Abstract
Obscurin is a multidomain protein composed of adhesion and signaling domains that plays key roles in the organization of contractile and membrane structures in striated muscles. Overexpression of the second immunoglobulin domain of obscurin (Ig2) in developing myotubes inhibits the assembly of A- and M-bands, but not Z-disks or I-bands. This effect is mediated by the direct interaction of the Ig2 domain of obscurin with a novel isoform of myosin binding protein-C slow (MyBP-C slow), corresponding to variant-1. Variant-1 contains all the structural motifs present in the known forms of MyBP-C slow, but it has a unique COOH terminus. Quantitative reverse transcription-polymerase chain reaction indicated that MyBP-C slow variant-1 is expressed in skeletal muscles both during development and at maturity. Immunolabeling of skeletal myofibers with antibodies to the unique COOH terminus of variant-1 demonstrated that, unlike other forms of MyBP-C slow that reside in the C-zones of A-bands, variant-1 preferentially concentrates around M-bands, where it codistributes with obscurin. Overexpression of the Ig2 domain of obscurin or reduction of expression of obscurin inhibited the integration of variant-1 into forming M-bands in skeletal myotubes. Collectively, our experiments identify a new ligand of obscurin at the M-band, MyBP-C slow variant-1 and suggest that their interaction contributes to the assembly of M- and A-bands.
INTRODUCTION
Obscurin-A (∼720 kDa) is the third and most recently discovered giant protein expressed in vertebrate striated muscle (Young et al., 2001). Like its fellow giants, titin and nebulin, it has a modular architecture composed of adhesion and signaling domains arranged in tandem (Russell et al., 2002; Fukuzawa et al., 2005; Kontrogianni-Konstantopoulos and Bloch, 2005). Its NH2 terminus contains repetitive immunoglobulin (Ig) domains that are 88-92 amino acids in length and joined without any apparent linker sequences (Young et al., 2001; Russell et al., 2002). In addition to the 59 Ig repeats present in its NH2 terminus, obscurin also contains two fibronectin-III (FnIII) domains, followed by a complex region consisting of four additional Ig domains, flanked by nonmodular sequences and several signaling domains, including an IQ motif, a Src homology (SH) 3 domain, and tandem Rho-guanine nucleotide exchange factor (Rho-GEF)/pleckstrin homology motifs, suggesting the protein's possible involvement in signaling pathways mediated by Ca2+ and Rho-GTPases. The extreme COOH terminus of the protein consists of two additional Ig repeats followed by a nonmodular region of ∼420 amino acids that contains several consensus phosphorylation motifs for extracellular signal-regulated kinase kinases.
Obscurin-A is encoded by the same gene cluster that produces obscurin-myosin light chain kinase (MLCK), a set of proteins that contain two serine-threonine kinase domains (Bang et al., 2001; Russell et al., 2002). Obscurin-MLCK may be expressed independently as smaller, alternatively spliced products that contain one (single MCLK; sMLCK) or both (tandem MLCK; tMLCK) kinase motifs or as part of the larger, ∼720-kDa form of obscurin-A, referred to as obscurin-B or giant MLCK (gMLCK; ∼860 kDa), in which the two MLCK motifs are arranged in tandem at the COOH terminus of the protein, replacing the nonmodular COOH terminus of obscurin-A (Russell et al., 2002; Fukuzawa et al., 2005; Borisov et al., 2008).
Unlike titin and nebulin, which are integral components of sarcomeres, obscurin is not present within sarcomeres but intimately surrounds them, primarily at the level of the M-band and Z-disk, where it is appropriately positioned to participate in their assembly and integration with other sarcoplasmic elements (Young et al., 2001; Kontrogianni-Konstantopoulos et al., 2003; Borisov et al., 2004; Kontrogianni-Konstantopoulos et al., 2006b; Bowman et al., 2007; Carlsson et al., 2008). Consistent with this, obscurin interacts with diverse protein partners located in distinct compartments within the cell, including members of the ankyrin superfamily, for example, small ankyrin 1 or Ank1.5, an integral component of sarcoplasmic reticulum (SR) membranes, Ran-binding protein 9, a scaffolding molecule that interacts with a variety of proteins, and the myofibrillar proteins titin and myomesin (Bang et al., 2001; Young et al., 2001; Bagnato et al., 2003; Kontrogianni-Konstantopoulos et al., 2003; Armani et al., 2006; Borzok et al., 2007; Bowman et al., 2008; Cunha and Mohler, 2008; Fukuzawa et al., 2008).
Using gain and loss of function approaches several laboratories, including ours, have begun to unravel the physiological roles of obscurin in muscle assembly and maintenance. Adenovirally mediated delivery of the nonmodular COOH terminus of obscurin-A in primary cultures of skeletal myotubes demonstrated that it is essential for the regular assembly and organization of sarcomeric myosin into A-bands (Kontrogianni-Konstantopoulos et al., 2004). Consistent with this, down-regulation of obscurin in primary cultures of skeletal myotubes or neonatal cardiocytes through small inhibitory RNA (siRNA) technology revealed a key role for obscurin in the formation and maintenance of sarcomeric A- and M-bands, and in the regular alignment of SR membranes around the contractile apparatus (Borisov et al., 2006; Kontrogianni-Konstantopoulos et al., 2006b). Depletion of obscurin in developing zebrafish embryos also resulted in marked disarray and failure of skeletal myofibrils to align laterally, disorganization of the SR membranes and abnormalities of cardiac structure and function (Raeker et al., 2006). Recently, a study by Bowman and colleagues assigned an additional role to obscurin through its Rho-GEF motif in the proper integration of the NH2 terminus of titin into the Z-disk (Bowman et al., 2008). In agreement with these observations, changes in the expression levels or mutations in the obscurin gene have been linked to the development of hypertrophic cardiomyopathy, which further support a critical role for obscurin in normal muscle development and physiology (Borisov et al., 2003; Arimura et al., 2007).
In the current study, we used adenovirally mediated gene transfer to overexpress the extreme NH2-terminal Ig domains of obscurin and examine their role in myofibrillogenesis. Our results indicate that overexpression of the second Ig domain of obscurin in primary cultures of skeletal myotubes results in the failure of two important structures to form, A-bands and their central M-bands. We provide evidence showing that this effect is mediated through the direct association of the Ig2 repeat of obscurin with a previously uncharacterized isoform of myosin binding protein-C slow, MyBP-C slow variant-1, a major component of the thick filaments with structural and regulatory roles (Flashman et al., 2004a; Oakley et al., 2007). MyBP-C slow variant-1 carries a unique COOH terminus, is abundantly expressed both during development and in adulthood, and preferentially localizes at the periphery of the myofibrillar M-band.
MATERIALS AND METHODS
Cultures of Skeletal Myotubes
Primary cultures of rat myotubes were prepared as reported previously (De Deyne et al., 1998). In brief, hindlimb muscles from postnatal day 1 rats were dissociated enzymatically and suspended at 106 cells/ml in DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) (Invitrogen). Cell aliquots (0.5 ml) were applied to sterile glass coverslips and supplemented with 1 ml of the same medium the next day. Medium was replaced 48 h later with medium containing 2 × 10−5 M cytosine arabinoside (Sigma-Aldrich, St. Louis, MO) to kill dividing cells. Cultures were infected with adenovirus 5 d after initial plating.
Generation of Recombinant Adenoviruses: Infections of Primary Cultures
NH2-terminal obscurin fragments containing amino acids 1-212 or 124-212 (XM_340807), encoding Ig repeats 1 and 2, or Ig repeat 2, respectively, were generated by polymerase chain reaction (PCR) amplification using RNA from adult rat skeletal muscle and the following set of primers: for Ig1/2: forward-1 (F1), 5′-ATGGACCACTCCTTC-3′, and reverse-2 (R2), 5′-TTAAGGTTCCGGGTGGTCC-3′; and for Ig2: forward-2 (F2), 5′-CCCACCTCCATTCGT-3′, and R2. The sense primers carried an XhoI recognition sequence and the antisense primers contained a HindIII site, for insertion into the pHcRed-C1 vector for production of red fluorescent fusion proteins (RFPs) (Clontech, Mountain View, CA). The pHcRed-C1-Obscurin-Ig1/2 and pHcRed-C1-Obscurin-Ig2 plasmids were subsequently used as templates for amplification of HcRed-Obscurin-Ig1/2 and HcRed-Obscurin-Ig2 by using the following primers: pHcRed, 5′-GTCGCCACCATGGTG-3′ and R2 that carried KpnI and HindIII restriction sites, respectively. The resulting fusion fragments were introduced into the pAdlox vector (Hardy et al., 1997), and the constructs were verified by sequencing analysis. Recombinant adenoviruses (Ad5) encoding RFP-Obscurin-Ig1/2, RFP-Obscurin-Ig2, or control RFP were created as described previously (Hardy et al., 1997). In brief, pAdlox-RFP-Obscurin-Ig1/2, pAdlox-RFP-Obscurin-Ig2, or pAdlox-RFP were cotransfected with their adenoviral DNA partner ψ5 into human embryonic kidney 293 cells that stably express cre recombinase (CRE8). Recombinant products were selected by repeated passage in CRE8 cells followed by two rounds of plaque purification by agarose overlay (Graham and Prevec, 1995). Viral titers were determined by measuring absorbance at A260, with 1.0 absorbance unit equivalent to ∼1012 viral particles/ml.
Postnatal day 1 (P1) myotubes were infected with 105-109 viral particles/ml either of Ad5-RFP-Obscurin-Ig1/2, Ad5-RFP-Obscurin-Ig2, or control Ad5-RFP in 1 ml of DMEM for 45 min at room temperature (RT). Infected cells were supplemented with 1 ml of DMEM plus 20% FBS and 4 × 10−5 M cytosine arabinoside. After 48 h, cultures were rinsed with phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde for 15 min at RT. Fixed cells were permeabilized with 0.1% Triton X-100 for 10 min at RT, rinsed with PBS, and processed for immunostaining and confocal imaging. Experiments were repeated three times, and ∼30 cells were analyzed from each.
Antibodies Used for Immunofluorescence Staining
Primary antibodies included rabbit antibodies to the COOH-terminal region of obscurin-A (obscurin-COOH, 3 μg/ml; Kontrogianni-Konstantopoulos et al., 2003), the first two NH2-terminal Ig domains of titin (titin-Z, 3 μg/ml; Kontrogianni-Konstantopoulos et al., 2006a; Bowman et al., 2008), the M-band region of titin (titin-M, 3 μg/ml; a generous gift of Dr. S. Labeit), and sAnk1 (3 μg/ml; Zhou et al., 1997). We also used mouse antibodies to the NH2 terminus of obscurin (obscurin-NH2, 3 μg/ml; Kontrogianni-Konstantopoulos and Bloch, 2005), adult slow myosin (NOQ7.5.4D, diluted 1:500; Sigma-Aldrich), α-actinin (diluted 1:500; Sigma-Aldrich), myomesin (tissue culture supernatant, diluted 1:10; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), the I-band region of titin (titin-I, clone T11, labels near the A/I junction, diluted 1:500; Sigma-Aldrich), and MyBP-C slow (3 μg/ml; to the C5 domain of slow forms of MyBP-C, Abnova, Walnut, CA). In addition, we used a guinea pig antibody to the Rho-GEF motif of obscurin (obscurin Rho-GEF, 3 μg/ml; Bowman et al., 2007). Secondary antibodies were goat anti-mouse, goat anti-rabbit, and goat anti-guinea pig IgGs conjugated to Alexa488, Alexa568, and Cy5, respectively (used at 1:200; Invitrogen).
A peptide (5′-CGGEQQQSLHNLDF-3′) containing sequences present only in the unique COOH terminus of MyBP-C slow variant-1 was used to immunize rabbits for production of polyclonal antibodies (Biosynthesis, Lewisville, TX). Specific antibodies were obtained through two rounds of affinity purification; in the first step, the anti-serum was purified against the unique peptide coupled to a SulfoLink column according to the manufacturer's instructions (SulfoLink kit; Pierce Chemical, Rockford, IL). In the second step, the eluted antibodies from the first step were incubated with 200 μg of blotted peptide at 4°C overnight. Bound antibodies were eluted with 2 M glycine, pH 2.2, and used at 3–5 μg/ml in subsequent immunofluorescent experiments. To test the specificity of the obtained antibodies, ∼5 ng of control maltose-binding protein (MBP) and MBP-MyBP-C 26aa (an MBP-fusion peptide containing only the unique COOH-terminal 26 amino acids of MyBP-C slow variant-1; see below) were solubilized in 2× SDS Laemmli buffer at 90°C for 5 min, separated by SDS-PAGE, transferred to nitrocellulose, and probed with the affinity-purified polyclonal antibodies to MyBP-C slow variant-1 (100 ng/ml). Immunoreactive bands were visualized by chemiluminescence (Tropix, Bedford, MA).
Immunofluorescence Staining and Confocal Microscopy
Rats were killed under anesthesia by cardiac perfusion either with buffered saline or buffered saline plus 2% paraformaldehyde, for preparation of unfixed or fixed tissue samples, respectively. Fixed soleus muscles were collected, snap frozen, and sectioned. Unfixed soleus was embedded in PBS with 7.5% gelatin, 15% sucrose and slowly frozen using 2-methylbutane. Fixed, permeabilized cultures of skeletal myotubes and frozen longitudinal and cross sections of adult rat soleus muscle were blocked in PBS, 0.1% bovine serum albumin (BSA), 10 mM NaN3 (PBS/BSA), for 1–2 h at RT, before immunolabeling with the appropriate primary antibodies, diluted in PBS/BSA, overnight at 4°C. Sections were washed, counterstained with the appropriate secondary antibodies in PBS/BSA, mounted with Vectashield (Vector Laboratories, Burlingame, CA), and analyzed with a 410 or 510 confocal laser scanning microscope (Carl Zeiss, Tarrytown, NY), both of which were equipped with a 63×, 1.4 numerical aperture objective (Carl Zeiss).
The fluorescence intensities and structural disruption of sarcomeric markers of the Z-disk (α-actinin), A-band (myosin), and M-band (the COOH terminus of titin), of equal numbers of myotubes infected with either pHcRed-Obscurin-Ig2 or pHcRed control virus, were quantified with ImageJ software (National Institutes of Health, Bethesda, MD). Fluorescence intensities were calculated by averaging the mean pixel intensity of individual myofibers. Statistical significance was determined with Student's t test, with significance set at p < 0.01. To quantify structural disruption, five regions of interest (ROI) of the same size and approximate location (i.e., close to the plasma membrane and at the middle and ends of each cell, avoiding nuclei) were randomly selected for each myotube. Fluorescence profiles for each ROI were measured, normalized to maximal pixel intensity, and plotted as a function of distance with respect to the longitudinal axis of the fibers. A fluorescence peak was defined as a change in normalized pixel intensity greater than 0.1 between a local maximum and its flanking troughs. Distances between adjacent peaks (in the cases of α-actinin and the COOH terminus of titin) or troughs (in the case of myosin) were used to generate average sarcomere lengths. Differences were evaluated with student's t test, with significance set at p < 0.01. Differences between the variances of peak-to-peak or trough-to-trough distances of control and experimental samples (our measure of structural disruption) were also calculated and evaluated with a two-sample F-test for variance, with significance set at p < 0.01 (OriginLab, Northampton, MA).
Yeast Two-Hybrid Screening
The Matchmaker two-hybrid system was used, as described by the manufacturer (Clontech). A fragment encoding the NH2-terminal Ig domains 1 and 2 of human obscurin was inserted into the pGBKT7 bait vector at EcoRI/XhoI sites with primers F1 and R2 that contained the recognition sites for the respective enzymes. After sequence verification the pGBKT7-Obscurin-Ig1/2 plasmid was transformed into Saccharomyces cerevisiae strain AH109 and mated with yeast pretransformed with a cDNA library from adult human skeletal muscle. Mated yeast was plated on SD-His/-Ade/-Leu/-Trp plates and true transformants were selected by plating on SD-His/-Ade/-Leu/-Trp plates in the presence of 80 mg/l X-α-gal. Positive plasmids were recovered by electroporation into Escherichia coli DH10B (Invitrogen) and sequenced.
For domain mapping, deletion constructs of the NH2 terminus of obscurin and the COOH terminus of MyBP-C slow variant-1 were generated by reverse transcription (RT)-PCR from human skeletal muscle RNA (Origene, Rockville, MD) and the SuperScript First Strand Synthesis System (Invitrogen). The following sets of primers were used for amplification of the Ig1 and Ig2 domains of obscurin (XM_340807): for Ig1, the sense primer F1 was used in combination with the antisense primer reverse-1 (R1), 5′-CCGCAGCAGAAAGTGG-3′; for generation of Ig2, the sense primer F2 was used along with the antisense primer R2. Similarly, for generation of the MyBP-C slow variant-1 deletion constructs, the following primer sets were used. For amplification of the partial C10 domain present in the yeast two-hybrid prey clone (referred to as MyBP-C slow C10Y2H), the sense primer forward-3 (F3), 5′-GATGATCCAAGATAC-3′, was used together with the antisense primer reverse-3 (R3), 5′-CACTTTCACCTCCAG-3′, and for amplification of the novel COOH-terminal 79 nucleotides, the sense primer forward-4 (F4), 5′-GTGATATATCAAGGAG-3′, was used in conjunction with the antisense primer reverse-4 (R4), 5′-TCAAAAATCCTTATTGTG-3′ (NM_002465). For both the obscurin and MyBP-C slow variant-1 deletion constructs, all the sense primers contained an EcoRI site, and all the anti-sense primers contained an XhoI site. After PCR amplification, the obscurin and MyBP-C slow variant-1 deletion products were introduced into the EcoRI/XhoI sites of the pGBTK7 bait and pGADT7 prey vectors, respectively. The authenticity of each construct was verified by sequence analysis. Subsequently, different combinations of bait and prey plasmids (see Results) were sequentially transformed into AH109 S. cerevisiae yeast cells, and transformants were processed as described above.
To compare the ability of the C10 domain of the slow, fast, and cardiac isoforms of MyBP-C to bind to the NH2 terminus of obscurin, the entire C10 domain of each variant was amplified from adult human skeletal or heart muscle RNA (Origene) and the following sets of primers; for the slow-C10, the sense primer forward-5 (F5), 5′-CCCATGTTTACTCAG-3′, was used in combination with R3 (NM_002465); for the fast-C10, the sense primer forward-6 (F6), 5′-CCCAAGTTCCTGACA-3′, was used along with the antisense primer reverse-6 (R6), 5′-TCACTGCGGCACTCG-3′ (NM_004533); and for the cardiac-C10, the sense primer forward-7 (F7), 5′-CCAAGCTTCACCCAG-3′, was used with the antisense primer reverse-7 (R7), 5′-TCACTGAGGCACTCG-3′ (X84075). Sense primers contained an EcoRI site, and antisense primers contained an XhoI site. The obtained PCR products were subcloned to the pGADT7 prey vector and transformed into AH109 S. cerevisiae along with the different obscurin bait constructs (see Results).
Generation and Purification of Glutathione Transferase (GST) and MBP Fusion Proteins
The MyBP-C slow variant-1 prey clone was used as template for PCR amplification of the following fragments: the partial C10 domain plus the novel 26 amino acid sequence (C10Y2H+26aa) with primers F3/R4, the partial C10 domain alone (C10Y2H) with primers F3/R3, and the unique 26 amino acid sequence alone (+26) with primers F4/R4. Similarly, the obscurin-Ig1/2 bait clone was used as template for generation of the following fragments: obscurin-Ig1/2, using primers F1/R2; obscurin-Ig1, using primers F1/R1; and obscurin-Ig2, using primers F2/R2. Moreover, for the generation of obscurin-Ig3, human skeletal muscle cDNA (Origene) was used as template with the following set of primers: sense primer, forward-10 (F10), 5′-TGCACGGTGACTGAAGGC-3′, and antisense primer, reverse-11 (R11), 5′-TCTGTGCTGGTCGTAGTG-3′. After amplification, the MyBP-C slow fragments were introduced into the pMAL-c2X vector at EcoRI/SalI sites (New England Biolabs, Ipswich, MA; XhoI and SalI have compatible ends) to produce MBP-fusion proteins, whereas the obscurin fragments were introduced into the pGEX4T-1 vector at EcoRI/XhoI sites (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) to generate GST-fusion proteins. Recombinant polypeptides were expressed by induction with 0.3 mM isopropyl β-d-thioglucopyranoside for 3 h and purified by affinity chromatography on amylose resin (MBP-fusion proteins) or glutathione-agarose (GST-fusion proteins) columns, following the manufacturer's instructions.
Generation of Protein Lysates from P1 Myotubes and Adult Rat Muscle
Homogenates of P1 myotubes cultured for 7 d as well as of soleus, quadriceps, and heart muscles of adult Sprague-Dawley rats (Zivic-Miller Laboratories, Zelienople, PA) were prepared at RT with a Brinkmann Polytron homogenizer at setting 3 (VWR, West Chester, PA) in 10 mM NaPO4, pH 7.2, 2 mM EDTA, 10 mM NaN3, 120 mM NaCl, 0.5% deoxycholate, and 0.5% NP-40, supplemented with Complete protease inhibitors (Roche, Indianapolis, IN). Each sample (∼50 μg) was solubilized in 2× SDS Laemmli buffer at 90°C for 5 min, fractionated by 10% SDS-PAGE using Tris/glycine running buffer, transferred to nitrocellulose, and probed with a pan-MyBP-C antibody (1:10,000 dilution; recognizing slow, fast, and cardiac forms of MyBP-C, a kind gift from Dr. E. Ehler). To obtain better resolution of the immunoreactive bands, the same samples were separated on a 4–12% BisTris gel using 2-(N-morpholino)ethanesulfonic acid running buffer (Invitrogen) and probed with an antibody specific for the slow forms of MyBP-C (300 ng/ml; Abnova). Immunoreactive bands were visualized with a chemiluminescence detection kit (Tropix).
GST ”Pull-Down“ Assay
Two types of GST pull-down assays were performed. In both, equal amounts of recombinant GST, GST-Obscurin-Ig1/2, GST-Obscurin-Ig1, GST-Obscurin-Ig2, and GST-Obscurin-Ig3 proteins were bound to glutathione-Sepharose beads. In type 1, the protein/beads mixture was incubated with 0.5 mg of adult rat soleus muscle homogenates in buffer A (PBS, pH 7.2, 10 mM NaN3, 0.1% Tween 20, and 1 mM dithiothreitol) and incubated at 4°C for 16 h. Beads were washed one time with buffer A and three times with buffer B (PBS, pH 7.2, 10 mM NaN3, and 0.1% Tween 20) in the cold. In type 2, the protein/beads mixture was incubated with 5 μg of recombinant MBP-MyBP-C slow-C10Y2H+26aa, MBP-MyBP-C slow-C10Y2H, or control MBP protein. Subsequently, the beads were washed three times in the cold with buffer B. After washing, beads were heated for 5 min at 90°C in 2× SDS Laemmli sample buffer. The soluble fraction was analyzed by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies specific for MyBP-C slow (Abnova) and MBP (New England Biolabs).
Blot Overlay
The blot overlay assays were performed as described previously, (Kontrogianni-Konstantopoulos and Bloch, 2003). In brief, aliquots (∼2.5 μg) of bacterially expressed, affinity-purified GST and GST-Obscurin-Ig1/2 proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose. Blots were first incubated in buffer C (50 mM Tris, pH 7.2, 120 mM NaCl, 3% BSA, 2 mM dithiothreitol, 0.5% NP-40, and 0.1% Tween 20) for 3 h at 25°C and then with ∼2.5 μg/ml MBP or MBP-MyBP-C slow C10Y2H+26aa diluted in buffer C for 16 h at 4°C. Blots were washed extensively with buffer C and once with buffer B (see above). Subsequently, they were blocked in buffer D (PBS, pH 7.2, 10 mM NaN3, 0.1% Tween 20, and 3% dry milk) and probed with antibodies to MBP (1:5000; New England Biolabs).
Reverse Transcription-Polymerase Chain Reaction
cDNA of human origin from fetal and adult skeletal muscles as well as adult left and right ventricular and atrial heart muscles was obtained commercially (Origene) and used for PCR amplification of MyBP-C slow transcripts with the oligonucleotide primers forward-8 (F8), 5′-GAGGGAAGGAGACTCTTT-3′, and reverse-8 (R8), 5′-GCCCACCTTAGATGATAT-3′, that anneal to sequences present in the 5′ and 3′ untranslated regions (UTRs), respectively.
Total RNA was isolated with TRIzol reagent (Invitrogen) from P1 rat myotubes cultured for 7 d and from adult rat soleus muscle. Aliquots containing ∼5 μg of RNA were reverse transcribed using the SuperScript First Strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer's instructions. PCR amplification of MyBP-C slow transcripts was performed with the F5/R8 primer set that anneals to sequences in the C10 domain and the 3′ UTR. Similarly, amplification of MyBP-C fast and MyBP-C cardiac transcripts was performed with primer sets F6/R6 and F7/R7, respectively, that flanked their C10 domains. All PCR products were analyzed by electrophoresis in 1% agarose gels, and their authenticity was verified by sequencing.
To evaluate the relative expression of MyBP-C slow variant-1 compared with variants 2–4 in P1 rat myotubes, we quantified the average intensities of the ∼325- and ∼250-bp bands that correspond to variant-1 and variants 2–4, respectively, from five independent experiments, using ImageJ software. Results were reported as percentage of the total population of MyBP-C slow transcripts.
Quantitative Reverse Transcription-Polymerase Chain Reaction
Adult human skeletal muscle RNA (Ambion, Austin, TX) was used in quantitative RT-PCR studies of the MyBP-C slow transcripts with the SYBR Green kit (Bio-Rad Laboratories, Hercules, CA) and a MyiQ Real Time PCR Detection System (Bio-Rad Laboratories). Two sets of primers were generated: the sense primer forward-9 (F9) 5′-CCAGGGAGTCTGTACCCTGG-3′ and the antisense primer reverse-9 (R9), 5′-GATATATCAAGGAGTAAATACC-3′, specifically amplified variant-1, whereas primer F9 in combination with the antisense primer reverse-10 (R10), 5′-GTGAAAGTCATTGCACAATAAGG-3′, amplified variants 2–4 but not variant-1. PCR products were analyzed by electrophoresis in 1% agarose gels and verified by sequencing. Experimental efficiency was calculated with a standard curve, as described by the manufacturer (Bio-Rad Laboratories) and was ∼95–105% for both sets of primers. Each experiment was performed in triplicate and repeated at least three times. Data are reported as an average cycle threshold (Ct) value and as -fold difference between the two populations (variant-1 vs. variants 2–4). Student's t test was used to assess the significance of the differences, with significance set at p < 0.01.
siRNA Adenoviral Constructs and Infection of Primary Cultures
A detailed description of the production and characterization of the siRNA adenovirus that targets the expression of obscurin as well as of the control siRNA virus has been reported previously (Kontrogianni-Konstantopoulos et al., 2006b). Primary cultures of skeletal myotubes were infected five days after initial plating with 109 viral particles/ml obscurin or control siRNA viruses in 1 ml of DMEM for 1 h at RT. Infected cells were supplemented with 1 ml of DMEM plus 20% FBS and 4 × 10−5 M cytosine arabinoside (Sigma-Aldrich). After 48 h, cultures were rinsed with PBS and fixed with 2% paraformaldehyde for 15 min at RT. Fixed cells were permeabilized with 0.1% Triton X-100 for 10 min at RT, rinsed with PBS, and processed for immunolabeling and confocal imaging as described before above. Experiments were repeated three times, and ∼15 cells were analyzed from each.
RESULTS
Overexpression of the Second Ig Repeat of Obscurin Results in Severe Disruption of A- and M-Bands
To study the role of the extreme NH2 terminus of obscurin in sarcomerogenesis, we generated adenoviruses containing the first and second immunoglobulin domains of obscurin (Ig1/2), fused with HcRed protein to facilitate direct visualization of the treated cells, and used them to infect primary cultures of P1 rat skeletal myotubes. Overexpression of the pHcRed-Obscurin-Ig1/2 adenovirus had detrimental effects on the cells, leading to their death. This result was independent of the viral dosage we administered, which ranged between 105 and 109 plaque-forming units as this was determined previously by our laboratory to be effective but not toxic for other parts of obscurin (Kontrogianni-Konstantopoulos and Bloch, 2005; Bowman et al., 2008). Consequently, we generated a different adenoviral construct that only contained the second Ig domain of obscurin, again fused with HcRed protein (pHcRed-Obscurin-Ig2) and infected primary cultures of P1 rat skeletal myotubes. These cells survived and so we studied them further. In control experiments, identical cultures were infected with control pHcRed virus.
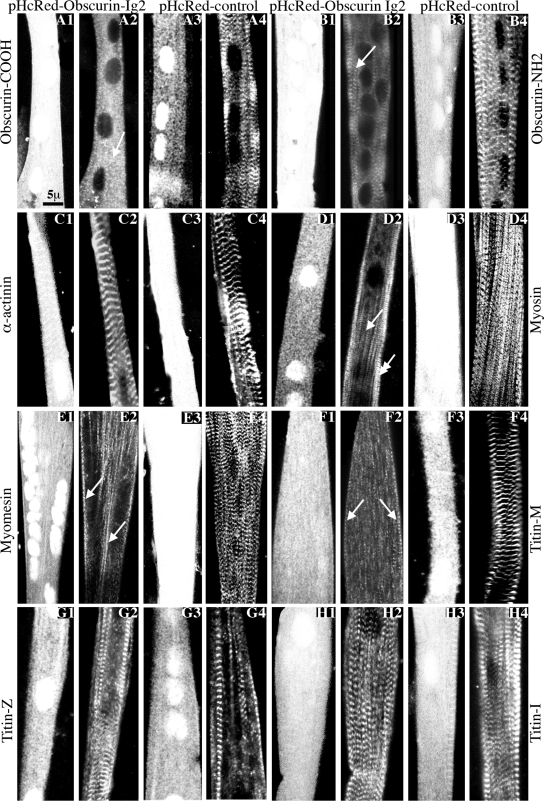
Immunoblots of homogenates prepared from cultures infected with pHcRed-Obscurin-Ig2 or control pHcRed virus showed that both proteins were abundantly expressed and in similar amounts (data not shown). Overexpression of pHcRed-Obscurin-Ig2 was not toxic but had dramatic effects on the assembly and organization of major sarcomeric structures (Figure 1). Examination of the subcellular distribution of endogenous obscurin 2 d after infection with antibodies to its COOH (Figure 1, A1 and A2) and NH2 termini (Figure 1, B1 and B2; the antibody recognizes epitopes present in Ig1; our personal observation) demonstrated that overexpression of the Ig2 domain inhibited the regular organization of the endogenous protein, most of which was primarily concentrated in puncta within the cytoplasm (Figure 1A2, arrow) or along fibrils at the cell periphery with occasional periodicity (Figure 1B2, arrow). By contrast, in cells treated with control pHcRed virus, endogenous obscurin, labeled with antibodies to its COOH (Figure 1, A3 and A4) and NH2 termini (Figure 1, B3 and B4) integrated normally at M-bands.
Figure 1.
Adenoviral overexpression of the Ig2 domain of obscurin in primary cultures of rat skeletal myotubes resulted in disorganized M- and A-bands but not Z-disks or I-bands. (A1–A4 and B1–B4) The striated distribution of endogenous obscurin was severely altered after overexpression of pHcRed-Obscurin-Ig2 virus (A1 and A2 and B1 and B2), but not control pHcRed virus (A3 and A4 and B3 and B4) as shown by immunostaining with antibodies to the COOH terminus (A2, arrow; and A4) or the NH2 terminus (B2, arrow; and B4) of obscurin (odd numbered panels show pHcRed fluorescence). (C1–C4) The regular organization of α-actinin at Z-disks was unaffected after overexpression of pHcRed-Obscurin-Ig2 (C1 and C2) or control pHcRed (C3 and C4) virus. (D1–D4) Sarcomeric myosin exhibited adiffuse cytoplasmic distribution (single arrow) with residual accumulation in striated structures at the cell periphery (double arrow) in cultures infected with pHcRed-Obscurin-Ig2 virus (D1-D2) but not in cultures infected with pHcRed virus where it assumed its typical periodic organization at A-bands (D3 and D4). (E1-E4 and F1-F4) Similar to sarcomeric myosin, myomesin (E1 and E2, arrows) and COOH-terminal epitopes of titin present at the M-band (F1 and F2, arrows) were primarily detected along fibrils showing occasional periodicity after overexpression of pHcRed-Obscurin-Ig2 but not control virus (E3 and E4 and F3 and F4, respectively). (G1-G4 and H1-H4) Strong periodic labeling of the NH2 terminus (G1-G4) and middle portion (H1-H4) of titin at the Z-disk and I-band, respectively, was observed in myotubes infected with either pHcRed-Obscurin-Ig2 (G1 and G2 and H1 and H2) or control pHcRed virus (G3 and G4 and H3 and H4).
As obscurin is a major component of both the M-band and Z-disk, we examined the effects of overexpressing the second Ig domain of obscurin on the distribution of other myofibrillar proteins. Immunolocalization of α-actinin indicated that its regular organization at Z-disks was not altered in cells infected with either pHcRed-Obscurin-Ig2 (Figure 1, C1 and C2) or control pHcRed (Figure 1, C3 and C4) virus. To the contrary, the great majority of infected cells (∼70%) failed to assemble sarcomeric myosin into periodic A-bands after overexpression of pHcRed-Obscurin-Ig2 and instead exhibited a diffuse staining within the cytoplasm (Figure 1, D1 and D2, single arrow) with occasional accumulations at striated fibrils at the cell periphery (double arrow). Similar to its effects on sarcomeric myosin, pHcRed-Obscurin-Ig2 virus also caused myomesin and COOH-terminal epitopes of titin, typically concentrated at M-bands, to be diffusely distributed, with occasional accumulations in filamentous structures (Figure 1, E1 and E2 and F1 and F2, arrows, respectively). Notably, in ∼90% of myotubes infected with control pHcRed virus, sarcomeric myosin concentrated in regular A-bands (Figure 1, D3 and D4), and myomesin (Figure 1, E3 and E4) and COOH-terminal epitopes of titin (Figure 1, F3 and F4) localized to M-bands. Contrary to the COOH terminus of titin, which failed to assemble into M-bands, the apparent organization of other regions of titin was not altered by overexpression of the Ig2 repeat of obscurin. Indeed, immunolabeling of treated cells with antibodies to the NH2 terminus of titin, which is present at Z-disks (Figure 1, G1 and G2), or the middle of the molecule, which resides at I-bands (Figure 1, H1 and H2), showed typical, periodic labeling, similar to cells infected with control pHcRed virus (Figure 1, G3 and G4 and H3 and H4, respectively).
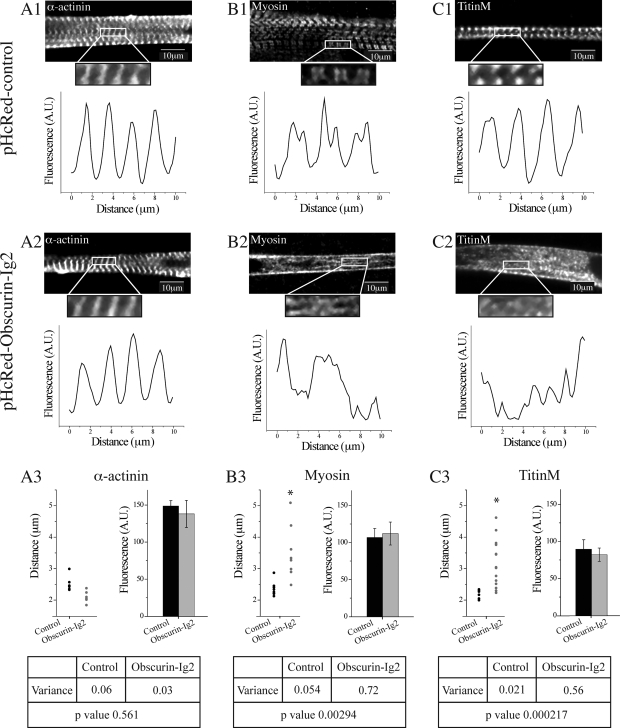
Next, we used ImageJ software to quantify the disruption of sarcomeric proteins of the A- and M-band after overexpression of the Ig2 domain of obscurin (see Materials and Methods). Analysis of the staining patterns of myosin at A-bands (Figure 2B2, top) and the COOH terminus of titin at M-bands (Figure 2C2, top) confirmed their severe disruption in cells infected with pHcRed-Obscurin-Ig2 virus (Figure 1, D2 and F2), as indicated by the irregular trough-to-trough and peak-to-peak distances (Figure 2, B2 and C2, bottom), compared with cells infected with control pHcRed virus (Figure 2, B1 and C1). Consistent with these findings, the variances of trough-to-trough or peak-to-peak distances between control and experimental cells were also significantly different for both myosin and the COOH terminus of titin, respectively (Figure 2, B3 and C3, left). By contrast, the distribution of α-actinin in myotubes overexpressing control HcRed protein (Figure 2A1, top) or the Ig2 domain of obscurin (Figure 2A2, top) verified its regular organization at Z-disks (Figure 2, A1 and A2, bottom, respectively) and showed no difference in the calculated variances of peak-to-peak distances between the two cell populations (Figure 2A3, left). However, myotubes overexpressing the Ig2 domain of obscurin contained smaller sarcomeres (∼2.1 μm) compared with cells expressing HcRed alone (∼2.5 μm), suggesting that although Z-disks may form normally in cells overexpressing the Ig2 domain, the absence of M- and A-bands affects their spacing. These results confirm that the overexpression of the Ig2 domain of obscurin leads to severe disruption of myofibrillar proteins of the A-band and M-band, but not of the Z-disk and I-band.
Figure 2.
The striated organization, but not the expression levels, of sarcomeric myosin and the COOH terminus of titin were significantly altered after overexpression of the Ig2 of obscurin in primary cultures of rat myotubes. (A1–C2) Representative examples of myotubes infected with control pHcRed (A1–C1) or pHcRed-Obscurin-Ig2 (A2–C2) viruses that were used to generate fluorescence profiles (A1–C2, bottom). α-Actinin, a Z-disk protein, shows regular fluorescent peaks in control (A1, bottom) and experimental (A2, bottom) samples; however, sarcomeric myosin at A-bands (B1 and B2) and the COOH terminus of titin at M-bands (C1 and C2) show complete lack of organization in cells infected with the obscurin-Ig2 virus (B2 and C2, bottom, respectively), compared with cells infected with control virus (B1 and C1, bottom, respectively). (A3–C3) Measurements of the average ”peak-to-peak“ distances for α-actinin (A3, left) indicated that they were slightly, yet significantly, decreased in myotubes treated with the obscurin-Ig2 virus (∼2.1 μm) compared with cells treated with control virus (∼2.5 μm); however, the variances between the two cell populations were not statistically different (A3, bottom). Likewise, ”trough-to-trough“ and peak-to-peak measurements for myosin (B3) and the COOH terminus of titin (C3) demonstrated significantly different distances (left) and variances (bottom) between experimental and control samples. Interestingly, the average fluorescent intensities for α-actinin (A3, right), myosin (B3, right), and the COOH terminus of titin (C3, right) were similar between cells treated with pHcRed-obscurin-Ig2 or pHcRed virus.
Although the majority of treated cells (∼80%) showed dramatic disorganization of myosin, myomesin, and titin at the M-band, quantification of their average fluorescence intensities, using ImageJ software, indicated that their expression levels were not significantly altered in cells treated with pHcRed-Obscurin-Ig2 compared with cells treated with control pHcRed virus (Figure 2, A3, B3, and C3, right). These findings suggest that failure of these proteins to integrate into the appropriate structures leads to their abnormal distribution in the myoplasm, but not to their degradation.
The NH2 Terminus of Obscurin Binds Directly to the Novel COOH Terminus of Myosin Binding Protein-C Slow Variant-1
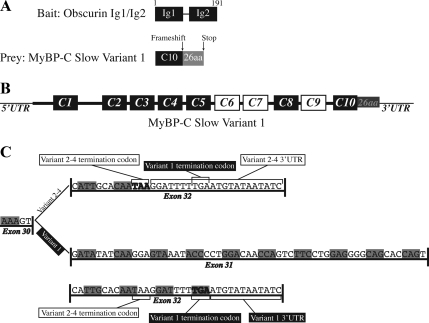
The severe phenotype that we observed after overexpression of the second Ig domain of obscurin, with or without the Ig1 domain, suggested that it might participate in the assembly and maintenance of A- and M-bands by binding directly to other, major components of these structures. To test this, we used the yeast two-hybrid screen to identify potential binding partners of the extreme NH2 terminus of obscurin. Because previous reports in the literature have documented that Ig repeats often function in pairs (Gregorio et al., 1998; Kontrogianni-Konstantopoulos and Bloch, 2003), we designed our ”bait“ to include both Ig1 and Ig2 and screened a cDNA library from adult human skeletal muscle (Figure 3A). Approximately 5 × 105 transformants were screened, of which five ”prey“ clones met the stringent criteria for positive interactions. These were further characterized by sequencing, which revealed that three of the five positive clones carried the same portion of the COOH-terminal region of MyBP-C slow and shorter or longer parts of its 3′ untranslated region. All three clones contained part of the last Ig domain of MyBP-C slow, C10 (amino acids 1071-1119; C10Y2H; NM_206821), but instead of the sequence of 12 nucleotides present in the prototypical forms of MyBP-C slow typically found after the C10 domain (variants 2–4; NM_206819, NM_206820, NM_206821; Furst et al., 1992; Weber et al., 1993), our clones had a unique sequence. Specifically, C10 was followed by a sequence of 59 novel nucleotides, contained within exon 31 that encoded 19 novel amino acids, followed by a stretch of 20 nucleotides, 10 of which are also present in the coding region of variants 2–4, and 10 of which are present in the 3′ UTR of these variants. Insertion of the 59 nucleotides introduces a frameshift mutation that results in the addition of another 7 novel amino acids, encoded by the 20-nucleotides long stretch, followed by a new termination codon. Together, these 79 nucleotides generate a novel COOH terminus containing a total of 26 new amino acids (Figure 3C) that to date is present in only one known form of MyBP-C slow, variant-1 (NM_002456). MyBP-C slow variant-1 encodes a protein of 1171 amino acid residues that shares the canonical domains of all members of the MyBP-C slow family, i.e., C1–C10 (Figure 3B).
Figure 3.
Yeast two-hybrid analysis identified MyBP-C slow variant-1 as a major ligand of the extreme NH2 terminus of obscurin. (A) Schematic representation of the NH2-terminal immunoglobulin domains Ig1 and Ig2 of obscurin that were used as bait to screen a human skeletal muscle library. Three of the five positive preys encoded part of the COOH-terminal immunoglobulin domain of MyBP-C Slow (C10; amino acids 1071-1119; NM_002465) followed by a novel sequence of 26 amino acids that results from a frameshift mutation and use of a novel termination codon. (B) Schematic representation of the structural organization of MyBP-C slow variant-1; the Ig and FnIII domains are shown as black and white boxes, respectively. (C) Sequence comparison of the COOH termini of the different MyBP-C slow isoforms. Variant-1 contains an additional exon, exon 31, between exons 30 and 32, which introduces a frameshift mutation that results in the addition of 26 new residues and an alternative stop codon.
Molecular Characterization of MyBP-C Slow Variant-1 in Skeletal Muscle
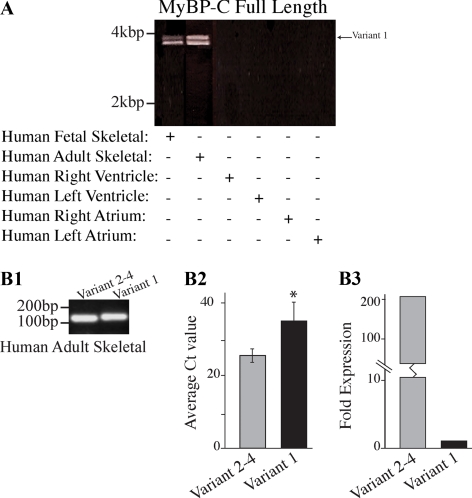
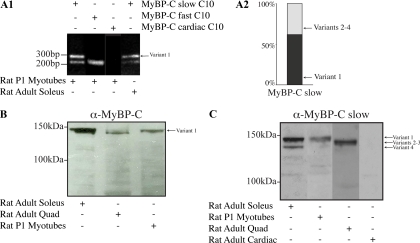
We used RT-PCR with human adult and fetal skeletal muscle cDNA and primers F8 and R8, designed to anneal to the 5′ and 3′ UTRs shared by the four MyBP-C slow variants (Figure 4A), to characterize the forms of MyBP-C present in developing and mature muscle. We observed two major products of ∼4 and ∼3.8 kbp in both fetal and mature samples. Sequence analysis showed that the top band corresponds to variant-1, whereas the bottom band represents a mixed population of variants 2–4. No amplification product was detected when human RNA from right and left ventricular or atrial heart tissue was used, demonstrating the specific and exclusive expression of MyBP-C slow transcripts in skeletal muscle.
Figure 4.
Molecular characterization of MyBP-C slow variant-1 in human skeletal muscle during development and at maturity. (A) RT-PCR was used to amplify full-length MyBP-C slow transcripts. Adult and fetal skeletal muscle cDNA of human origin was amplified using primers, forward-8 and reverse-8 that flanked the 5′ and 3′ UTR regions, respectively. Two major bands of ∼4 and ∼3.8 kbp were obtained in both samples. Notably, no amplification product was detected when cDNA from right or left ventricles and atria was used. Sequence analysis of the fetal and adult products indicated that the top band corresponds to MyBP-C slow variant-1 (NM_002456), whereas the lower band corresponds to a mixed population of variants 2–4 (NM_206819, NM_206820, and NM_206821). (B1) RT-PCR using human skeletal muscle RNA and primer sets designed to specifically amplify the unique COOH terminus of variant-1 (primers F9/R9) or the common COOH terminus of variants 2–4 (primers F9/R10). (B2) Quantification of the mRNA levels of MyBP-C slow variant-1 and variants 2–4 expressed as average Ct values (n = 5; p < 0.01). (B3) Average Ct values from five independent experiments were used to calculate the -fold difference of the expression levels of variant-1 compared with variants 2–4; these indicated that variant-1 is expressed in lower amounts (∼200-fold difference) compared with variants 2–4.
To compare the expression levels of MyBP-C slow variant-1 to the other MyBP-C slow variants, we performed quantitative RT-PCR using adult human skeletal muscle RNA obtained from a mixture of fast and slow twitch muscles and two different sets of primers that flanked the COOH termini of the MyBP-C slow variants. We specifically amplified the unique COOH terminus of variant-1 (primers F9/R9; 146 nucleotides), as well as the common COOH terminus of variants 2–4 (primers F9/R10; 138 nucleotides) (Figure 4B1, lanes 1 and 2, respectively). We optimized the annealing temperatures and reaction efficiencies for each set of primers to yield one specific product with efficiencies between 95 and 105% as determined by standard curves of Ct values, using a 10-fold dilution series of template RNA. To ensure the selective amplification of specific transcripts, we analyzed the melting curve for each reaction and separated all the PCR products by agarose gel electrophoresis (data not shown). These experiments revealed that MyBP-C slow variant-1 is expressed in significantly lower amounts, compared with variants 2–4, in adult human skeletal muscle (∼200 fold lower; Figure 4B3), as evidenced by its higher Ct value (Figure 4B2).
To study the concurrent expression of MyBP-C slow transcripts that contain (i.e., variant-1) or lack (i.e., variants 2–4), the novel 79-nucleotide COOH-terminal coding sequence in specific skeletal muscles, we performed RT-PCR with adult rat soleus muscle RNA and primers designed to anneal to sequences in the C10 domain and the 3′ UTR of MyBP-C slow (Figure 5A1, lane 4). Two PCR products were obtained with sizes of ∼325 base pairs and ∼250 base pairs. Sequencing indicated that the ∼325-base pairs product encoded the C10 domain and the unique COOH-terminal 79-nucleotide sequence of variant-1, where as the ∼250-base pair product encoded the C10 domain followed by the 12-nucleotide COOH terminus of variants 2–4. Similar analysis using RNA from P1 rat skeletal myotubes, cultured for 7 d gave similar results (Figure 5A1, lane 1). To measure the relative expression of these two mRNA populations in rat myotubes, we used ImageJ software to quantify the average intensities of the top and bottom bands from five independent experiments and expressed each as the percentage of the total population of MyBP-C slow transcripts. Notably, the mRNA encoding variant-1 is the predominant form in developing rat myotubes (∼60% of the total MyBP-C slow population; Figure 5A2) and adult soleus muscle. The same cells also express MyBP-C transcripts typical of fast twitch, but not cardiac muscle, which, as expected, are restricted to the developing and mature heart (Figure 5A1, lanes 2 and 3, respectively).
Figure 5.
Molecular characterization of MyBP-C slow variant-1 in rat skeletal muscle during development and at maturity. (A1) RT-PCR was performed using rat cDNA from adult soleus muscle or cultures of P1 skeletal myotubes and primers forward-5 and reverse-8 located in the C10 domain and the 3′ UTR, respectively. Two PCR products were generated with sizes of ∼325 and ∼250 nt. Sequence analysis indicated that the longer product (∼325 nt) included the C10 domain together with the unique COOH-terminal 79 nucleotides, corresponding to variant-1, whereas the smaller product (∼250 nt) contained the C10 domain followed by the 12-nucleotide-long COOH terminus found in variants 2–4. cDNA prepared from primary cultures of skeletal myotubes also contained transcripts of the fast isoform (∼220 nt) but not of the cardiac isoform (for both the fast and cardiac isoforms the primers sets were designed to anneal to sequences present in the respective C10 repeats and their 3′ UTRs). (A2) Semiquantitative analysis, using five independent experiments, of the relative expression levels of MyBP-C slow variants in cultures of rat skeletal myotubes demonstrated that variant-1 composes ∼60% of the total MyBP-C slow population. (B) Protein homogenates of rat origin were prepared from adult soleus and quadriceps skeletal muscles and P1 skeletal myotubes cultured for 7 d. These were immunoprobed with a MyBP-C antibody that recognizes all forms of MyBP-C (slow, fast, and cardiac; pan MyBP-C; B) or an antibody specific for the slow isoforms (C). Two closely migrating bands of ∼126–129 and ∼132 kDa were detected in homogenates prepared from slow twitch soleus muscle whereas a main band of ∼126–129 kDa was detected in homogenates prepared from fast twitch quadriceps muscle. Notably, homogenates prepared from P1 myotubes contained a main band of ∼132 kDa. (C) Use of a different gel system (see Materials and Methods) that allowed better separation of the closely migrating bands detected with the pan-MyBP-C antibody, and of an antibody that specifically recognizes the slow forms of MyBP-C also demonstrated the presence of a main band of ∼132 kDa in soleus and P1 myotube homogenates and a less prominent band of ∼126 kDa in soleus, which correspond to variant-1 and variant-4, respectively. To the contrary, a prominent immunoreactive band of ∼128–129 kDa was detected in quadriceps homogenates, which was absent from soleus and P1 lysates, and may represent a mixed population of variants 2 and 3. No immunoreactive band was detected in lysates prepared from heart muscle, which confirmed the specificity of the antibody used.
Next, we prepared immunoblots of protein lysates from adult rat soleus and quadriceps skeletal muscles as well as P1 skeletal myotubes, probed with a pan-MyBP-C antibody (Figure 5B; this antibody recognizes all the different forms of MyBP-C). We detected two immunoreactive bands in homogenates of slow twitch soleus muscle, that migrated very closely at ∼126–129 and ∼132 kDa, respectively (Figure 5B, lane 1). Interestingly, a main band of ∼126–129 kDa was detected in lysates prepared from fast twitch quadriceps skeletal muscle (Figure 5B, lane 2) that comigrated with the lower band present in soleus homogenates, whereas a faint band of ∼132 kDa was also observed. Notably, in lysates from P1 myotubes a single immunopositive band of ∼132 kDa was present (Figure 5B, lane 3) that aligned with the larger of the two bands seen in homogenates of soleus muscle. As the inclusion of the novel 26-amino acids long COOH terminus in MyBP-C slow variant-1 gives rise to a protein with an estimated molecular mass of ∼132 kDa, and variants 2–4 give rise to proteins ranging in size from ∼126 kDa (slow variant-4) to 128 kDa (slow variant-3 and fast isoform) to ∼129 kDa (slow variant-2), the top band (∼132 kDa) most likely corresponds to variant 1, whereas the bottom band (∼126–129 kDa) most likely includes a mixed population of slow variants 2–4 and MyBP-C fast.
To obtain better separation of the MyBP-C slow immunoreactive bands, we used a different gel system (see Materials and Methods) and an antibody that specifically recognizes the slow forms of MyBP-C (Figure 5C). This antibody is specific for the slow isoforms, as it does not react with homogenates from adult rat cardiac muscle (Figure 5C, lane 4). We detected two major bands of ∼132 and ∼126 kDa in homogenates of adult rat soleus muscle, which seem to correspond to slow variants 1 and 4, respectively (Figure 5C, lane 1), with the top one (∼132 kDa; variant-1) being the most prominent. Again, the ∼132 kDa band predominated in the myotubes, whereas other variants of ∼128–129 kDa, probably a mixture of slow variants 2 and 3, predominated in quadriceps muscle (Figure 5C, 2 and 3 lanes, respectively).
Together, the results of our RT-PCR and immunoblotting studies suggest that the top band in rat soleus and P1 lysates corresponds to MyBP-C slow variant-1 (∼132 kDa; NM_002456), whereas the lower band in soleus muscle may correspond to variant-4 (∼126 kDa; NM_206821). Quadriceps skeletal muscle expresses a band of ∼128–129 kDa, which may be a mixture of MyBP-C slow variants 2 and 3 (∼129 kDa, NM_206819 and ∼128 kDa, NM_206820, respectively) and MyBP-C fast (∼128 kDa; NM_004533).
The Ig2 Repeat of Obscurin and the C10 Domain of MyBP-C Slow Contain the Minimal Binding Sites Required for Their Interaction
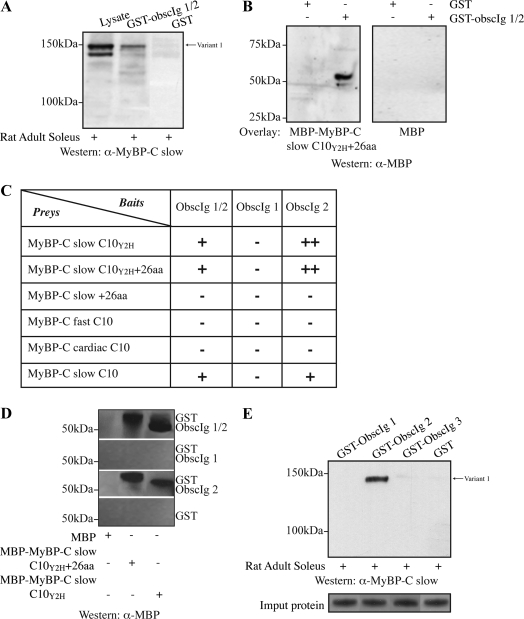
To confirm the interaction of MyBP-C slow with the NH2 terminus of obscurin, we used a GST pull-down assay. Recombinant GST-Obscurin-Ig1/2 adsorbed native MyBP-C slow variant-1 from adult rat soleus muscle homogenates as detected by Western blotting with the MyBP-C slow specific antibody (Figure 6A). This binding was specific for the NH2-terminal Ig domains of obscurin, because no binding occurred with GST alone.
Figure 6.
Characterization of the interaction between the NH2 terminus of obscurin and the COOH terminus of MyBP-C slow variant-1. (A) Equivalent amounts of GST-Obscurin-Ig1/2 and GST were bound to glutathione matrices and incubated with protein homogenates prepared from adult rat soleus muscle. Total lysates were also included in the Western blot for comparison purposes. Recombinant GST-Obscurin-Ig1/2, but not GST, efficiently adsorbed native MyBP-C slow variant-1 (top band), and to a lesser degree, variant-4 (bottom band), as shown by SDS-PAGE and immunoblotting with the MyBP-C slow antibody. (B) The specific and direct interaction between the NH2 terminus of obscurin and the COOH terminus of MyBP-C slow variant-1 was further verified in vitro by overlay assays. Equal amounts of bacterially expressed GST and GST-obscurin-Ig1/2 were separated by SDS-PAGE and overlaid with recombinant MBP-MyBP-C slow-C10Y2H+26aa or control MBP. MBP-MyBP-C slow-C10Y2H+26aa, but not MBP, bound to GST-Obscurin-Ig1/2, but not to GST. (C) The minimal regions of obscurin and MyBP-C slow variant-1 involved in binding were identified using the yeast system. A series of deletion constructs were generated for both obscurin and MyBP-C slow variant-1 and introduced into the bait and prey vector, respectively. Yeast two-hybrid analysis indicated that the Ig2 domain of obscurin and the C10 domain of MyBP-C slow are both necessary and sufficient to support their direct association in vitro. This interaction is specific for the C10 domain of MyBP-C slow because neither the C10 domain of the fast isoform nor the C10 domain of the cardiac isoform were able to interact with the NH2 terminus of obscurin in the yeast system. (D) To confirm the ability of the minimal interacting domains of obscurin and MyBP-C slow variant-1 identified in the yeast system to support their direct association, we also used an in vitro pull-down assay. Equivalent amounts of bacterially expressed GST-Obscurin-Ig1/2, GST-Obscurin-Ig1, GST-Obscurin-Ig2, and GST were bound to glutathione matrices and tested for their ability to retain recombinant MBP, MBP-MyBP-C slow-C10Y2H+26aa, and MBP-MyBP-C slow C10Y2H in a pull-down assay. GST-Obscurin-Ig1/2 and GST-Obscurin-Ig2, but not GST-Obscurin-Ig1 or GST, were able to efficiently pull down MBP-MyBP-C slow-C10Y2H+26 and MBP-MyBP-C slow-C10Y2H. (E) GST fusion proteins of obscurin (GST-Obscurin-Ig1, GST-Obscurin-Ig2, and GST-Obscurin-Ig3) were incubated with protein homogenates prepared from adult rat soleus muscle and examined for their ability to adsorb endogenous MyBP-C slow variant-1; GST-Obscurin-Ig2 adsorbed native MyBP-C slow variant-1, whereas neither GST-Obscurin-Ig1 nor GST-Obscurin-Ig3 was able to precipitate endogenous MyBP-C slow variant-1. Coomassie Blue staining of the input protein is shown in the bottom panel to indicate equal loading.
We confirmed the ability of the NH2-terminal region of obscurin to bind directly and specifically to the COOH-terminal region of MyBP-C slow variant-1 in blot overlay assays. Equivalent amounts of affinity purified GST and GST-Obscurin-Ig1/2 proteins were separated by SDS-PAGE, transferred to nitrocellulose, and overlaid with recombinant MBP-MyBP-C slow C10Y2H+26aa or MBP alone. Binding was visualized by probing with antibodies to MBP. MBP-MyBP-C slow C10Y2H+26aa specifically and directly bound to GST-Obscurin-Ig1/2 but not to GST-protein (Figure 6B, left). No specific binding occurred with an identical blot overlaid with MBP alone (Figure 6B, right).
To identify more precisely the sequences within the NH2 terminus of obscurin and the COOH terminus of MyBP-C slow variant-1 that mediated their binding, we generated a series of deletion constructs, which we used in the yeast two-hybrid system. For obscurin, bait constructs encoding Ig1 (amino acids 1-114), Ig2 (amino acids 115-210), or Ig1/2 (amino acids 1-210) were coexpressed in yeast with prey constructs encoding the portion of the C10 domain of MyBP-C slow present in our original prey clones (amino acids 1071-1119; C10Y2H, NM_002456), the novel 26 amino acid sequence of variant 1, or the partial C10 repeat with the 26 COOH-terminal amino acids (C10Y2H+26; Figure 6C). The minimal sequences required for binding of obscurin to MyBP-C slow variant-1 are confined within repeats Ig2 and C10Y2H, respectively. To learn whether the C10 domains of the fast and cardiac MyBP-C isoforms can also bind to the Ig2 repeat of obscurin, we repeated our assays with the equivalent portions of their C10 domains. Neither interacted with obscurin's Ig2 in the yeast system (Figure 6C).
We further confirmed the interaction between the minimal domains of obscurin and MyBP-C slow (i.e., Ig2 and C10Y2H, respectively) using pull-down assays (Figure 6D). Equivalent amounts of bacterially expressed GST-Obscurin-Ig1/2, GST-Obscurin-Ig1, GST-Obscurin-Ig2, and control GST-protein were bound to glutathione matrices and incubated with recombinant MBP, MBP-MyBP-C slow C10Y2H+26, or MBP-MyBP-C slow C10Y2H (Figure 6D). Both GST-Obscurin-Ig1/2 and GST-Obscurin-Ig2, but not GST-Obscurin-Ig1 or GST, were able to specifically retain MBP-MyBP-C slow-C10Y2H+26 and MBP-MyBP-C slow-C10Y2H but not MBP protein. Using dot blot overlays of synthetic peptides of ∼15 residues each, we were unable to identify shorter sequences within the C10 domain of MyBP-C slow capable of binding the NH2-terminal region of obscurin (Supplemental Figure 1). Together, these data suggest that the minimal binding site on MyBP-C slow for the second Ig domain of obscurin is the last ∼50 residues of the C10 domain.
Last, to confirm that the second Ig domain of obscurin is capable of retaining native MyBP-C slow variant-1 from adult rat soleus muscle homogenates, we performed a GST-pull down assay. Sequence comparison of the flanking Ig domains, Ig1 and Ig3, indicated that they share an ∼50% homology with Ig2. Consequently, we generated recombinant Ig1 and Ig3 peptides and examined their ability to precipitate endogenous MyBP-C slow variant-1 along with Ig2 (Figure 6E, top). Equivalent amounts of GST, GST-Obscurin-Ig1, GST-Obscurin-Ig2, and GST-Obscurin-Ig3 attached to glutathione beads (Figure 6E, bottom) were incubated with protein homogenates from soleus and analyzed by immunoblotting with the MyBP-C slow specific antibody. Only, GST-obscurin-Ig2 adsorbed native MyBP-C slow variant-1, whereas control GST-protein, GST-Obscurin-Ig1, and GST-Obscurin-Ig3 failed to precipitate endogenous MyBP-C slow variant-1 (Figure 6E, top). Furthermore, GST-Obscurin-Ig2 specifically adsorbed variant-1, but not variant-4, from soleus extracts (Figure 6E), suggesting that although the 26 novel amino acids present in the COOH terminus of variant-1 are neither necessary nor sufficient for binding, they are required for the specificity of the interaction between obscurin and variant-1.
MyBP-C Slow Variant-1 Concentrates at the M-Band in Developing and Adult Skeletal Myofibers
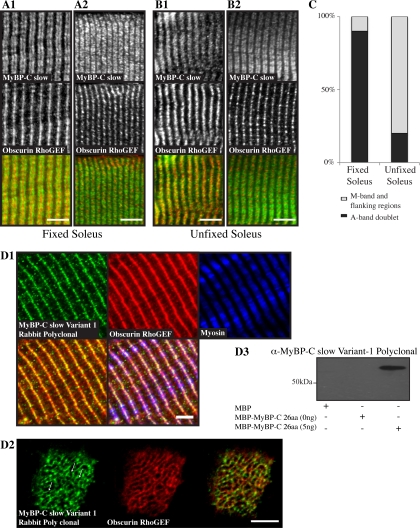
To study the subcellular distribution of MyBP-C slow in skeletal muscle, we stained longitudinal sections of adult rat soleus muscle with an antibody that specifically recognizes the slow forms of MyBP-C (Figure 7, A and B, green, top panels). Sections were also labeled with an antibody to the Rho-GEF domain of obscurin, that preferentially labels M-bands (Figure 7, A and B, middle, red).
Figure 7.
MyBP-C slow variant-1 preferentially accumulates at the M-band. Fixed (A1 and A2) and unfixed (B1 and B2) adult rat soleus fibers were costained with antibodies specific for the MyBP-C slow isoforms (A1–B2, top, green) and the Rho-GEF domain of obscurin (A1–B2, middle, red), which specifically labels M-bands. (A1 and A2) In ∼90% of myofibers in fixed soleus muscle, MyBP-C slow assumed its typical staining at A-bands (top) and obscurin at M-bands (middle), as clearly shown in the overlay (bottom). In ∼10% of myofibers, however, MyBP-C slow was also present midway of A-bands, at M-bands (top), as shown by colabeling with antibodies to the Rho-GEF domain of obscurin (middle); areas of overlap between MyBP-C slow and obscurin look yellow in the overlay (bottom). (B1 and B2) By contrast, in unfixed soleus muscle, in ∼80% of myofibers the MyBP-C slow antibody stained the entire A-band, including the central M-band (top), as shown by costaining with antibodies to the Rho-GEF domain of obscurin (middle); areas of coincident distribution between MyBP-C slow and obscurin look yellow in the overlay (bottom). In the remaining ∼20% of myofibers, MyBP-C slow assumed its typical organization at A-bands (top) as no overlapping staining with obscurin (middle) was observed in the color overlay (bottom). Bar (A and B), 10 μm. (C) Graph presenting the percentage of cells showing labeling at A- or A- and M-bands in fixed and unfixed soleus muscle. (D1) Labeling of rat adult soleus muscle with affinity-purified rabbit antibodies raised against the unique COOH terminus of MyBP-C slow-variant-1 (top left, green) and antibodies to the Rho-GEF domain of obscurin (top middle, red) that label the M-band and to the slow isoform of sarcomeric myosin (top right, blue) revealed the presence of MyBP-C slow variant-1 in the middle of A-bands (bottom right, triple staining overlay), at M-bands and possibly flanking regions (bottom left, double staining overlay). (D2) Cross sections of adult rat soleus muscle were costained with antibodies to the unique COOH terminus of MyBP-C slow variant-1 (left, green) and the Rho-GEF domain of obscurin (middle, red). Similarly to obscurin, MyBPC slow variant-1 is present in a reticular pattern at the level of the M-band, as shown in the color overlay (right; and arrows). Bar, 5 μm. (D3) Western blot analysis indicated that the affinity-purified antibodies for MyBP-C slow variant-1, used in D1, specifically recognize recombinant MBP-MyPB-C-26aa.
Previous work from our laboratory and others has suggested that some epitopes are sensitive to fixation, especially in the dense myofibrillar cytoskeleton of striated muscles (Barth and Elce, 1981; Flucher et al., 1999; Williams and Bloch, 1999; Williams et al., 2000). Consequently, we examined the distribution of MyBP-C slow in longitudinal sections from both fixed and unfixed skeletal muscles (Figure 7). In the fixed sections, the MyBP-C slow antibody labeled doublets typical of A-bands in ∼90% of the fibers analyzed (Figure 7A1), but in ∼10% of the fibers, it labeled a single broad structure consisting of A-bands and their central M-bands (Figure 7A2). Interestingly, this pattern was reversed in unfixed soleus muscle, with ∼80% of fibers labeling with the MyBP-C slow antibody across whole A-bands, including M-bands (Figure 7B1), and ∼20% labeling in A-band doublets (Figure 7B2). These findings suggested that MyBP-C slow is sensitive to fixation and that some form(s) of it concentrates at M-bands in both fixed and unfixed muscle where the NH2 terminus of obscurin also resides (Kontrogianni-Konstantopoulos and Bloch, 2005).
We next compared the distribution of variant-1 of MyBP-C slow with all the variants of this protein, by generating three different antibodies specific to the unique COOH terminus of variant-1, prepared in rabbits (Figure 7) and in mice (Supplemental Figure 2). Although all three antibodies were ineffective in recognizing native MyBP-C slow variant-1 in immunoblots (data not shown), they were successful in binding specifically to a recombinant peptide that contained the 26 novel amino acids of variant-1 (Figure 7D3; data not shown) and were also active in immunofluorescent studies. After affinity purification of the rabbit serum, the resultant antibodies were tested in triple immunofluorescent experiments using fixed adult rat soleus muscle and antibodies to the Rho-GEF domain of obscurin that localizes at M-bands and to the slow form of sarcomeric myosin (Figure 7D1). Polyclonal antibodies specific for variant-1 showed strong staining at M-bands in longitudinal sections of soleus muscle (Figure 7D1, top left, green), which coincided with obscurin (Figure 7D1, top middle, red) and labeled midway of myosin (Figure 7D1, top right, blue; in the overlay, bottom right, areas of coincident localization look white); notably, this pattern was not detected with nonimmune primary antibodies (data not shown). Moreover, colabeling of cross sections of soleus muscle with antibodies to the Rho-GEF domain of obscurin (Figure 7D2, middle, red) and MyBP-C slow variant-1 (Figure 7D2, left, green) revealed that similarly to obscurin, MyBP-C slow variant-1 assumes a reticular distribution, suggesting that it surrounds myofibrils at the level of the M-band (Figure 7D2, arrows). Together, these results indicate that MyBP-C slow variant-1 localizes at the periphery of M-bands, where it codistributes with obscurin, further supporting their interaction in situ.
Manipulation of the Expression Levels of Obscurin Disrupts the Organization of MyBP-C Slow Variant-1 in Developing Myotubes
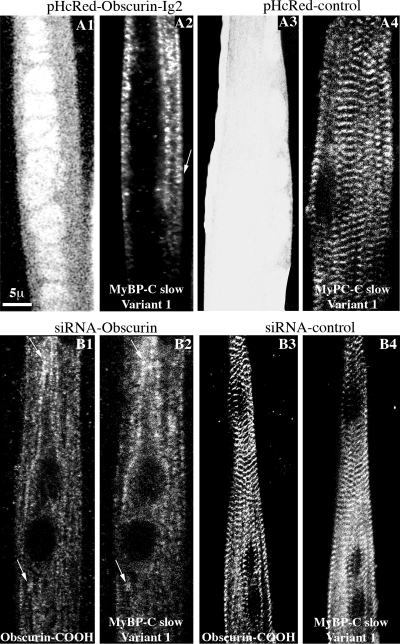
We next examined the effects of altering the levels of obscurin on the subcellular organization of MyBP-C slow variant-1 by using adenoviral vectors to overexpress its Ig2 domain (Figure 8A1) or to reduce the expression of obscurin with targeted siRNA (Figure 8A2) in primary cultures of skeletal myotubes. Like other proteins of the M-band (Figure 1), MyBP-C slow variant-1 failed to incorporate into developing M-bands in myotubes that express pHcRed-Obscurin-Ig2 (Figure 8A1); instead, it concentrated in structures present at the cell periphery showing occasional periodicity (Figure 8A2, arrows). By contrast, MyBP-C slow variant-1 assumed a striated distribution at M-bands (Figure 8A4) in myotubes infected with control pHcRed virus (Figure 8A3). Suppression of the synthesis of obscurin caused the amounts of MyBP-C slow variant-1 detected by immunofluorescence to be dramatically reduced, whereas any remaining protein failed to assemble into regular M-bands (Figure 8B2) and instead concentrated in the same nonstriated filamentous structures as residual obscurin (Figure 8B1). This was not the case in cells infected with control siRNA virus in which both obscurin (Figure 8B3) and MyBP-C slow variant-1 (Figure 8B4) occupied regular M-bands. These experiments suggest that the levels of expression of variant-1 of MyBP-C slow and its distribution in myotubes are dependent on the expression and incorporation of obscurin into M-bands.
Figure 8.
MyBP-C slow variant-1 failed to assemble into M-bands in primary cultures of skeletal myotubes after manipulation of the expression of obscurin. (A1–A4) Confocal images of P1 myotubes treated with pHcRed-Obscurin-Ig2 virus (A1) and stained with antibodies specific to MyBP-C slow variant-1 (A2); MyBP-C slow variant-1 failed to organize at M-bands, after overexpression of the Ig2 repeat of obscurin and concentrated along fibrillar structures showing occasional periodicity (A2, arrow). This was not the case in cells infected with control pHcRed virus (A3) in which MyBP-C slow variant-1 showed a regular distribution at M-bands. (B1–B4) Confocal images of P1 myotubes treated with a siRNA virus that specifically targets obscurin and labeled with antibodies to the COOH terminus of obscurin (B1) and antibodies specific for MyBP-C slow variant-1 (B2); MyBP-C slow variant-1 failed to assemble into M-bands when obscurin was knocked down and concentrated in the same structures as residual obscurin did (arrows). In cells infected with control siRNA virus, both obscurin (B3) and MyBP-C slow variant-1 (B4) assumed their typical organization at M-bands.
We also prepared adenoviral constructs that overexpressed the C10 domain of MyBP-C slow, the C10 domain plus the 26 COOH-terminal residues of variant-1, or the 26 residues alone that were all toxic to developing myotubes and thus could not be used to learn whether, conversely, MyBP-C slow influences the organization of obscurin. The most plausible explanation for this severe effect is that the three constructs provide binding sites for other sarcomeric proteins, in addition to obscurin. Consistent with this, the C10 domain is also the binding site for sarcomeric myosin and contributes to the binding site for titin (Okagaki et al., 1993a; Freiburg and Gautel, 1996; Obermann et al., 1998; Miyamoto et al., 1999; Welikson and Fischman, 2002).
DISCUSSION
We report the direct interaction of the second Ig domain of obscurin with variant-1 of MyBP-C slow, a protein that typically associates with thick filaments. MyBP-C slow variant-1 shares a common primary sequence and domain architecture with the prototypical forms of MyBP-C slow (variants 2–4), but it contains a unique COOH terminus, consisting of 26 amino acids after the last immunoglobulin domain (C10) present in all MyBP-C slow isoforms. MyBP-C slow variant-1 preferentially concentrates at M-bands in both developing and adult skeletal myofbers, in which it codistributes with obscurin (Kontrogianni-Konstantopoulos et al., 2003, 2004; Bowman et al., 2007). The second immunoglobulin domain of obscurin (Ig2) and the last immunoglobulin domain of MyBP-C slow variant-1 (C10) are both necessary and sufficient to support their interaction, which is, however, significantly enhanced by the presence of the novel 26 amino acids in the COOH terminus of variant-1. Overexpression of the Ig2 domain of obscurin severely disrupted the formation of M- and A-bands but not of Z-disks or I-bands. Our results suggest that binding of the NH2 terminus of obscurin to the COOH terminus of MyBP-C slow variant-1 at the periphery of the M-band plays a key role in the integrity of the M-band and the assembly of thick filaments into A-bands in developing skeletal myofibers.
Obscurin is expressed early during myofibrillogenesis. It first becomes organized at developing M-bands and only later at mature Z-disks, suggesting that it may play important roles in the initial assembly of M-bands but not of Z-disks (Borisov et al., 2004; Kontrogianni-Konstantopoulos et al., 2006b). Altering the expression levels of obscurin, either by siRNA-mediated down-regulation or by adenoviral overexpression of portions of the molecule, has dramatic effects on the assembly and stabilization of both M- and A-bands (Kontrogianni-Konstantopoulos et al., 2004, 2006), but the mechanisms of its actions are still elusive. Our results suggest that obscurin participates in the formation of A- and M-bands by binding directly to MyBP-C slow variant-1 that selectively concentrates at M-bands.
MyBP-C of striated muscles is the product of three genes that map to different chromosomes of the human genome: the cardiac form to chromosome-11, the fast-twitch skeletal form to chromosome-19 and the slow-twitch skeletal form to chromosome-12 (Yasuda et al., 1995). The forms of MyBP-C found in skeletal muscle consist of a linear array of 10 globular domains, termed C1–C10, seven of which are immunoglobulin I-like domains (C1–C5, C8 and C10; Figure 3B), with the remaining three resembling fibronectin-III repeats (C6, C7, and C9; Figure 3B). The fast and slow isoforms can be coexpressed in some skeletal muscle types and can even coexist within the same sarcomere; expression of the cardiac isoform, however, is restricted to the developing and mature heart (Fougerousse et al., 1998; Gautel et al., 1998).
Four different MyBP-C slow transcripts are expressed in human skeletal muscle and are referred to as variants 1–4. The different variants contain 1171, 1148, 1141, and 1123 amino acids corresponding to proteins with molecular masses of ∼132, ∼129, ∼128, and ∼126 kDa, respectively. Only transcript variant-1 contains the unique 79-nucleotide long 3′ sequence, which generates a novel 26-amino acid-long COOH terminus. The other three variants (i.e., variants 2–4) share the same 3′ sequence consisting of 12 nucleotides that encode four amino acids adjacent to the C10 domain.
Using quantitative RT-PCR, we found that in human adult skeletal muscle RNA the expression levels of MyBP-C slow variant-1 transcripts are ∼200-fold lower compared with those of variants 2–4. To the contrary, semiquantitative RT-PCR indicated that MyBP-C slow variant-1 accounts for ∼60% of MyBP-C slow transcripts in developing rat myotubes and slow twitch soleus muscle. This apparent difference in the amounts of variant-1 between the two species may stem from the RNA source that we used. Specifically, the human RNA was obtained from a mixture of fast and slow twitch muscles, therefore masking potential muscle- or fiber-type differences in the expression levels of variant-1, whereas the rat RNA was extracted from slow twitch soleus or developing hindlimb muscle. Notably, our immunoblotting studies support the high transcript levels of variant-1 in developing myotubes and adult soleus with corresponding high levels of protein expression.
Use of three different antibodies directed against the unique COOH terminus of variant-1 demonstrated that, unlike other forms of MyBP-C that localize at the C-zone of the A-band, MyBP-C slow variant-1 preferentially concentrates at M-bands and possibly flanking regions. As variant-1 shares the same primary sequence, with the exception of its unique COOH terminus, with the other three variants of MyBP-C slow, it was surprising that an antibody that recognizes all four slow isoforms only labeled the M-band region in ∼10% of myofibers in fixed soleus muscle, whereas labeled the A-bands in all. This is likely to be due to fixation, however, as the same antibody labeled M-bands (and A-bands) in ∼80% of myofibers in unfixed soleus muscle. Thus, our results with an antibody specific for all variants of MyBP-C slow agree with our findings with antibodies specific for the unique COOH terminus of MyBP-C slow variant-1 and indicate that at least variant-1 selectively concentrates at the periphery of M-bands, where it codistributes with obscurin (Figure 9).
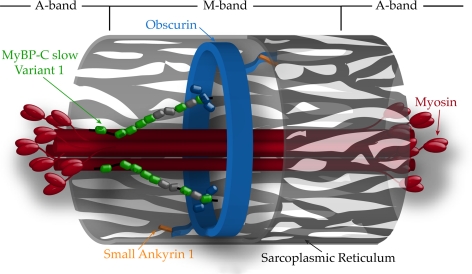
Figure 9.
Model of MyBP-C slow variant-1 at the M-band. MyBP-C slow variant-1 (shown as an array of green, Ig, and gray, FnIII, ovals) is located at the periphery of the M-band where it interacts with the NH2-terminal Ig2 domain of obscurin (shown in blue) via its C10 repeat. Variant-1 may also associate with the S2 portion of superficial myosin filaments (shown in red) through its MyBP-C motif (shown as a black line), located between repeats C1 and C2. Although the NH2-terminal region of obscurin is oriented to facilitate its interactions with sarcomeric proteins at the surface of the M-band, like variant-1, its COOH-terminal region faces the surrounding SR membranes (shown as light gray structure with a series of fenestrae), in which it can interact with sAnk1 (shown in orange), an integral component of the SR. Myosin head groups are shown to denote the boundaries between A- and M-bands. For reasons of simplicity, other ligands of obscurin at the M-band (e.g., titin and myomesin; Fukuzawa et al., 2008) are not depicted.
To date, all the known isoforms of MyBP-C have been shown to concentrate at the C-zones of A-bands, where they are located in seven to nine transverse lines spaced at 43-nm intervals, perpendicular to the long axis of the myosin thick filament (Winegrad, 1999; Flashman et al., 2004b, 2008). Consistent with their localization, biochemical studies have demonstrated that all canonical forms of MyBP-C bind to the light meromyosin (LMM) portion of the myosin rod through their C10 domain, although binding is significantly enhanced in the presence of the C8 and C9 repeats, and to subfragment 2 (S2) of the heavy meromyosin (HMM) portion of myosin through their MyBP-C motif, located between repeats C1 and C2 (Moos et al., 1975; Starr and Offer, 1978; Okagaki et al., 1993b; Gilbert et al., 1996, 1999; Alyonycheva et al., 1997). Although the C10 domains of the cardiac, slow and fast isoforms bind to the LMM portion of sarcomeric myosin, the cardiac and fast isoforms do not bind to the Ig2 repeat of obscurin, suggesting that distinct amino acid residues are involved in their respective sites of interaction. Thus, MyBP-C slow variant-1 may associate with the LMM portion of myosin filaments located at the surface of M-bands via its C10 domain, although such an association would likely be dynamic and depend on the contractile state of the muscle cell. Conversely, it may associate with the S2 subfragment of superficial myosin filaments at the level of M-bands via its MyBP-C motif (Figure 9). In either case, MyBP-C slow variant-1 may associate with thick filaments, similar to the canonical forms of MyBP-C, although in a unique arrangement, around myofibrillar M-bands, and with unique roles. Interestingly, skelemin (also known as myomesin-I), a major component of the myofibrillar M-band, has been also shown to concentrate at the periphery of M-bands (Price, 1987), in which it directly interacts with the rod portions of thick myosin filaments (Obermann et al., 1997; Auerbach et al., 1999).
The distinct localization of MyBP-C slow varint-1 is not without a precedent. A novel alternatively spliced form of cardiac MyBP-C, referred to as cardiac MyBP-C(+), contains a stretch of 10 additional amino acids in the COOH-terminal C9 domain, which promotes binding to titin (Sato et al., 2003). Cardiac MyBP-C(+) does not localize to A-bands in cardiac myocytes, but exhibits a diffuse distribution, has a markedly decreased affinity for myosin and titin, and is the predominant form of cardiac MyBP-C in aged atrial muscle.
While this work was in preparation, a study by Fukuzawa and colleagues (Fukuzawa et al., 2008) demonstrated that the Ig1 repeat of obscurin interacts directly with the last Ig domain of titin (the M10 domain) located in the M-band and that the Ig3 domain of obscurin interacts directly with the linker region between the FNIII-like domains, My4 and My5, of myomesin. Overexpression of Ig1 or Ig3 repeats of obscurin and the M10 domain of titin in cultured cardiocytes had a subtle effect in the organization of M-bands, whereas overexpression of the My4-linker-My5 domains of myomesin had a more severe effect, suggesting that in cardiocytes myomesin is required for the integration of both titin's M-band region and obscurin at M-bands (Fukuzawa et al., 2008). Interestingly, the same investigators found that overexpression of any of these motifs in C2C12 myoblasts caused massive cell death, suggesting a possible role for each of them in de novo myofibrillogenesis (Fukuzawa et al., 2008). Consistent with this, our previous and current studies show that elimination of obscurin (Kontrogianni-Konstantopoulos et al., 2006b) or overexpression of its Ig2 domain in primary cultures of skeletal myotubes has a dramatic and specific effect on the formation of M-bands and on the incorporation of myosin into A-bands. The differences in the activities of obscurin in developing skeletal and cardiac muscle cells is likely to stem from its distinct binding partners. Consistent with this, we found that the second Ig domain of obscurin, binds well to the COOH-terminal region of MyBP-C slow variant-1, but poorly, if at all, to the same region of cardiac MyBP-C.
Thus, a multimolecular complex consisting of the M-band portion of titin, myomesin, obscurin, and MyBP-C slow variant-1 seems to play a critical role in the formation and maintenance of the M-band during myofibrillogenesis in skeletal myotubes. Whether all these interactions take place simultaneously or in all different fiber types is not known. The topological constraints, however, introduced by the three adjacent Ig domains that support these interactions most likely favor dynamic, rather than static, interactions among these proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. H. Catino and S. S. Hirsch for technical assistance in the initial stages of this work and B. L. Prosser for helpful discussions. We also thank Drs. S. Labeit (Institut für Anästhesiologie und Operative Intensivmedizin, Universitätsklinikum, Mannheim, Germany) and E. Ehler (King's College London, London, United Kingdom) for the titin-M and pan-MyBP-C antibodies, respectively. Our research has been supported by National Institutes of Health grant R01 AR52768 and a grant from the Muscular Dystrophy Association (to A.K.K.) and National Institutes of Health grant R01 HL64304-08 and the Muscular Dystrophy Association (to R.J.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1251) on April 29, 2009.
REFERENCES
- Alyonycheva T. N., Mikawa T., Reinach F. C., Fischman D. A. Isoform-specific interaction of the myosin-binding proteins (MyBPs) with skeletal and cardiac myosin is a property of the C-terminal immunoglobulin domain. J. Biol. Chem. 1997;272:20866–20872. doi: 10.1074/jbc.272.33.20866. [DOI] [PubMed] [Google Scholar]
- Arimura T., Matsumoto Y., Okazaki O., Hayashi T., Takahashi M., Inagaki N., Hinohara K., Ashizawa N., Yano K., Kimura A. Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2007;362:281–287. doi: 10.1016/j.bbrc.2007.07.183. [DOI] [PubMed] [Google Scholar]
- Armani A., Galli S., Giacomello E., Bagnato P., Barone V., Rossi D., Sorrentino V. Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp. Cell Res. 2006;312:3546–3558. doi: 10.1016/j.yexcr.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Auerbach D., Bantle S., Keller S., Hinderling V., Leu M., Ehler E., Perriard J. C. Different domains of the M-band protein myomesin are involved in myosin binding and M-band targeting. Mol. Biol. Cell. 1999;10:1297–1308. doi: 10.1091/mbc.10.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang M. L., Centner T., Fornoff F., Geach A. J., Gotthardt M., McNabb M., Witt C. C., Labeit D., Gregorio C. C., Granzier H., Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Barth R., Elce J. S. Immunofluorescent localization of a Ca2+-dependent neutral protease in hamster muscle. Am. J. Physiol. 1981;240:E493–E498. doi: 10.1152/ajpendo.1981.240.5.E493. [DOI] [PubMed] [Google Scholar]
- Borisov A. B., Kontrogianni-Konstantopoulos A., Bloch R. J., Westfall M. V., Russell M. W. Dynamics of obscurin localization during differentiation and remodeling of cardiac myocytes: obscurin as an integrator of myofibrillar structure. J. Histochem. Cytochem. 2004;52:1117–1127. doi: 10.1369/jhc.3A6183.2004. [DOI] [PubMed] [Google Scholar]
- Borisov A. B., Martynova M. G., Russell M. W. Early incorporation of obscurin into nascent sarcomeres: implication for myofibril assembly during cardiac myogenesis. Histochem. Cell Biol. 2008;129:463–478. doi: 10.1007/s00418-008-0378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov A. B., Raeker M. O., Kontrogianni-Konstantopoulos A., Yang K., Kurnit D. M., Bloch R. J., Russell M. W. Rapid response of cardiac obscurin gene cluster to aortic stenosis: differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem. Biophys. Res. Commun. 2003;310:910–918. doi: 10.1016/j.bbrc.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Borisov A. B., Sutter S. B., Kontrogianni-Konstantopoulos A., Bloch R. J., Westfall M. V., Russell M. W. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem. Cell Biol. 2006;125:227–238. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- Borzok M. A., Catino D. H., Nicholson J. D., Kontrogianni-Konstantopoulos A., Bloch R. J. Mapping the binding site on small ankyrin 1 for obscurin. J. Biol. Chem. 2007;282:32384–32396. doi: 10.1074/jbc.M704089200. [DOI] [PubMed] [Google Scholar]
- Bowman A. L., Catino D. H., Strong J. C., Randall W. R., Kontrogianni-Konstantopoulos A., Bloch R. J. The rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Mol. Biol. Cell. 2008;19:3782–3792. doi: 10.1091/mbc.E08-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A. L., Kontrogianni-Konstantopoulos A., Hirsch S. S., Geisler S. B., Gonzalez-Serratos H., Russell M. W., Bloch R. J. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L., Yu J. G., Thornell L. E. New aspects of obscurin in human striated muscles. Histochem. Cell Biol. 2008;130:91–103. doi: 10.1007/s00418-008-0413-z. [DOI] [PubMed] [Google Scholar]
- Cunha S. R., Mohler P. J. Obscurin targets ankyrin-B and protein phosphatase 2A to the cardiac M-line. J. Biol. Chem. 2008;283:31968–31980. doi: 10.1074/jbc.M806050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Deyne P. G., O'Neill A., Resneck W. G., Dmytrenko G. M., Pumplin D. W., Bloch R. J. The vitronectin receptor associates with clathrin-coated membrane domains via the cytoplasmic domain of its beta5 subunit. J. Cell Sci. 1998;111:2729–2740. doi: 10.1242/jcs.111.18.2729. [DOI] [PubMed] [Google Scholar]
- Flashman E., Korkie L., Watkins H., Redwood C., Moolman-Smook J. C. Support for a trimeric collar of myosin binding protein C in cardiac and fast skeletal muscle, but not in slow skeletal muscle. FEBS Lett. 2008;582:434–438. doi: 10.1016/j.febslet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Flashman E., Redwood C., Moolman-Smook J., Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 2004a;94:1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- Flashman E., Redwood C., Moolman-Smook J., Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 2004b;94:1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- Flucher B. E., Conti A., Takeshima H., Sorrentino V. Type 3 and type 1 ryanodine receptors are localized in triads of the same mammalian skeletal muscle fibers. J. Cell Biol. 1999;146:621–630. doi: 10.1083/jcb.146.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougerousse F., Delezoide A. L., Fiszman M. Y., Schwartz K., Beckmann J. S., Carrier L. Cardiac myosin binding protein C gene is specifically expressed in heart during murine and human development. Circ. Res. 1998;82:130–133. doi: 10.1161/01.res.82.1.130. [DOI] [PubMed] [Google Scholar]
- Freiburg A., Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- Fukuzawa A., Idowu S., Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J. Muscle Res. Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- Fukuzawa A., Lange S., Holt M., Vihola A., Carmignac V., Ferreiro A., Udd B., Gautel M. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J. Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- Furst D. O., Vinkemeier U., Weber K. Mammalian skeletal muscle C-protein: purification from bovine muscle, binding to titin and the characterization of a full-length human cDNA. J. Cell Sci. 1992;102:769–778. doi: 10.1242/jcs.102.4.769. [DOI] [PubMed] [Google Scholar]
- Gautel M., Furst D. O., Cocco A., Schiaffino S. Isoform transitions of the myosin binding protein C family in developing human and mouse muscles: lack of isoform transcomplementation in cardiac muscle. Circ. Res. 1998;82:124–129. doi: 10.1161/01.res.82.1.124. [DOI] [PubMed] [Google Scholar]
- Gilbert R., Cohen J. A., Pardo S., Basu A., Fischman D. A. Identification of the A-band localization domain of myosin binding proteins C and H (MyBP-C, MyBP-H) in skeletal muscle. J. Cell Sci. 1999;112:69–79. doi: 10.1242/jcs.112.1.69. [DOI] [PubMed] [Google Scholar]
- Gilbert R., Kelly M. G., Mikawa T., Fischman D. A. The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J. Cell Sci. 1996;109:101–111. doi: 10.1242/jcs.109.1.101. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Prevec L. Methods for construction of adenovirus vectors. Mol. Biotechnol. 1995;3:207–220. doi: 10.1007/BF02789331. [DOI] [PubMed] [Google Scholar]
- Gregorio C. C., et al. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell Biol. 1998;143:1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M. L. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Bloch R. J. The hydrophilic domain of small ankyrin-1 interacts with the two N-terminal immunoglobulin domains of titin. J. Biol. Chem. 2003;278:3985–3991. doi: 10.1074/jbc.M209012200. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Bloch R. J. Obscurin: a multitasking muscle giant. J. Muscle Res. Cell Motil. 2005;26:419–426. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Bloch R. J. De novo myofibrillogenesis in C2C12 cells: evidence for the independent assembly of M bands and Z disks. Am. J. Physiol. Cell Physiol. 2006a;290:C626–C637. doi: 10.1152/ajpcell.00442.2005. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Randall W. R., Bloch R. J. Obscurin regulates the organization of myosin into A bands. Am. J. Physiol. Cell Physiol. 2004;287:C209–C217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Sutter S., Borisov A. B., Pumplin D. W., Russell M. W., Bloch R. J. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006b;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Jones E. M., Van Rossum D. B., Bloch R. J. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto C. A., Fischman D. A., Reinach F. C. The interface between MyBP-C and myosin: site-directed mutagenesis of the CX myosin-binding domain of MyBP-C. J. Muscle Res. Cell Motil. 1999;20:703–715. doi: 10.1023/a:1005513312939. [DOI] [PubMed] [Google Scholar]
- Moos C., Offer G., Starr R., Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J Mol. Biol. 1975;97:1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- Oakley C. E., Chamoun J., Brown L. J., Hambly B. D. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int. J. Biochem. Cell Biol. 2007;39:2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Obermann W. M., Gautel M., Weber K., Furst D. O. Molecular structure of the sarcomeric M band: mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16:211–220. doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann W. M., van der Ven P. F., Steiner F., Weber K., Furst D. O. Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band. Mol. Biol. Cell. 1998;9:829–840. doi: 10.1091/mbc.9.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki T., Weber F. E., Fischman D. A., Vaughan K. T., Mikawa T., Reinach F. C. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 1993a;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki T., Weber F. E., Fischman D. A., Vaughan K. T., Mikawa T., Reinach F. C. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 1993b;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. G. Skelemins: cytoskeletal proteins located at the periphery of M-discs in mammalian striated muscle. J. Cell Biol. 1987;104:1325–1336. doi: 10.1083/jcb.104.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeker M. O., Su F., Geisler S. B., Borisov A. B., Kontrogianni-Konstantopoulos A., Lyons S. E., Russell M. W. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev. Dyn. 2006;235:2018–2029. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- Russell M. W., Raeker M. O., Korytkowski K. A., Sonneman K. J. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–246. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Sato N., Kawakami T., Nakayama A., Suzuki H., Kasahara H., Obinata T. A novel variant of cardiac myosin-binding protein-C that is unable to assemble into sarcomeres is expressed in the aged mouse atrium. Mol. Biol. Cell. 2003;14:3180–3191. doi: 10.1091/mbc.E02-10-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr R., Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem. J. 1978;171:813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F. E., Vaughan K. T., Reinach F. C., Fischman D. A. Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. Eur. J Biochem. 1993;216:661–669. doi: 10.1111/j.1432-1033.1993.tb18186.x. [DOI] [PubMed] [Google Scholar]
- Welikson R. E., Fischman D. A. The C-terminal IgI domains of myosin-binding proteins C and H (MyBP-C and MyBP-H) are both necessary and sufficient for the intracellular crosslinking of sarcomeric myosin in transfected non-muscle cells. J. Cell Sci. 2002;115:3517–3526. doi: 10.1242/jcs.115.17.3517. [DOI] [PubMed] [Google Scholar]
- Williams M. W., Bloch R. J. Extensive but coordinated reorganization of the membrane skeleton in myofibers of dystrophic (mdx) mice. J. Cell Biol. 1999;144:1259–1270. doi: 10.1083/jcb.144.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. W., Resneck W. G., Bloch R. J. Membrane skeleton of innervated and denervated fast- and slow-twitch muscle. Muscle Nerve. 2000;23:590–599. doi: 10.1002/(sici)1097-4598(200004)23:4<590::aid-mus19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Winegrad S. Cardiac myosin binding protein C. Circ. Res. 1999;84:1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]
- Yasuda M., Koshida S., Sato N., Obinata T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J. Mol. Cell Cardiol. 1995;27:2275–2286. doi: 10.1016/s0022-2828(95)91731-4. [DOI] [PubMed] [Google Scholar]
- Young P., Ehler E., Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Birkenmeier C. S., Williams M. W., Sharp J. J., Barker J. E., Bloch R. J. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J. Cell Biol. 1997;136:621–631. doi: 10.1083/jcb.136.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.