Summary of recent advances
Bacterial marine natural products are an important source of novel lead structures for drug discovery. The cytotoxic properties of many of these secondary metabolites are of particular interest for the development of new anti-cancer agents. Tremendous advances in marine molecular biology, genome sequencing, and bioinformatics have paved the way to fully exploit the biomedical potential of marine bacterial products. In addition, unique biosynthetic enzymes discovered from bacteria from the sea have begun to emerge as powerful biocatalysts in medicinal chemistry and total synthesis. The increasingly interdisciplinary field of marine natural product chemistry thus strongly impacts future developments in medicine, chemistry, and biology.
Introduction
Investigations on bioactive natural products have yielded many of the most important chemotherapeutic agents for the treatment of human diseases known to date (1). Beginning with the discovery of the first marine-derived natural products, spongouridine and spongothymidine from the sponge Cryptetethia crypta by Bergmann and Feeney and of prostaglandin derivatives in the Caribbean horn coral Plexaura homomalla by Weinheimer and Spraggins, the quest for new bioactive agents was extended from terrestrial organisms into the marine environment. Marine invertebrates have since served as a tremendous source of structurally unprecedented bioactive secondary metabolites (2). The use of these potential drugs, however, has largely been hampered by the low abundance of the natural producers and/or the low concentrations of the compounds of interest. Furthermore, the highly complex structures of many of these metabolites, makes synthetic approaches for their development at least economically daunting.
The biomedical potential of marine bacterial agents, which should be amenable to biotechnological production, might help overcome the problems of supply and sustainability encountered with macroorganisms from the sea. Marine cyanobacteria continue to be an important source of bioactive metabolites (3). Prominent examples of compounds isolated from cyanobacteria comprise the anti-microtubule agents dolastatin A and curacin A, which entered preclinical and clinical trials and served as lead structures for the development of a number of synthetic analogs with pronounced antitumor activity. In addition to this well established source of microbial metabolites, marine-derived actinomycetes, such as species belonging to the genus Salinispora (4), have attracted considerable attention in recent years (5,6). The most promising example of a metabolite derived from these marine-dwelling bacteria is salinosporamide A, a highly potent irreversible inhibitor of the 20S proteasome, which entered clinical evaluation only three years after its discovery (7).
The development of advanced analytical and preparative methods helps extend the value of marine bacteria, the products they synthesize, and the genes they carry. While improvements of analytical tools facilitate the identification of the natural source of a certain compound even from complex microbial consortia, the application of modern microbial genetics allows for the rational design of recombinant organisms (8) to produce novel “unnatural” natural products by combinatorial biosynthesis (9) and total in vitro (bio)synthesis (10). This review highlights recent advances and strives to illustrate the dynamics of the highly active field of modern microbial marine natural product chemistry.
Discovery of novel bioactive compounds from marine bacteria
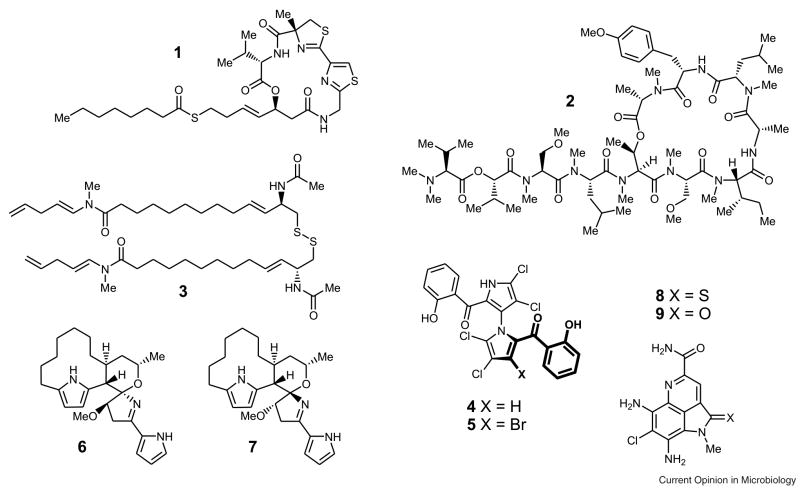
Marine bacteria still represent an emerging source of new natural products. The majority of compounds isolated from marine cyanobacteria originates from the filamentous order Nostocales, in particular from members of the genera Lyngbya, Oscillatoria, and Symploca. Many of these metabolites are modified peptides or peptide–polyketide hybrids exhibiting antitumor activities (3). Despite the common occurrence of this structure class, fascinating new molecules with potent biological activities, such as largazole (1), coibamide (2), and somocystinamide A (3) (Figure 1), continue to be identified. Largazole (1) was isolated from a cyanobacterial sample of a Symploca sp. collected from Key Largo, Florida Keys (11). Its structure is remarkable as it is the first cyanobacterial natural product containing a thioester. The sulfur-containing side chain of 1, the 3-hydroxy-7-mercaptohept-4-enoic acid residue, constitutes a new structural feature in marine natural product chemistry. The antiproliferative 1 exhibits an enhanced biological activity against transformed human mammary epithelial and fibroblastic osterosarcoma cells as compared to untransformed cells, which suggests preferentially targeting cancer cells (11). Its impressive biological activity has already led to the development of a number of total syntheses (12–14), providing sufficient material to establish its mode of action as a histone deacetylase inhibitor (13,14). Coibamide A (2), which was isolated from a Leptolyngbya sp. (Coiba National Park, Panama), is likewise a novel peptidic cytotoxin (15). This highly N-methylated cyclic depsipeptide stands out due to its as yet unprecedented selectivity profile in the NCI 60 cell line panel, indicating inhibition of cancer cell proliferation through a novel mechanism. Compound 2 thus represents a valuable lead for drug development. Another very promising candidate for cancer therapy is the structurally atypical lipopeptide somocystinamide A (3), which had originally been described in 2002 by the Gerwick lab and was most recently reported to initiate caspase-8-dependent apoptosis in a number of human tumor cell lines (16). Interestingly, modifications of either the disulfide or the lipopeptide moiety in 3 led to a complete loss of activity. Integration of 3 into nanosomes, however, was shown not to affect the biological activity. Application of targetable nanoplatforms (17) harboring 3, might thus solve stability and solubility problems of the compound and facilitate transport of 3 exclusively into the desired tumor cells.
Figure 1.
Highly bioactive natural products recently isolated from marine cyanobacteria and actinomycetes: largazole (1) from Symploca sp., coibamide (2) from Leptolyngbya sp., somocystinamide A (3) from a Lyngbya majuscula/Schizothrix mixed assemblage, as well as marinopyrroles A (4) and B (5), marineosins A (6) and B (7), and ammosamides A (8) and B (9), all isolated from Streptomyces spp.
The structural and biosynthetic variations of secondary metabolites isolated from marine actinomycetes, as exemplified by the marinopyrols, the marineosins, and the ammosamides, exceeds the breadth displayed by cyanobacterial compounds. The marinopyrroles A (4) and B (5), produced by a Streptomyces sp. isolated from a marine sediment sample collected in La Jolla, California, are heavily halogenated bispyrroles, whose monomeric building blocks are connected via a unique, rotationally stable N,C-biaryl axis (18). Both compounds exhibit promising activities against methicillin-resistant Staphylococcus aureus and might thus be highly interesting as antibiotic lead structures. While the monomeric units of 4 and 5 have long been known as natural products, the two streptomycete-derived diastereomers marineosins A (6) and B (7) possess a previously unknown carbon skeleton which contains a spiro-tetrahydropyran-dihydropyrrole aminal moiety (19). The spiro center is likely formed by a non-stereoselective hetero Diels-Alder reaction of a linear precursor, which is in part derived from prodigiosin biosynthesis. Most interestingly, 7 is approximately 100-fold less active than 6. The latter showed broad cytotoxicity in the NCI 60 cell line panel, in addition to considerable selectivity against leukemia and melanoma cell lines. The ammosamides A (8) and B (9), derived from a deep-sea sediment sample Streptomyces strain collected in the Bahamas (20), are chlorinated tricyclic pyrroloquinoline alkaloids, structurally related to the microbial and sponge metabolites lymphostin and batzelline A, respectively. The slow conversion of the thioamide in 8 to the amide in 9 upon standing on air suggests a non-enzymatic formation of the latter in the natural producer. Both compounds exhibited strong in vitro cytotoxicity against HCT-116 colon carcinoma and pronounced selectivity against a number of other cancer cell lines, thus indicating a specific mode of action. The cellular target of these compounds was identified using a semisynthetic immunoaffinity fluorescent probe (21) composed of ammosamide B (9) coupled to 7-dimethylaminocoumarin-4-acetic acid (22). The simple one-step preparation of such an ammosamide-based fluorescent probe that selectively binds myosin facilitates future investigations on a variety of myosin-regulated cellular processes and may thus significantly impact further developments of chemotherapeutics targeting this protein.
Investigations on the origin of marine microbial natural products
The biosynthetic origin of marine natural products isolated from samples obtained by the collection of a certain organism from its natural habitat is often far less obvious than in the above examples. This is in particular true for invertebrates. The marine environment consists of highly complex microbial communities and many invertebrates are associated with large amounts of epi- and endobiotic microorganisms (23,24). The structural homology of a number of metabolites isolated from marine macroorganisms with natural products also found in microbial sources thus imposes questions about the true biosynthetic origin of these molecules. Early experimental approaches to address this problem were based on the separation of invertebrate and microbial cells and independent analysis of the metabolites of the respective purified cell samples. Recent technological advances in mass spectrometry leading to increased resolution and mass accuracy facilitate the detection and – in part – even structural assignment of minute quantities of compounds from crude extracts of a potential producer of interest (25). The application of MALDI-TOF-MS even allows the analysis of complex bacterial mixtures and to taxonomically identify individual microorganisms based on their mass fingerprints (phyloproteomics), a technique which has already been applied to probe the phylogenetic and chemical diversity of bacteria in marine sponges (26). Expanding on previous work by the von Döhren lab, natural product MALDI-TOF-imaging (npMALDI-I) for the identification and spatial detection of secondary metabolites in marine cyanobacteria and sponges was recently introduced (25,27). While this approach has been used before in other fields (28,29), it had never before been applied to natural product analysis. This new imaging technique has now been utilized to locate phytochemical marker molecules within certain cyanobacteria, even in complex mixtures with other cyanobacteria and to resolve the spatial distribution of molecules in heterogenous assemblages, such as the sponge Dysidea herbacea, thereby illuminating the existence of micro-environments within sponge tissues. Furthermore, npMALDI-I was applied to rapidly identify unknown metabolites in a specific producer (27). Future applications of npMALDI-I might thus have a high impact on directed drug-discovery programs. Nevertheless, npMALDI-I and all other methods mentioned above can only confirm the presence of a secondary metabolite in a particular cell or extract. The mere presence of a compound in a certain organism within a biocoenosis, however, can also be the result of active or passive accumulation and does not necessarily correlate with the site of its biosynthetic production. An unambiguous assignment of the biosynthetic origin of a natural product derived from a complex assemblage of marine organisms thus has to originate at the genomic level. This is particularly true for bacterial symbionts, which have to date eluded laboratory cultivation.
Gene-based strategies to access marine bacterial natural products
The first experimental demonstration of the power of microbial genetics to answer yet unsolved questions on the origin of natural products was provided by Piel et al. by cloning the pederin biosynthesis gene cluster from the symbiotic bacterium of the Paederus beetle (30). The Piel group further showed that the antitumor polyketide onnamide, which is structurally related to pederin and was previously assigned to be produced by the sponge Theonella swinhoei, is likewise of bacterial origin (31). Following these milestone publications, two independent studies reported the discovery of a microcin-like ribosomal pathway for the biosynthesis of ‘ascidian-derived’ cyclic peptides, the patellamides, e.g. patellamide C (10), in their cyanobacterial symbionts Prochloron didemni (Figure 2) (32, 33). In addition, this work established the first example of the cloning of a marine natural product biosynthetic gene cluster derived from a bacterial symbiont and its functional expression in the heterologous host Escherichia coli. Comparative analysis of biosynthesis genes isolated from different Prochloron spp. involved in the formation of structurally diverse cyclic peptides revealed a highly conserved genetic background (99% identity) within these clusters, only differing in short, interchangeable, hypervariable cassettes (patE) encoding the precursor peptide, which directly yields the backbone of the respective products (34). Simple mutations of the amino acid sequences in patE in these Prochloron spp. is reported to give rise to the accumulation of natural combinatorial peptide libraries in the bacterial hosts, the ascidians. The assembly line for cyanobactin biosynthesis thus should offer the opportunity to genetically engineer biomedically relevant cyclic peptides with predictable structures. The practicability and ease of this approach has already been demonstrated by mutating the ulithiacyclamide (11) amino acid sequence from PatE2 to heterologously produce the novel peptide eptidemnamide (12) (34), and by the generation of the additionally prenylated antitumor preclinical candidate trunkamide using a similar procedure (35). Very recently, the pathway specific proteases PatA and PatG were shown to catalyze proteolytic cleavage of the precursor peptide with subsequent cyclization to give cyanobactins (36). In particular the cyclization reaction catalyzed by PatG might prove useful to produce a structurally diverse library of cyclic peptides with potential biomedical value from simple linear synthetic precursors. The global occurrence of cyanobactin-type ribosomal peptides was further validated by the identification of two related clusters from the freshwater cyanobacterium Microcystis aeroginosa encoding microcyclamide (37) and microviridin (38,39) biosynthesis.
Figure 2.
Engineered production of eptidemnamide (12) in the heterologous host E. coli by exchanging the ulithiacyclamide (11) sequence (green) in natural PatE2 with the amino acid sequence specifying 12 (yellow). Patellamide C (10, grey) is encoded by both, PatE2 and the recombinant PatEdm.
The establishment of the cyanobactin biosynthetic family impressively demonstrates the power of microbial genetics to identify the origin of secondary metabolites and to subsequently utilize the respective biosynthetic machinery to produce new therapeutically interesting derivatives. The detection of biosynthetic gene clusters by construction of clone libraries from genomic DNA of an assumed metabolic producer of a compound of interest, however, can be tedious and time consuming. Improvements in genome sequencing techniques and, consequently, dramatically reduced prices and greatly enhanced speed for such projects renders whole genome mapping a worthwhile alternative. Furthermore, having the entire genome sequence of a promising bacterial secondary metabolite producer in hand not only allows one to identify and manipulate a single biosynthetic pathway leading to a known natural product, but also allows for the opportunity to mine new chemical entities associated with orphan gene clusters, to control primary metabolic precursors that also support secondary metabolism, and to explore the ecological relevance and evolutionary history of the respective secondary metabolome.
Genome sequencing as a tool to discover novel biosynthetic pathways
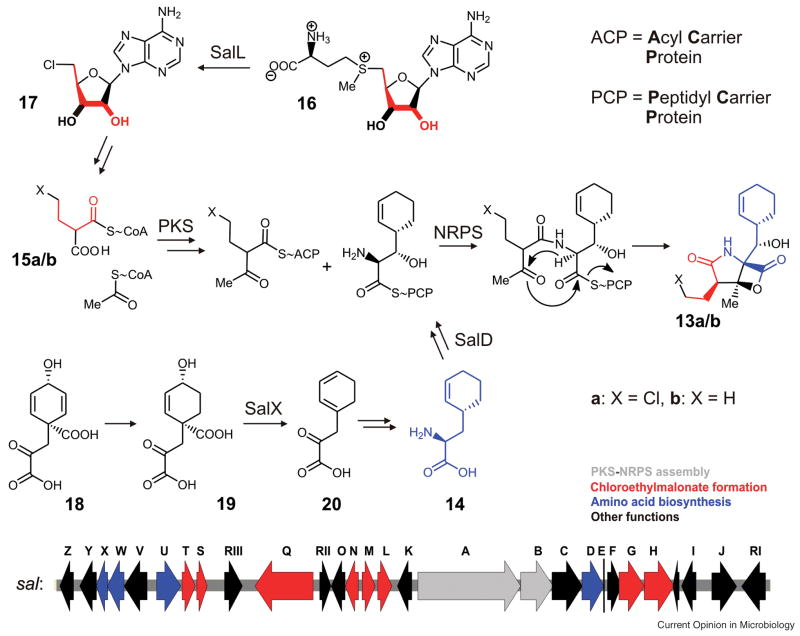
The first example of a fully sequenced genome of an obligate marine actinobacterium is that of the deep-sea dwelling actinomycete Salinispora tropica (40). Bioinformatic analysis of the sequence data revealed an unusual high percentage of the 5,183,331-bp circular chromosome to be dedicated to secondary metabolism (ca. 10%). The strain harbors at least 19 biosynthetic loci that encode a broad variety of natural product classes, including siderophores, polyketides, melanins, nonribosomal and ribosomal peptides, terpenoids, and aminocyclitols. Most striking is the high diversity of polyketide pathways found, including modular type I polyketide synthases (PKSs), iterative enediyne type I PKSs, hybrid type I PKS-NRPSs (nonribosomal peptide synthetases), heterodimeric type II PKSs, and homodimeric type III PKSs. Despite this large number of biosynthetic pathways and their remarkable diversity, only three different classes of secondary metabolites had been identified from S. tropica prior to the release of the sequence data, represented by the salinosporamides, the sporolides, and lymphostatin. The genomic data was used to predict and – in combination with classical isolation and structural elucidation techniques – fully characterize a fourth metabolite from S. tropica, the polyene macrolactam salinilactam A (40), thus clearly proving the high potential of the combination of genome mining and natural product chemistry for future drug discovery programs. In addition, bioinformatic analyses of the S. tropica genome led to the discovery of the pathways yielding the enediyne-derived sporolides (spo), which involves the biosynthesis of a highly functionalized cyclohexenone building block (41), and furthermore provided valuable insights into the formation of the salinosporamides. The biosynthesis of the latter had previously been investigated by feeding experiments using isotopically labeled precursors (42), which indicated a mixed PKS–NRPS biosynthetic origin. While salinosporamide A (13a) and its des-chloro analog salinosporamide B (13b) share two metabolic building blocks, namely acetate and the unprecedented amino acid 3-(cyclohex-2-enyl)alanine (14), they incorporate different PKS extension units, chloroethylmalonyl-CoA (15a) and ethylmalonyl-CoA (15b), respectively (Figure 3). Bioinformatic analysis of the salinosporamide cluster (sal) revealed the deduced pathway to be largely consistent with the proposed biosynthesis purely based on the feeding studies. Close examination of the biochemistry leading to the novel PKS extension unit 15a gave rise to the discovery and characterization of the unique SAM-dependent chlorinase SalL (43). This enzyme catalyzes the nucleophilic substitution of methionine from SAM (16) by a chloride ion to give 5′-chloro-5′-deoxyadenosine (17), which is further metabolized to yield 5′-chloro-5′-deoxyribose phosphate and ultimately chloroethylmalonyl-CoA (15a). Together with the fluorinase from Streptomyces cattleya and the duf-62 enzymes (44), SalL forms a new class of SAM-cleaving biocatalysts and represents a new biological chlorination reaction. The biosynthesis of the salinosporamide-specific amino acid building block involves a novel shunt in the shikimic acid pathway likely at the stage of prephenate (18) in which the pathway specific enzyme SalX, a prephenate dehydratase homolog, putatively catalyzes the decarboxylative dehydration of 19 to the diene 20 to finally give 3-(cyclohex-2-enyl)alanine (14).
Figure 3.
Proposed biosynthesis of salinosporamide A (13a) and B (13b) involving a novel chlorination pathway initialized by the conversion of 16 to 17 and the generation of a new amino acid building block 14.
Engineering of unnatural natural products in marine actinomycetes – salinosporamide A
The therapeutic significance of the salinosporamides triggered further efforts to genetically engineer novel γ-lactam-β-lactone derivatives to gain structure-activity relationship data. Like many proteasome inhibitors, salinosporamides form a covalent adduct with the nucleophilic Thr1 residue of the 20S proteasome through its electrophilic β-lactone unit (45). The added advantage of 13a over other inhibitors is the strategically placed chloro group that gets displaced from the ethyl side chain to give a stable, irreversibly bound adduct (46). The generation of analogs differing in the leaving group chemistry at the ethyl side chain of 13a was thus of particular interest. The respective bromine and iodine substituted derivatives were initially produced by precursor-directed biosynthesis (47) and semi-synthesis (48), respectively. Both halide analogs retained the biological activity and binding mechanism of 13a, owing to the similar reactivity of the leaving groups. Due to the significantly different leaving group properties of fluorine, the generation and biological evaluation of a fluorosalinospormide derivative 21 was sought by taking advantage of the newly discovered SalL-initiated chlorination pathway (Figure 4). Elimination of background biosynthesis of the chloroethylmalonyl building block (15a) by PCR-targeted gene replacement of the chlorinase salL facilitated the introduction of fluorine into the natural product by supplementing the blocked mutant with synthetic 5′-fluoro-5′-deoxyadenosine (22) (49). Investigations on the biological activity of fluorosalinosporamide (21) revealed its potent proteasome binding properties in which the fluoro group significantly extends its residence time on the proteasome.
Figure 4.
Inactivation of key enzymatic processes in salinosporamide biosynthesis, i.e. chlorination (SalL), oxidation (SalD), and precursor amino acid biosynthesis (SalX) in combination with complementary feeding of precursors for the production of novel salinosporamide derivatives with different halogen substituents, oxygenation patterns, and alkyl-substituents, respectively.
A similar mutasynthetic strategy was employed to structurally alter the cyclohexenyl group of salinosporamide A, which is key for its selectivity and binding to the proteasome β5-subunit. Chemical complementation of the salX disruption mutant with natural and unnatural amino acids 23a–c led to the formation of unnatural salinosporamide derivatives 24a–c (50), including the hybrid salinosporamide/omuralide compound antiprotealide (24a), which had previously been prepared by total synthesis (51) (Figure 4). Biological testing of these analogs delivered important data on the impact of ring size and saturation of the amino acid-derived alkyl substituent of salinosporamide on proteasome inhibition and will help in the intelligent design of future derivatives. In addition, the importance of the hydroxy substituent at the cycloalkenyl side chain was explored by deletion of the cytochrome P450 hydroxylase salD, which catalyzes the installation of this group in the biosynthesis of salinosporamide A (13a), and subsequent isolation and biomedical characterization of the less active deshydroxy analog 25 (50).
The above experiments did not only help illuminate structure-activity relationship data on salinosporamide-type compounds, but also led to the discovery of a number of novel biosynthetic transformations. Due to their fascinating chemical structures, marine microbial natural products continue to serve as the inspiration for the discovery of new biosynthetic enzymes (52–54) that may serve as valuable biocatalysts for the fast and effective generation of derivative libraries important in the drug discovery process. Enhanced use of such remarkable biosynthesis catalysts may ultimately influence modern natural product total synthesis.
Harnessing marine bacterial enzymes to total (bio)synthesis
An impressive example for the utilization of biosynthesis enzymes to access highly complex biologically active metabolites in vitro is the multienzyme total synthesis of the bacteriostatics enterocin (26) and wailupemycin (55). Their formation was achieved exclusively using proteins as biocatalysts, in case of 26 nine recombinant and three commercially available enzymes. The chemical starting material consisted of benzoic acid (27), the starter unit of the enterocin pathway, and malonyl-CoA (28) for PKS extension, as well as ATP and SAM (Figure 5). Using these components, enterocin total biosynthesis succeeded in a single reaction vessel. The enzymatic transformation led to the formation of ten C-C bonds, five C-O bonds, and the generation of seven chiral centers with perfect stereocontrol in 25% chemical yield. These numbers highlight the superiority with regard to cost and efficiency of such an ex vivo biosynthesis when compared to classical chemical synthesis. The extensive work associated with cloning, heterologous expression and purification of a large number of biosynthetic enzymes, however, constitutes a major drawback of the total biosynthesis of complex natural products. The combination of modern synthetic methods with powerful biocatalysts, in particular when dealing with (stereo)chemically demanding reactions, might be the most effective future approach to synthesize natural products in the chemical laboratory, as recently exemplified by the first stereoselective total synthesis of the antiproliferative polyketide (R)-aureotin (56).
Figure 5.
Ex vivo total biosynthesis of the bacteriostatic polyketide enterocin (26).
Conclusions
The biomedical potential of products derived from obligate marine bacteria is just beginning to be realized. The remarkable capabilities of these microbes to produce chemically unique bioactive molecules are supported by the characterization of a wealth of intriguing new structures through the application of classical screening and isolation techniques. In addition, an increasing number of secondary metabolites believed to originate from marine invertebrates are instead proving to be biosynthesized by mutualistic bacteria. These microbes generally organize their biosynthetic genes for each secondary metabolite in compact clusters, making it practical to trace and clone the respective pathways in order to create effective heterologous producers. The assembly line logic of the biosynthetic enzymes furthermore facilitates the manipulation of the encoding genes, allowing the generation of even increased metabolic diversity. Using this metabolic engineering approach, the rational design of novel analogs for biomedical applications has become possible. Furthermore, the powerful and meanwhile quite affordable genome sequencing of the most promising bacterial strains paves the way to discover and mine orphan biosynthetic clusters encoding as yet unexpressed novel chemistry. As an example, the genome of the marine bacterium Salinispora arenicola has most recently been completed (GenBank accession no. CP000850) and revealed the presence of approximately 39 different biosynthetic loci, a tremendous source of new chemistry awaiting discovery and evaluation. The strong interplay of classical natural product chemistry with modern microbial genetics and bioinformatics will thus help to overcome supply and sustainability issues from the past and to promote marine bacterial secondary metabolites to a well recognized alternative for future drug discovery programs.
Acknowledgments
The authors thank A.S. Eustaquio and E. Gontang for providing a photograph of S. tropica. This work was generously supported by grants from the National Institute of Health (CA127622 and AI47818 to B.S.M.). T.A.M.G. is a postdoctoral fellow of the German Academic Exchange Service (DAAD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Newmann DJ, Cragg GM. Natural Products as Sources of New Drugs over the Last 25 Years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Blunt JW, Copp BR, Hu W-P, Munro MHG, Northcote PT, Prinsep MR. Marine natural products. Nat Prod Rep. 2008;25:35–94. doi: 10.1039/b701534h. [DOI] [PubMed] [Google Scholar]
- 3.Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Jensen PR, Williams PG, Oh D-C, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 6.Bull AT, Stach JEM. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorg Med Chem. 2009;17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortman JL, Sherman DH. Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. ChemBioChem. 2005;6:960–978. doi: 10.1002/cbic.200400428. [DOI] [PubMed] [Google Scholar]
- 9.Menzella HG, Reeves CD. Combinatorial biosynthesis for drug development. Curr Opin Microbiol. 2007;10:238–245. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 11•.Taori K, Paul VJ, Luesch H. Structure and activity of largazole, a potent antiproliferatvie agent from the Floridian marine cyanobacterium Symploca sp . J Am Chem Soc. 2008;130:1806–1807. doi: 10.1021/ja7110064. Marine cyanobacteria continue to produce structurally fascinating and biologically important natural products as exemplified by the macrolide thioester largazole. [DOI] [PubMed] [Google Scholar]
- 12.Nasveschuk CG, Ungermannova D, Liu X, Phillips AJ. A concise total synthesis of largazole, solution structure, and some preliminary structure activity relationships. Org Lett. 2008;10:3595–3598. doi: 10.1021/ol8013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying Y, Taori K, Kim H, Hong J, Luesch H. Total synthesis and molecular target of largazole, a histone deacetylase inhibitor. J Am Chem Soc. 2008;130:8455–8459. doi: 10.1021/ja8013727. [DOI] [PubMed] [Google Scholar]
- 14.Bowers A, West N, Taunton J, Schreiber SL, Bradner JE, Williams RM. Total synthesis and biological mode of action of largazole: A potent Class I histone deacetylase inhibitor. J Am Chem Soc. 2008;130:11219–11222. doi: 10.1021/ja8033763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina RA, Goeger DE, Hills P, Mooberry SL, Huang N, Romero LI, Ortega-Barría E, Gerwick WH, McPhail KL. Coibamide A, a potent antiproliferative cyclic depsipeptide from the Panamanian marine cyanobacterium Leptolyngbya sp. J Am Chem Soc. 2008;130:6324–6325. doi: 10.1021/ja801383f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrasidlo W, Mielgo A, Torres VA, Barbero S, Stoletov K, Suyama TL, Klemke RL, Gerwick WH, Carson DA, Stupack DG. The marine lipopeptide somocystinamide A triggers apoptosis via caspase 8. Proc Natl Acad Sci USA. 2008;105:2313–2318. doi: 10.1073/pnas.0712198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torchilin VP. Multifunctional nanocarriers. Adv Drug Deliv Rev. 2006;58:1532–1555. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Hughes CC, Prieto-Davo A, Jensen PR, Fenical W. The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org Lett. 2008;10:629–631. doi: 10.1021/ol702952n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boonlarppradab C, Kauffman CA, Jensen PR, Fenical W. Marineosins A and B, cytotoxic spiroaminals from a marine-derived actinomycete. Org Lett. 2008;10:5505–5508. doi: 10.1021/ol8020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes CC, MacMillan JB, Gaudêncio SP, Jensen PR, Fenical W. The ammosamides: structures of cell cycle modulators from a marine-derived Streptomyces sp. Angew Chem Int Ed. 2009;48:725–727. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander MD, Burkart MD, Leonard MS, Portonovo P, Liang B, Ding X, Joullie MM, Gulledge BM, Aggen JB, Chamberlin AR, et al. A central strategy for converting natural products into fluorescent probes. ChemBioChem. 2006;7:409–416. doi: 10.1002/cbic.200500466. [DOI] [PubMed] [Google Scholar]
- 22•.Hughes CC, MacMillan JB, Gaudêncio SP, Fenical W, La Clair JJ. Ammosamides A and B target myosin. Angew Chem Int Ed. 2009;48:728–732. doi: 10.1002/anie.200804107. This interesting study reports the mode of action of the newly discovered ammosamides, furthermore providing a molecular probe to study myosin-involving cellular processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egan S, Thomas T, Kjelleberg S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr Opin Microbiol. 2008;11:219–225. doi: 10.1016/j.mib.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Grozdanov L, Hentschel U. An environmental genomics perspective on the diversity and function of marine sponge-associated microbiota. Curr Opin Microbiol. 2007;10:215–220. doi: 10.1016/j.mib.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Simmons TL, Coates RC, Clark BR, Engene N, Gonzales D, Esquenazi E, Dorrestein PC, Gerwick WH. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc Natl Acad Sci USA. 2008;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieckmann R, Graeber I, Kaesler I, Szewzyk U, von Döhren H. Rapid screening and dereplication of bacterial isolates from marine sponges of the Sula Ridge by Intact-Cell-MALDI-TOF mass spectrometry (ICM-MS) Appl Microbiol Biotechnol. 2005;67:539–548. doi: 10.1007/s00253-004-1812-2. [DOI] [PubMed] [Google Scholar]
- 27••.Esquenazi E, Coates C, Simmons L, Gonzales D, Gerwick WH, Dorrestein PC. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging . Mol BioSyst. 2008;4:562–570. doi: 10.1039/b720018h. This paper describes a novel imaging technique with high value for drug discovery and ecological studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 29.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spec Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 30.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci USA. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 35••.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. This manuscript firmly establishes the biosynthetic diversity of the cyanobactins, which are highly modified, ribosomally-derived peptides from cyanobacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziemert N, Ishida K, Quillardet P, Bouchier C, Hertweck C, de Marsac NT, Dittmann E. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl Environ Microbiol. 2008;74:1791–1797. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chem Int Ed. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 39.Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. Post-translational modification in microviridin biosynthesis. ChemBioChem. 2008;9:3066–3073. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 40••.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete . Salinispora tropica Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. This study reports the first complete, genome sequence of a marine obligate actinomycete and its impressive secondary metabolic capabilities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGlinchey RP, Nett M, Moore BS. Unraveling the biosynthesis of the sporolide cyclohexenone building block. J Am Chem Soc. 2008;130:2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- 42.Beer LL, Moore BS. Biosynthetic convergence of salinosporamides A and B in the marine actinomycete Salinispora tropica. Org Lett. 2007;9:845–848. doi: 10.1021/ol063102o. [DOI] [PubMed] [Google Scholar]
- 43••.Eustáquio AS, Pojer F, Noel JP, Moore BS. Discovery and characterization of a marine bacterial SAM-dependent chlorinase. Nat Chem Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. This paper reports a new enzymatic reaction for the assimilation of chlorine into a natural product that is orthogonal to the known oxidative mechanisms for creating organochlorides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng H, O’Hagan D. The flurinase, the chlorinase and the duf-62 enzymes. Curr Opin Chem Biol. 2008;12:582–592. doi: 10.1016/j.cbpa.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 45.Moore BS, Eustáquio AS, McGlinchey RP. Advances in and applications of proteasome inhibitors. Curr Opin Chem Biol. 2008;12:434–440. doi: 10.1016/j.cbpa.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Groll M, Huber R, Potts BC. Crystal structures of salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of β-lactone ring opening and a mechanism for irreversible binding. J Am Chem Soc. 2006;128:5136–5141. doi: 10.1021/ja058320b. Important and highly interesting study illuminating the mechanism of action and binding of the salinosporamides to the 20S proteasome. [DOI] [PubMed] [Google Scholar]
- 47.Reed KA, Manam RR, Mitchell SS, Xu J, Teisan S, Chao T-H, Deyanat-Yazdi G, Neuteboom ST, Lam KS, Potts BC. Salinosporamides D-J from the marine actinomycete Salinispora tropica, bromosalinosporamide, and thioester derivatives are potent inhibitors of the 20S proteasome. J Nat Prod. 2007;70:269–276. doi: 10.1021/np0603471. [DOI] [PubMed] [Google Scholar]
- 48.Macherla VR, Mitchell SS, Manam RR, Reed KA, Chao T-H, Nicholson B, Deyanat-Yazdi G, Mai B, Jensen PR, Fenical WF, et al. Structure-activity relationship studies of salinosporamide A (NPI-0052), a novel marine derived proteasome inhibitor. J Med Chem. 2005;48:3684–3687. doi: 10.1021/jm048995+. [DOI] [PubMed] [Google Scholar]
- 49••.Eustáquio AS, Moore BS. Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew Chem Int Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. This report describes the first genetic engineering of a new salinosporamide analog, which is the most active “reversible” proteasome inhibitor of this structure class known to date. [DOI] [PubMed] [Google Scholar]
- 50.McGlinchey RP, Nett M, Eustáquio AS, Asolkar RN, Fenical W, Moore BS. Engineered biosynthesis of antiprotealide and other unnatural salinosporamide proteasome inhibitors. J Am Chem Soc. 2008;130:7822–7823. doi: 10.1021/ja8029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy LR, Fournier J-F, Reddy BVS, Corey EJ. An efficient, stereocontrolled synthesis of a potent omuralide-salinosporin hybrid for selective proteasome inhibition. J Am Chem Soc. 2005;127:8974–8976. doi: 10.1021/ja052376o. [DOI] [PubMed] [Google Scholar]
- 52•.Winter JM, Moffit MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS. Molecular basis for chloronium-mediated meroterpene cyclization: cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J Biol Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. This paper describes the cloning of the first vanadium-dependent chloroperoxidase from a marine bacterium involved in the biosynthesis of a natural product. [DOI] [PubMed] [Google Scholar]
- 53.Gu L, Geders TW, Wang B, Gerwick WH, Hakansson K, Smith JL, Sherman DH. GNAT-like strategy for polyketide chain initiation. Science. 2007;318:970–974. doi: 10.1126/science.1148790. [DOI] [PubMed] [Google Scholar]
- 54.Moore BS. Extending the biosynthetic repertoire in ribosomal peptide assembly. Angew Chem Int Ed. 2008;47:9386–9388. doi: 10.1002/anie.200803868. [DOI] [PubMed] [Google Scholar]
- 55••.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Enzymatic total synthesis of enterocin polyketides. Nat Chem Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. An impressive first example of a total biosynthesis of a marine microbial polyketide. [DOI] [PubMed] [Google Scholar]
- 56.Werneburg M, Hertweck C. Chemoenzymatic total synthesis of the antiproliferative polyketide (+)-(R)-aureothin. ChemBioChem. 2008;9:2064–2066. doi: 10.1002/cbic.200800301. [DOI] [PubMed] [Google Scholar]