E. M., Heun P., Laroche T., Pillus L., Gasser S. M. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr. Biol. 2000;10:373–382. doi: 10.1016/s0960-9822(00)00413-9. [DOI] [PubMed] [Google Scholar]
- Straube A., Weber I., Steinberg G. A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 2005;24:1674–1685. doi: 10.1038/sj.emboj.7600644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P. The fine structure of pronuclear fusion in the coenocytic marine alga Bryopsis hypnoides Lamouroux. J. Cell Biol. 1969;42:606–611. doi: 10.1083/jcb.42.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E. A., Hiller M. A., Scherson T. Y., Rose M. D. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J. Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan S., Yang X. M., Chan C. S., Dobson M., Jayaram M. Partitioning of the 2-microm circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded rep protein distribution. J. Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Du Y., Pypaert M., Ferro-Novick S., Novick P. Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Mol. Biol. Cell. 2003;14:4770–4782. doi: 10.1091/mbc.E03-04-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.