Summary
We examined genome-wide expression datasets from human frontal cortex of normal and schizophrenic individuals ranging from 19 to 81 years of age. We found that changes in gene expression that are correlated with aging in normal subjects differ dramatically from those observed with aging in schizophrenic subjects. Only 2.5% of genes were correlated with age in both groups. Surprisingly, we also found a significant overlap (29−34%) between those genes whose expression was correlated with aging in normal subjects and those significantly altered in subjects with early-stage schizophrenia (within 4 years of diagnosis). This suggests that schizophrenia onset anticipates the normal aging process, and further, that some symptoms of aging, i.e. dementia and psychosis, might be explained by these common molecular profiles.
Keywords: Age, aging, psychiatric, microarray, real-time PCR, antipsychotic, Alzheimer's
Schizophrenia afflicts 1% of the general population, with onset in the late teens or early adulthood. In his original report, Kraepelin described schizophrenia as a chronic deteriorating psychiatric disorder characterized by rapid cognitive disintegration, calling it “dementia praecox” (premature dementia) (Kraepelin 1971 (original 1919)). While the degenerative nature of schizophrenia is controversial, studies have demonstrated that brain structural features, as well as predominant symptomatology change through the course of illness (Lieberman 1999; Hulshoff Pol and Kahn 2008). These events may involve an active, progressive pathology that continues after onset, or a response to a developmental insult(s) that changes with the aging process. Several studies have reported genome-wide-expression changes in schizophrenia [reviewed in (Mirnics et al. 2006)], although none have examine the molecular features of aging in this disease.
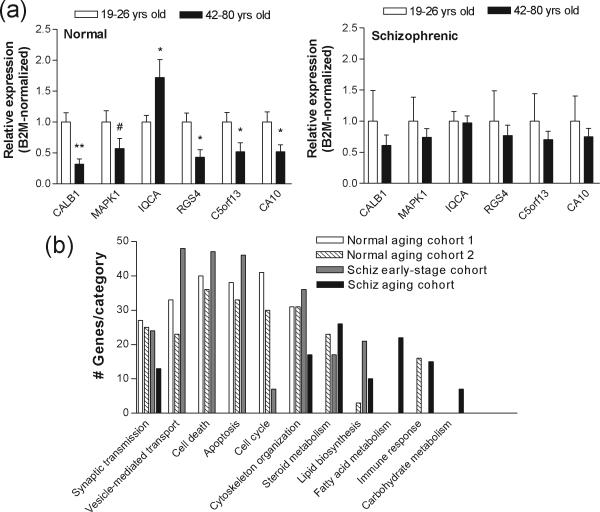
In our previous studies, we generated gene expression profiles from post-mortem human prefrontal cortex (BA46) of 30 normal and 29 schizophrenic subjects ranging from 19 to 81 years of age (Supplementary Table 1) (Narayan et al. 2008) using Affymetrix Human Genome U133-Plus 2.0 arrays [see (Narayan et al. 2008) for detailed descriptions of RNA preparations and microarray hybridizations]. We re-analyzed these raw data using linear regression analyses and computed Pearson Product-Moment Correlation coefficients for age against the log2 expression values for 14,439 genes in each subject (Supplementary Methods). After adjusting for multiple statistical testing and the effects of tissue covariables, pH and PMI, the expression of 643 genes in controls and 343 genes in individuals with schizophrenia was found to be significantly correlated with age (Pearson|r|≥0.5; p<0.05; Suppl. Table 2). Surprisingly, there was very little overlap between the two lists of genes, with the expression of only 10 genes being correlated with age in both schizophrenic and control subjects. Real-time PCR analysis confirmed expression differences for the top correlated genes (Pearson|r|≥0.8) in young (19−26 yrs) vs. older (42−81 yrs) aged control groups, changes that were not observed in schizophrenic subjects (Fig. 1a).
Figure 1.
A. Real-time PCR analysis of expression levels for the indicated genes in human prefrontal cortical samples (BA46) from normal and schizophrenic individuals. The relative abundance of each gene expression was normalized by beta-2 microglobulin (B2M) and beta-tubulin (TUBB) in young vs. old human samples, respectively. Data are depicted as fold-change of the mean expression level ± SEM (n=8−12 normal and/or schizophrenic subjects per). Student's t tests were used to determine significant differences in gene expression levels. Genes are denoted by their official Unigene gene symbol IDs. * denotes significantly different from control at p<0.05, **, p<0.01, two-tailed t test; # denotes significantly different from controls at p<0.05, one-tailed t test. B. Gene Ontology categories significantly represented in each group of subjects, as indicated. Gene Ontology classification was performed using the DAVID database. The numbers of genes in each category is shown on the y-axis.
We next performed the same analysis on microarray expression data from a second cohort of normal subjects (n=30; 26−106 years of age)(Lu et al. 2004), using data freely available on the GEO/NCBI website (record #GDS707), and compared the resulting correlated genes to those obtained from subgroups of our normal population. Using the Chi-Squared Test for Independence to compare frequencies of overlapping genes, we found that the expression of a significantly greater number of genes was correlated with age between both normal cohorts compared to those genes correlated with age between normal and schizophrenic cohorts (n=130.0±75 vs. 6.6±3.33, respectively; χ2=139.8−1647; p=0.00). Thus, the relative similarities among the normal populations indicate that the lack of similarity in age-correlated variation between schizophrenics and normals is not due to general heterogeneity within the population or between microarray datasets.
“Functions” analyses of our gene lists using The Database for Annotation, Visualization and Integrated Discovery (DAVID) database indicated that the normal aging process was significantly linked to abnormalities in pathways related to synaptic function, cell cycle/DNA damage and apoptosis (Fig. 1b), consistent with previous microarray studies investigating aging in normal humans (Erraji-Benchekroun et al. 2005; Yankner et al. 2008). In contrast, aging in schizophrenia was significantly associated with fatty acid and steroid metabolism, but not with those functions associated with normal aging (Fig. 1b).
The different age-related expression profiles detected in schizophrenic subjects might result from a progressive pathogenic process, a response to pathology or, possibly, drug treatment, considering that a confounding factor in post-mortem research on schizophrenia is the unknown effect of antipsychotic drugs, which are known to alter gene expression (Thomas 2006). Arguments against a strong treatment effect in our data include the facts that the expression of only 1 of the 343 age-correlated genes was correlated with the patients’ recorded drug doses, and that we found no changes in expression of a subset of these genes in the brains of mice treated with haloperidol (2 mg/kg) or fluphenazine (2.5 mg/kg) (Suppl. Fig. 1), the same drugs with which most of the schizophrenic subjects were treated (Suppl. Table 1). However, it remains possible that antipsychotic drug treatment might affect transcriptome profiles in aging schizophrenics.
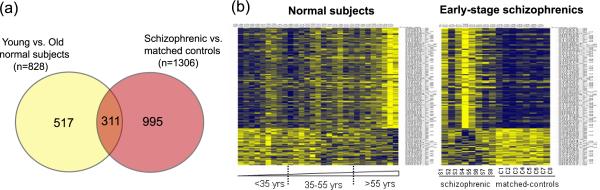
Given Kraepelin's description of premature dementia in schizophrenia, it is perhaps not surprising that, at disease onset, schizophrenia is associated with a decline in cognition and adaptive functioning, similar to that observed in normal aging. Recent studies have also detected microglia activation, which is associated with normal aging (Miller and Streit 2007), in recent-onset schizophrenia (van Berckel et al. 2008). Furthermore, normal aging has been linked to alterations in white matter density and volume, gray matter volume decline, cognitive dysfunction and psychotic symptoms (van der Werf et al. 2007; Yankner et al. 2008), which also characterize schizophrenia at first episode (Lieberman 1999; Steen et al. 2006; Witthaus et al. 2008). Given these commonalities, we hypothesized that early-stage schizophrenia and normal aging might share common molecular underpinnings. We compared transcriptome variation between schizophrenic subjects in early stages of illness (up to 4 years from initial diagnosis; n=8) versus matched controls from our previous studies (see Suppl. Table 3; (Narayan et al. 2008)) with that generated from linear regression correlation through aging in normal subjects described above. Surprisingly, we found a substantial overlap between these two gene lists: 189 of the genes (29.3%) whose expression levels were correlated with age in normal subjects was concordantly dysregulated in subjects with early-stage schizophrenia compared to age- and sex-matched controls. A further group-wise comparison of young (<38 years of age) versus aged normal subjects (>50 years of age) using ANOVA (Suppl. Methods) revealed 811 differentially expressed genes (p<0.05; Suppl. Table 3), 34.2% of which also significantly varied in subjects with early-stage schizophrenia (Fig. 2a). These overlaps were actually greater than those observed between our two control populations and significantly greater than that predicted from two independent samples (χ2=13.9; p=0.0009). Heatmap depictions of these transcriptome profiles during normal aging and early-stage schizophrenia are shown in Fig. 2b. We also found that pathways/functions associated with early-stage schizophrenia identified by DAVID searches were similar to those related to normal aging and totally different from those associated with aging in schizophrenia (Fig. 1b).
Figure 2.
A). Overlap of genes differentially expressed in early-stage schizophrenia with those occurring in normal aging (p<0.05). Gene lists used for these comparisons are provided in Suppl. Table 3. B). Heatmap visualization of the expression values (Log2-transformed; unclustered) of the overlapping genes shown in (A) in normal subjects throughout aging (n=30 subjects) and in schizophrenic subjects ≤4 yrs from diagnosis (n=8 schizophrenic subjects [S1-S8] and matched controls [C1-C8]). Each colored pixel represents an individual gene expression value from a single subject. Relative decreases in gene expression are indicated by yellow and increases in expression by blue.
These data demonstrate that the molecular correlates for aging differ between schizophrenic and normal subjects. In addition, normal aging and early-stage schizophrenia share common molecular signatures, suggesting that the onset of schizophrenia anticipates the normal aging process. In addition, we suggest that some symptoms of aging, i.e. dementia and psychosis, might be explained by these common molecular profiles.
Supplementary Material
Acknowledgements
This study was funded by the National Institutes of Health grant MH069696 to E.A.T.
References
- Buckner RL. Memory and executive function in aging and AD: multiple factors that cause decline and reserve factors that compensate. Neuron. 2004;44:195–208. doi: 10.1016/j.neuron.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJSibille E. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–58. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HEKahn RS. What happens after the first episode? A review of progressive brain changes in chronically ill patients with schizophrenia. Schizophr Bull. 2008;34:354–66. doi: 10.1093/schbul/sbm168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepelin E. Dementia praecox and paraphrenia. Kreiger R. E.; Melbourne: 1971. (original 1919) [Google Scholar]
- Lieberman JA. Is schizophrenia a neurodegenerative disorder? A clinical and neurobiological perspective. Biol Psychiatry. 1999;46:729–39. doi: 10.1016/s0006-3223(99)00147-x. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan JYankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Miller KRStreit WJ. The effects of aging, injury and disease on microglial function: a case for cellular senescence. Neuron Glia Biol. 2007;3:245–53. doi: 10.1017/S1740925X08000136. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Levitt PLewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–76. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean BThomas EA. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RG, Mull C, McClure R, Hamer RMLieberman JA. Brain volume in first-episode schizophrenia: systematic review and meta-analysis of magnetic resonance imaging studies. Br J Psychiatry. 2006;188:510–8. doi: 10.1192/bjp.188.6.510. [DOI] [PubMed] [Google Scholar]
- Thomas EA. Molecular profiling of antipsychotic drug function: convergent mechanisms in the pathology and treatment of psychiatric disorders. Mol Neurobiol. 2006;34:109–28. doi: 10.1385/MN:34:2:109. [DOI] [PubMed] [Google Scholar]
- van Berckel BN, et al. Microglia activation in recent-onset schizophrenia: a quantitative (R)-[11C]PK11195 positron emission tomography study. Biol Psychiatry. 2008;64:820–2. doi: 10.1016/j.biopsych.2008.04.025. [DOI] [PubMed] [Google Scholar]
- van der Werf M, van Boxtel M, Verhey F, Jolles J, Thewissen Vvan Os J. Mild hearing impairment and psychotic experiences in a normal aging population. Schizophr Res. 2007;94:180–6. doi: 10.1016/j.schres.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Witthaus H, et al. White matter abnormalities in subjects at ultra high-risk for schizophrenia and first-episode schizophrenic patients. Schizophr Res. 2008;102:141–149. doi: 10.1016/j.schres.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Yankner BA, Lu TLoerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.