Abstract
Primary liver cancer is the fifth most common malignancy in the world and is a leading cause of cancer-related mortality. Available treatment for hepatocellular carcinoma (HCC), the commonest primary liver cancer, is rarely curative and there is a need to develop therapy that is more effective. Specific and powerful gene silencing that can be achieved by activating RNA interference (RNAi) has generated enthusiasm for exploiting this pathway for HCC therapy. Many studies have been carried out with the aim of silencing HCC-related cellular oncogenes or the hepatocarcinogenic hepatitis B virus (HBV) and hepatitis C virus (HCV). Proof of principle studies have demonstrated promising results, and an early clinical trial assessing RNAi-based HBV therapy is currently in progress. Although the data augur well, there are several significant hurdles that need to be overcome before the goal of RNAi-based therapy for HCC is realized. Particularly important are the efficient and safe delivery of RNAi effectors to target malignant tissue and the limitation of unintended harmful non-specific effects.
Keywords: RNA interference, Hepatocellular carcinoma, Hepatitis B virus, Hepatitis C virus, Molecular pathogenesis, Delivery vectors
INTRODUCTION
Globally, hepatocellular carcinoma (HCC) is a major cause of mortality[1,2]. It is the commonest primary liver cancer and accounts for 80%-90% of this class of malignancy. HCC is characterized by a very poor prognosis and is associated with high mortality. Annual mortality from HCC is virtually the same as its annual incidence, attesting to its rapid course and grave prognosis. Moreover, although HCC is the fifth most common cancer worldwide, it is the third most common cause of cancer-related mortality[3]. Recent observations have shown that incidence and mortality from liver cancer in the United States is the fastest growing of all cancers[2]. This is despite a decline in the overall cancer mortality rate that has occurred during the past 20 years. Currently available treatments of HCC are largely inadequate. However, with better understanding of the molecular pathogenesis of the cancer and significant advances in gene silencing technology, improved approaches are being devised to counter the malignancy. In particular, harnessing the RNA interference (RNAi) pathway to inhibit expression of genes that are implicated in transformation of hepatocytes is an exciting new approach to treating HCC. Therapeutic gene silencing technology is however at an early stage of development and there are several important hurdles that need to be overcome before this approach becomes a reality for treating HCC. In this review, we summarize HCC molecular pathogenesis as a background to discussing the interesting prospects of RNAi-based drugs for treating HCC.
CAUSES AND MOLECULAR PATHOGENESIS OF HCC
There are several etiological agents and risk factors that have been implicated in causing HCC[2]. Typically, HCC arises within a diseased liver and chronic liver injury per se, usually cirrhosis is thought to be causative of HCC. Thus, agents that result in chronic liver disease, although perhaps not directly carcinogenic, may be risk factors for HCC. Liver cancer is not evenly distributed throughout the world and is a consequence of unequal prevalence of major causative factors of the malignancy[2]. HCC has a particularly high incidence in sub Saharan Africa, East and South East Asia where chronic hepatitis B virus (HBV) infection is common. Of the factors that have been identified to cause HCC, persistent HBV infection has the strongest association with the malignancy[1]. The long term risk of HCC in HBV carriers has been reported to be in the range of 25%-40%. Infection with hepatitis C virus (HCV) is also directly causative of liver cancer[4]. The long-term risk for HCC in HCV-infected individuals is estimated to be 1%-3% after 30 years, although in cases of established HCV-related cirrhosis, the annual rate of HCC development may be as high as 7%. Globally it is estimated that there are 387 million carriers of HBV[1,5] and 170 million people persistently infected with HCV[6]. These enormous numbers make HBV and HCV chronic infections the major HCC-predisposing factors. A primary focus of preventing HCC is thus aimed at eliminating these viruses.
Several other hepatocarcinogenic factors, which may cause transformation directly or indirectly, have been identified[2]. These include excessive alcohol intake, aflatoxin ingestion, vinyl chloride exposure, obesity, diabetes mellitus, dietary iron overload, cigarette smoking and use of oral contraception. Regardless of the study population, males have a higher risk for the malignancy than females. The male to female ratio of HCC varies between 2:1 and 4:1. A reason for this gender bias is a higher risk for exposure to HCC-causing agents such as alcohol and tobacco amongst males. Additionally, androgens per se may contribute to hepatocarcinogenesis.
HBV
HBV is the prototype member of the hepadnaviridae family of hepatotropic viruses. It is a small, non-cytopathic, enveloped partly double stranded or relaxed circular DNA (rcDNA) virus with a genome size of 3.2 kb[7–9]. Viral rcDNA is converted to covalently closed circular DNA (cccDNA) within an infected hepatocyte. cccDNA then serves as template for expression of viral genes, formation of pregenomic RNA and ultimately the production of progeny viruses. The viral genome is remarkably compact and encodes four overlapping open reading frames (ORFs): core (C), polymerase (P), envelope (surface, S) and X (HBx), which collectively encompass the entire genome. Viral cis elements that control transcription and aspects of replication are thus embedded within protein coding sequences. This remarkably economical use of the small genome limits HBV sequence plasticity and the virus is thus a good target for therapy that is based on nucleic acid hybridization.
The exact mechanism of HBV-mediated hepatocarci-nogenesis is not completely understood. Integration of viral DNA into the host cellular genome with resultant effects on chromosome stability and surrounding genes may play a role. Several lines of evidence also implicate HBx in the transformation of hepatocytes. This protein has wide ranging effects on cellular processes that are involved in regulating cell differentiation, apoptosis and proliferation. The mechanism of action includes indirect transcriptional activation of cellular cis elements, effects on cell signalling pathways as well as modulation of apoptosis[10].
HCV
HCV is a member of the Flaviviridae family. The virion is enveloped and has an RNA genome comprising 9.6 kb of uncapped RNA with sense polarity[11]. The internal ribosomal entry site (IRES), which is located within the 5' NTR of the HCV genome, initiates translation of a large precursor polyprotein. This precursor is processed by cellular and viral proteinases to form 10 viral proteins, namely core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B. E1 and E2 encode the envelope proteins and the viral core is derived from the C sequence. Non-structural proteins are responsible for assembling the viral proteinase machinery (NS2, NS3 and NS4A), forming the specialized membrane compartment for viral replication (NS4B) and RNA-dependent RNA polymerase (NS5B). The hydrophobic p7 protein, required for the late stages of viral assembly, is thought to function as a viroporin. The frameshift (F) protein, which is expressed during natural HCV infection, is yet to be fully characterized[12–14]. The entire HCV lifecycle is cytoplasmic and involves the formation of minus RNA and dsRNA intermediates within the membranous web. HCV dsRNA activates the innate immune response, but cellular antiviral effects are inhibited by the E2, NS3 and NS5A proteins[15–17]. Unlike with HBV, sequence heterogeneity and rapid evolution of quasispecies are characteristic of HCV infection. However, structural characteristics of the 5' NTR, which determine its function as an IRES impart sequence conservation in this region[11].
Research suggests that HCV exerts its oncogenic effect through the actions of its viral proteins. The amino terminal of the NS3 protein[18] and the core protein[19] have been shown to influence various cellular functions including the enhancement[20–22] or inhibition[23–25] of apoptosis. The core protein may activate transcription of the proto-oncogene c-myc and also has an effect on apoptosis through effects on Fas, tumor necrosis factor (TNF-)[26] and a mitogen activated protein kinase or extracellular signal regulated kinase (MAPK/ERK) signaling cascade. In addition, the NS4B protein functions as a transcriptional activator of intracellular signals that play roles in cell proliferation and inflammation. NS3 and NS5A proteins may also contribute to hepatocarcinogenesis by blocking the action of p53[27,28].
Aflatoxin and ethanol
Aflatoxin exposure and excessive alcohol intake are important causes of liver cancer, which may have synergistic action when occurring together with chronic HBV and HCV infection[29]. Aflatoxins are mycotoxins generated by certain Aspergillus species and are potent natural carcinogens[30]. When converted to exo-8,9-epoxides in the liver they are capable of damaging guanine nucleotides in hepatocytes DNA to form mutagenic and potentially carcinogenic DNA adducts. Importantly, aflatoxin-induced mutation hot spots in the liver include a transversion that occurs in the third position of codon 249 of the p53 tumor suppressor gene[31,32]. Alcohol metabolism in the liver has also been implicated in mutagenesis and hepatocarcinogenesis[33]. The mechanism is through generation of reactive oxygen species (ROS) and increasing hepatic oxidative stress. Acetaldehyde accumulation also contributes to the formation of protein and DNA adducts which result from ROS generation. Excessive alcohol intake is also a cause of chronic liver injury and cirrhosis, which is itself indirectly hepatocarcinogenic.
CURRENT TREATMENT OF HCC
Although there have been advances in the detection of HCC, improvement in the management of the disease has not been significant[34]. Inadequacy of current therapy and presentation of patients with advanced disease have meant that the prognosis of individuals with HCC remains poor. Available treatment modalities include surgical intervention, percutaneous and arterial attempts at tumor ablation as well as drug-based treatment. If patients have well localized malignancies and good underlying liver function, surgical resection does have some success in eradicating tumors. However, this is clinically atypical, and patients with HCC usually present with cancers that have spread extensively within the liver and to distant sites. Treatment in these cases is palliative and the prognosis is extremely grave. Very early tumor detection and novel effective therapy are important objectives for improving management of patients with HCC. The potent targeted gene silencing that can be achieved by activation of RNAi has prompted investigations that apply this approach to direct treatment of HCC as well as through eliminating risk factors for the malignancy (e.g. HBV and HCV).
HARNESSING RNAI TO TREAT HUMAN DISEASE
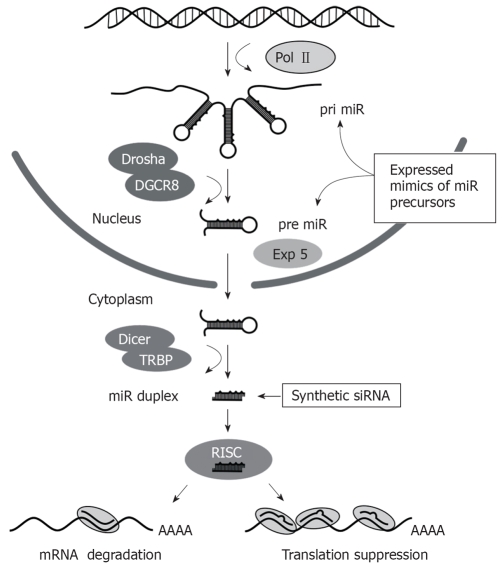
RNAi is a powerful gene silencing mechanism that operates in most eukaryotic cells[35]. The effecter molecules comprise short duplex RNA sequences of 21-23 bp that direct inhibition of homologous genes (Figure 1). Naturally, the pathway is important for processing of regulatory micro RNAs (miRs)[36,37]. These non-coding cellular sequences have roles in several pathways such as cell differentiation, metabolism, proliferation and malignant transformation. The miR processing pathway is initiated by transcription of miR-encoding cellular genes by RNA polymerase II (Pol II) to produce hairpin-containing primary miRs (pri miRs). These pri miRs may be derived from intronic sequences and may be polycistronic. Within the nucleus, pri miRs are processed to form precursor miR (pre-miR) hairpins of 60-80 nt in length. This step is catalyzed by the microprocessor complex, which contains Drosha and di George Critical Region 8 (DGCR8) proteins. Drosha functions as an RNAse III enzyme and DGCR8 is its double stranded RNA binding protein partner. Pre-miRs are exported from the nucleus to the cytoplasm by the RanGTP-dependent Exportin 5 transporter. Pre-miRs are then processed by Dicer with associated TAR RNA-binding protein (TRBP) to form a staggered RNA duplex of 21-24 bp with 2 nt 3’ overhangs. This duplex is handed on to the RNA induced silencing complex (RISC), which includes several components such as Argonaute 1 (AGO1), Argonaute 2 (AGO2) and Fragile X proteins. One strand of the RNA duplex, which is designated the passenger strand, is cleaved within RISC and is then released from the complex. The remaining intact single stranded guide RNA activates RISC to direct target-specific silencing. Mature cellular miRs are usually not entirely complementary to their targets and bind to the 3' untranslated regions of cognate mRNA to induce translational suppression. Hybridization between target and nucleotides 2-8 from the 5' end of the guide strand, termed the seed sequence, are all that is required to cause translational suppression[38]. When base pairing between entire guide and target is perfectly matched, the AGO2 component of RISC exerts silencing through site-specific cleavage (‘slicing’) of the guide complement[39,40].
Figure 1.
Schematic illustration of the RNAi pathway showing the essential steps, with nuclear or cytoplasmic location, involved in micro RNA processing. Exogenous activators of the pathway, which may be synthetic siRNA or expressed mimics of miR precursors, are shown.
Exogenous activators of RNAi may be used for therapeutic application by silencing specific pathology-causing sequences. These activators are typically expressed or synthetic sequences and they activate RNAi at different stages of the pathway (Figure 1). Exogenous small interfering RNAs (siRNAs) are usually synthetic mimics of the dsRNA duplexes that are formed after Dicer processing, whereas expressed RNAi effectors are typically designed to simulate pri miR or pre miR sequences. The commonest form of expression cassette comprises a short hairpin RNA (shRNA)-encoding sequence that is inserted downstream of a Pol III promoter. When introduced into a cell, the shRNA acts as a mimic of pre miR and is processed by Dicer to form a siRNA. Synthetic or expression cassette-derived siRNAs are structurally symmetrical molecules but stability at the 5' ends of the siRNA duplex has an influence on the bias of guide strand selection[41]. The strand which is less tightly paired (A/U rich) to its complement at its 5' end is preferentially incorporated into RISC. Other factors that influence the effectiveness of siRNAs against their cognate targets have been described and have been incorporated into predictive algorithms used for design of exogenous silencing sequences. The U6 and H1 Pol III promoters have commonly been used to generate expressed shRNAs and they are capable of efficient transcription of short defined sequences. Importantly, Pol III promoters are usually constitutively active, which means that regulation of intracellular concentration of shRNAs is difficult to achieve[42]. Related to this, recent demonstration that U6 Pol III-expressed shRNAs may saturate the endogenous miR pathway to cause serious toxicity in vivo is an important concern[43]. Since naturally occurring pri miRs are expressed from Pol II promoters[44], shuttle cassettes incorporating features of pri miRs may improve the efficiency of Pol II-expressed RNAi sequences.
HCC AND MIRS
There are several studies that have implicated disruption of miR expression in carcinogenic mechanisms[45,46]. miR concentrations may be elevated or depressed in HCC, which suggests that by interacting with their cognates, these sequences may act as oncogenes or suppressors of hepatocyte transformation. Recent studies using miR microarrays, showed high expression of miR-21[47] and low levels of miR-122a[48] in HCC. miR-21 contributes to hepatocyte transformation, growth and spread by inhibiting phosphatase and tensin homolog (PTEN) tumor suppressor. Modulation of cyclin G1, a cell cycle modulator, is the mechanism by which decreased miR-122a expression was reported to be hepatocarcinogenic. Another study analyzing the role of peroxisome proliferator-activated receptor alpha (PPARalpha) in hepatocarcinogenesis showed that this protein is an important regulator of hepatic miRNA expression[49]. Of particular significance was PPARalpha inhibition of let-7C-mediated signaling and c-myc induction, which leads to hepatocyte proliferation and transformation. Other studies have implicated different alterations in miR expression that may be important for HCC[50,51]. A recent study, which analyzed sequence variation of 59 miRs in HCC and adjacent non-malignant tissue, revealed four variations in three microRNAs[52]. These miRs included miR-106b, miR-192 and let-7a-2. The significance of the variations for hepatocarcinogenesis was however not clear as the same sequence differences were found in non-malignant cells but not in eight liver cancer-derived cell lines. The conclusion from this study was that miR mutation is a rare event in HCC and is unlikely to represent the main mechanism of hepatocarcinogenesis. Collectively the data show considerable heterogeneity in HCC-related altered miR expression. This complicates developing a generic approach to HCC treatment that is based on modulation of miR expression.
RNAI TARGETS TO COUNTER HEPATOCARCINOGENESIS
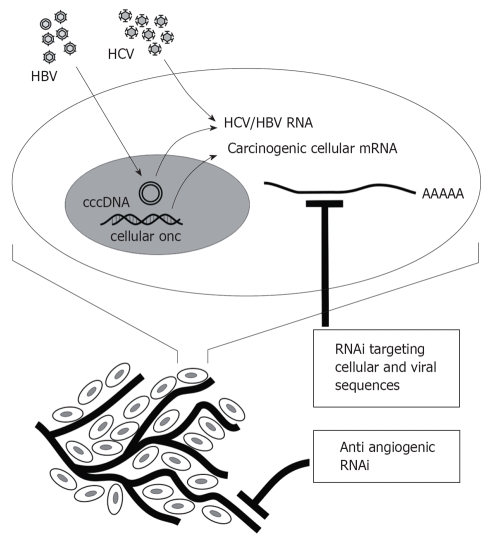
Suitable targets for development of RNAi-based treatment of HCC include cellular oncogenic sequences, angiogenic factors, as well as HBV and HCV genes (Figure 2). Studies to date, which have mainly demonstrated proof of principle, are summarized below. A difficulty of assessing usefulness of RNAi against HCC, HBV and HCV has been the lack of convenient small animal models. Advances have recently been made in addressing this obstacle. HCV replicons[11], cells in culture that are permissive for HBV[53] and HCV[54] infection, the hydrodynamic injection procedure[55], transgenic mice and xenografted mice[56] have, and will be, particularly useful.
Figure 2.
RNAi targets that may be silenced to counter HCC. In addition to HBV and HCV genes, cellular sequences that are involved in hepatocyte transformation may be silenced to inhibit growth of malignant liver cells. Growth of HCC is also dependent on angiogenesis and inhibition of this process is expected to limit tumor growth.
HBV
As with most investigations aimed at developing RNAi-based therapy, synthetic and expressed sequences have been used to activate RNAi and counter replication of HBV. Most studies to date have provided proof of principle in cell culture and small animal models of HBV replication. Recently however, FDA approval has been granted for an investigational new drug license to test the use of expressed RNA sequences against HBV (http://www.nucleonicsinc.com/news/pdfs/FDAClearanceHepBTrial_may2007.pdf).
One of the first studies aimed at assessing efficacy of RNAi-activating sequences against HBV tested a panel of six U6 promoter cassettes that encoded shRNAs[57]. Profound inhibition of HBV surface antigen secretion from transfected cultured cells and in vivo in the murine hydrodynamic injection model was observed. Inhibitory effects were found to be due to a direct effect that is not dependent on an antigen-dependent immune response. Others have recently demonstrated varying degrees of inhibition of HBV replication by expressed shRNA sequences[58–64] and one study showed antiviral synergy between lamivudine and shRNA sequences in a cell culture model[61]. To address concerns about the emergence of RNAi escape mutants, Weinberg and colleagues[65] used long hairpin RNA (lhRNA) expression cassettes to target the HBx ORF of HBV. These sequences were designed to generate multiple siRNAs from the 62 bp duplex region of the hairpin. Although impressive knockdown was achieved, the efficacy was not equal across the span of the duplex. siRNAs generated from the stem base were produced more efficiently and effected better silencing than the loopside siRNA template. Limited processivity by Dicer, which initiates its RNase III activity at the hairpin stem base, is thought to be the cause of this effect. Using an approach that obviates this problem by cleavage of hairpin RNA in vitro, endoribonuclease-prepared siRNAs (esiRNAs) targeting all four HBV ORFs were recently used to inhibit HBV replication with high efficiency[66].
Since there is no convenient small animal model of HBV infection, transgenic mice have been used in some studies as a stringent model to simulate virus replication that occurs in HBV chronic carriers. Expressed shRNAs have been tested in 4 studies on HBV transgenic mice[43,62,63,67]. Hepatotropic recombinant adenovirus vectors expressing shRNAs from Pol III promoters caused sustained inhibition of viral replication over a 2-4 wk period after administration of a single dose of the vector[62,63]. The importance of optimizing the dose of expressed anti HBV shRNA sequences was highlighted in a recent study that showed fatality in HBV transgenic mice that were treated with recombinant adeno-associated virus (AAV) type 8 vectors that over expressed shRNAs[43]. Recombinant vectors expressing a 25-mer anti-HBV shRNA consistently caused death of treated mice, whereas low dose of a 19-mer shRNA vector efficiently suppressed markers of viral replication. These studies provide important evidence that HBV is susceptible to RNAi-based gene silencing in a model that simulates established ongoing replication that occurs in HBV chronic carriers.
Effective knockdown of HBV replication by synthetic siRNA has also been shown in vitro and in vivo. A siRNA duplex that targeted sequences immediately upstream of the surface ORF initiation codon was found to be particularly effective against HBV without a requirement for HBV DNA synthesis[68]. This property is distinct from anti-HBV nucleoside or nucleoside analogues, which act on the viral DNA polymerase to have their therapeutic effect. Efficacy of surface ORF-targeted siRNAs was reported in other studies[69,70]. Improved knockdown by repeated siRNA transfection of cells in culture was also observed[71] as well as effectiveness against a HBV target that includes the sequences encoding the lamivudine-resistance YMDD polymerase gene mutation[72]. Recently, Morrissey and colleagues showed that chemically modified siRNAs caused potent and persistent anti-HBV activity in vivo[73]. Sequences were administered intravenously within a stable nucleic acid lipid particle (SNALP) formulation. Efficiency of the complexes is likely to be a result of increased stability in vivo and also diminished non specific immunostimulatory effects. Apolipoprotein A-1-derived nanoparticles have also been used successfully in vivo to deliver synthetic anti HBV sequences[74]. Importantly, these vectors are liver specific and are efficient at low doses. As well as being developed as an independent therapy, RNAi effectors may be used in combination with established licensed drugs to improve efficacy. Such synergy, which is likely to result from different mechanisms of drug action, has been demonstrated when using anti-HBV sequences in conjunction with lamivudine[61].
HCV
Since it is an RNA virus that replicates in the hepatocyte cytoplasm, HCV is considered a prime candidate for RNAi-based treatment. However, a high degree of heterogeneity of viral sequences has been a particularly significant obstacle to developing antiviral RNAi effectors. For this reason, the 5' NTR has been the favored HCV target of several studies[75–79]. Also, the lack of suitable models of HCV reproduction in vivo has hampered development of RNAi-based approaches to therapy. Extensive use of subgenomic replicon systems has been used successfully to study efficacy of antiviral therapeutic agents in cell culture and this approach has provided valuable insights[80,81]. Currently available models of HCV infection in vivo are limited and include the chimpanzee[82] and chimaeric immunodeficient mice that are grafted with human hepatocytes[83].
In an early study that employed RNAi against HCV[79], both synthetic and expressed RNAi effectors against the 5' NTR caused significant reduction of markers of HCV replication in a replicon model. Subsequent investigations targeting the 5' NTR showed suppression of viral gene expression by naked shRNAs[78] and also synthetic siRNAs[84]. Domain IV regions were found to be a particularly good target for RNAi-based HCV gene silencing[79]. However, a report by Takigawa and colleagues[77] showed that expressed shRNAs targeted to NS3-1 and NS5B were more effective than sequences against the 5' NTR. Other studies using both synthetic and expressed siRNAs also achieved significant inhibition of virus gene expression when targeting NS3 and NS5B sequences[85,86]. Although good efficacy against HCV has been demonstrated, a major concern remains the ability of the virus to accumulate evading nucleotide sequence mutations. Not surprisingly, emergence of resistant HCV replicons has been shown in cultured cells after repeated treatments with siRNA targeted against the NS5B coding region[87]. Point mutations within the siRNA target sequence were observed, but resistant replicons were sensitive to a siRNA that targets another part of the genome. As with HBV[65], vectors that express lhRNAs have been shown to be effective against HCV[88,89]. Although these sequences have the theoretical advantage of generating multiple siRNAs, the approach may be limited by incomplete Dicer processing of the hairpin (see above and[65]). Although approaches that produce polycistronic miR shuttles or esiRNAs may be preferable, a concern of using multiple siRNAs for therapy is the increased likelihood of silencing non-targeted genes as well as disruption of the endogenous miRNA pathway.
Other studies have aimed to circumvent the problem of viral escape by targeting host genes that are required for HCV replication[90–92]. Synthetic siRNA and adenovirus-delivered effectors specific for La autoantigen (La), polypyrimidine tract-binding protein (PTB), cyclophilins and human VAMP-associated protein of 33 kDa (hVAP-33) substantially blocked HCV replication. Endogenous hepatic miR-122 has recently been shown to suppress haem oxygenase-1 (HO-1) and facilitate HCV replication[93,94]. Upregulation of HO-1 or suppression of miR-122 thus represents an interesting strategy to counter HCV infection. Caspase 8 and the Fas cell death receptor, which mediate T-cell hepatocyte toxicity caused by viral infection, were efficiently silenced using RNAi[95,96]. Although promising, it remains to be established whether silencing of these endogenous genes causes toxicity.
Cellular oncogenic sequences
As the molecular mechanism of hepatocarcinogenesis becomes better understood, so the number of potential targets that can be inhibited using RNAi improves. There are many oncogenes that have been described, which are implicated in the disruption of control of normal hepatocyte proliferation. Although this is encouraging, the targeting of specific cellular sequences is hampered by two main factors: (1) heterogeneity of gene expression in liver cancer cells from different sources, and (2) difficulty of achieving sufficient transfer of RNAi effectors to be of therapeutic benefit against the malignancy. The possibilities for therapeutic application are nevertheless intriguing.
A recent study aimed to analyze the effect of silencing the pituitary tumor transforming gene (PTTG) family on hepatocarcinogenesis[97]. PTTG is a recently discovered group of oncogenes that plays a role in the genesis of several types of cancer through effects on cell division, apoptosis and DNA repair[98]. PTTG1, but not PTTG2 and PTTG3, is frequently over expressed in patient liver cancer tissue as well as in established HCC lines[97]. Infecting cells with a recombinant adenovirus expressing an anti PTTG1 RNAi effecter depleted cells of PTTG1 and resulted in the activation of p53 with consequent increased p21 expression and apoptosis. Inhibition of tumor growth in a nude mouse xenograft model of HCC further supported the notion that PTTG1 is a good candidate for RNAi-mediated HCC therapy.
Other studies aimed at silencing the serine protease urokinase-type plasminogen activator (u-PA) demonstrated similar proof of principle efficacy[99,100]. Signalling through u-PA and its receptor (uPAR) have been implicated in cell invasion, survival, and metastasis of a variety of cancers[101,102]. Silencing of u-PA using RNAi-based approaches has been used successfully in tumor models of prostate cancer[103] and gliomas[104]. To assess efficacy of RNAi-based u-PA silencing on HCC, Salvi and colleagues[100] demonstrated that stable expression of a shRNA effectively knocked down u-PA. Moreover, silencing of u-PA resulted in attenuation of malignancy-associated cellular properties, such as migration, invasion and proliferation. In a follow up study, the effects of stable inhibition of u-PA on xenografted tumor cells were assessed[99]. Cells that stably produce silencing sequences targeted to u-PA were injected subcutaneously into nude mice. Knockdown of both u-PA protein and mRNA concentrations was observed, which lasted for a period of 11 weeks. A delay in xenografted tumor growth was observed in cells expressing anti u-PA sequences, which indicates that u-PA silencing may be beneficial for HCC therapy.
A further study aimed at developing RNAi-based HCC therapy, assessed inhibition of function of human gankyrin gene product (p28GANK)[105]. This novel oncogenic protein is ubiquitously over expressed in HCC and plays a role in cell cycle progression in normal hepatocytes and liver regeneration[106–109]. After screening for susceptible target sites, a shRNA expression cassette was incorporated into an adenoviral vector. This was used to determine silencing of p28GANK and assess antitumor properties of the viral vectors[105]. Effective silencing of approximately 80% was achieved. This depletion of p28GANK enhanced dephosphorylation of Rb1 decreased transcriptional activity of E2F-1 in cultured liver-derived cells and inhibited cell growth. Moreover, tumor cell growth was retarded in xenografted nude mice, which was thought to be a result of increased caspase-8- and caspase-9-mediated apoptosis caused by p28GANK inhibition.
RNAi-based approaches to the inhibition of vasculari-zation of tumors has recently received attention[110–115]. HCC growth is dependent on neovascularization and inhibition of factors that are required for angiogenesis should therefore be effective in countering the growth of this cancer. Most work has focused on the silencing of vascular endothelial growth factor (VEGF) to reduce the formation of new vessels that are required for tumor development. RNAi-based inhibition of VEGF in cases of the wet form of macular degeneration has reached an advanced stage of clinical trial (http://www.agingeye.net/maculardegen/maculardegennewdevelopments.php). Although this disease is not a malignancy, demonstration that VEGF can be inhibited successfully in vivo using RNAi indicates that this target may also be suitable for clinical application to HCC therapy. Alnylam and Inex Pharmaceuticals, leaders in the field of developing RNAi-based human therapy, have recently advanced a combination systemic drug for the treatment of liver cancer (http://www.alnylam.com/therapeutic-programs/programs.asp). The therapeutic, which is a liposomal formulation termed ALN-VSP-1, contains siRNAs that target VEGF and kinesin spindle protein (KSP). siRNA-mediated inhibition of KSP production leads to cell cycle arrest and death in malignant hepatocytes.
CHALLENGES FACING USE OF RNAI-B ASED THERAPY FOR TREATMENT OF HCC
Although the studies summarized above indicate that RNAi could be used for preventative or curative HCC treatment, there are several important hurdles that need to be overcome before clinical application. These include activation of the innate immune response, limitation of unintended interaction of RNAi effectors with cellular sequences, dosage regulation and optimizing delivery methods. These topics are briefly summarized below.
Innate immune response activation
Duplex RNA within cells is sensed as unwanted gene activity and may result in unintended harmful effects caused by activation of inflammatory cytokines and the interferon (IFN) response[116–118]. Stimulation of cytoplasmic pattern recognition receptors, such as dsRNA dependent protein kinase (PKR), retinoic acid inducible gene-I(RIG-I) and Toll-like receptors (TLRs), leads to a cascade of events, which culminates in activation of transcription factors such as NF-®, IRF3 and IRF7. This in turn causes increased expression of genes that include inflammatory cytokines and interferons. The response may be further amplified by autostimulatory positive feedback that involves JAK-STAT pathway activation. IFN pathway activation may also lead to inhibition of protein synthesis and degradation of cellular mRNA with consequent programmed cell death (apoptosis). The type of effecter sequence that is used to activate RNAi also has a bearing on immunostimulation[119]. Synthetic siRNAs that are longer than 30 bp[120], possess 5' triphosphates[121] and lack 2 nucleotide 3' overhangs[122] stimulate the innate immune system. Also, ‘danger’ motifs (e.g. GU rich sequences, 5'GUCCUUCAA3' and 5'UGUGU3') may activate endosomal TLR3, TLR7 and TLR8[123]. RNAi effectors derived from expression cassettes that are transcribed from the nucleus do not pass through the endosomal compartment to activate these TLRs. However, unmethylated CpG islands within RNAi-activating DNA expression cassettes may be immunostimulatory. Recently, chemical modifications of siRNAs have been shown to attenuate immunostimulatory effects[124], and this has been used successfully in vivo to counter HBV replication without release of interleukins and inflammatory cytokines[73].
Non-specific interaction of silencing molecules with cellular sequences
Cross-hybridization of siRNAs with transcripts that have partial sequence identity[125], particularly in the seed region of the intended target, may contribute to non-specific silencing effects. An interesting recent observation has been that 2’ O-methyl ribosyl modification at position 2 of the siRNA guide strand reduces off target silencing at the seed site[126]. This effect was independent of the target and did not influence knockdown efficiency of perfectly matched sequences. Incorporation of HCC-specific transcriptional regulatory elements (e.g. the alpha fetoprotein promoter) may be helpful to improve specificity of expressed RNAi effecter by limiting transcription to malignant hepatocytes. This approach has been used successfully to accomplish tumor-specific transgene expression[127–129]. In the long term, to address the problem of off-target silencing it is likely that detailed microarray analysis of cellular transcripts will need to be undertaken as part of developing RNAi-based HCC therapy.
Optimizing delivery vectors
One of the most difficult challenges impeding the advancement of RNAi-based HCC therapy is efficient and safe delivery of effecter sequences. Ideally, vectors should deliver silencing molecules selectively to most if not all the malignant hepatocytes. Synthetic siRNAs are smaller than DNA expression cassettes. They do not require delivery to the nucleus and activate RNAi within the cytoplasmic compartment. This makes regulated non-viral vector (NVV)-mediated delivery of synthetic siRNAs easier to achieve than it is for larger DNA expression cassettes. Viral vectors incorporate expression cassettes of necessity, and are generally more efficient vehicles in vivo than NVVs. Ease of scalable synthesis and chemical modification to influence biological properties are important properties of NVVs that, with improved delivery efficiency, are likely to contribute to their gaining favor for clinical application.
Recombinant adenoviruses and adeno-associated viruses (AAVs) are capable of transducing liver cells with high efficiency and have been used successfully to deliver sequences that silence HBV or HCV gene expression[43,62,63,90]. Despite recent concerns[130], recombinant AAVs are attractive vectors as they appear to be safe and capable of long-term gene expression. They are also relatively easy to propagate and capsid sequences from naturally occurring hepatotropic AAV serotypes have been used to confer liver-targeting properties on the recombinant viruses[131]. Although efficient delivery vehicles, adenoviruses are strong stimulators of innate and adaptive immune responses. This may cause toxicity and limit repeated administration[132]. An added problem is that malignant hepatocytes may be refractory to infection with recombinant adenovirus vectors[128]. To modify tropism and reduce immune responses, recent studies have used surface modified[133] or helper-dependent ‘gutless’ vectors[134]. Surface modification of adenovirus vectors with synthetic polymers such as poly-N-(2-hydroxypropyl) methacrylamide (poly-HPMA) and polyethylene glycol (PEG) has been used to facilitate tissue targeting and diminish immunostimulatory protein-protein interactions. Helper dependent and chemically modified vectors may have an improved safety profile that could be better suited to clinical application. An interesting effect of adenoviruses on miR biogenesis, which may have an influence on their suitability of use as vectors for RNAi-based therapy, is mediated by the virus associated RNA (VA1)[135]. This RNA sequence of approximately 160 nt folds into a structure that mimics miR and has the effects of reducing nuclear export of pre miRs by saturating exportin 5 and acting as a competitive inhibitor of Dicer.
NVV nanoparticles have been used effectively to target HCC cells and also to deliver anti-HBV/HCV sequences. Modifications to confer HCC specificity include incorporation of HBV L protein[136] as well as epidermal growth factor (EGF)[137] into the complexes. Proof of principle has been demonstrated in xenografted models of HCC, but efficiency of these vectors may not yet be adequate for therapeutic gene transfer. SNALPs containing synthetic siRNAs have been used successfully to inhibit HBV replication in a murine model of virus replication[73] and also to silence endogenous hepatic gene expression in primates[138]. A recent study investigated the use of a lactosylated cationic liposome 5 (CL-LA5) vectors to deliver anti HCV siRNAs to cultured cells and in vivo[139]. The complexes accumulated in the liver and specifically suppressed intrahepatic HCV gene expression in transgenic mice. SNALP technology, CL-LA5 vectors and the recently described siRNA Dynamic PolyConjugates[140] are interesting new vectors that offer exciting possibilities for future clinical application to HCC treatment.
Regulating dose
Lethal toxicity in vivo, which was caused by saturation of the endogenous hepatocyte miRNA pathway has come as an important warning for the development of RNAi-based treatment[43]. Grimm et al showed that expressed shRNA sequences disrupted essential endogenous miRNA-mediated cell functions by saturating the rate limiting activity of exportin-5. Synthetic siRNAs should bypass this step and this has recently been shown to be the case[141]. Dosage and intracellular copy of shRNAs is difficult to achieve with the commonly used constitutively active Pol III promoters, such as U6 and H1. Use of tissue-specific and inducible Pol II promoters may go some way to refining transcription control and improving dosage of expressed RNAi sequences.
CONCLUSIONS AND PROSPECTS
The enormous therapeutic potential of RNAi-based specific gene silencing has prompted enthusiasm for advancement of novel therapies for difficult to treat diseases, such as HCC. Developments have been exciting, and clinical trials are now in progress for treating a variety of diseases[142]. However, use of gene silencing technology to treat established HCC faces major difficulties. These include identification of optimal targets, efficient and safe delivery of RNAi sequences and limitation of unintended off target effects. Hepatocarcinogenesis is a multistep process and because of the considerable heterogeneity underlying the molecular mechanisms of hepatocyte transformation, identification of ideal RNAi targets to treat the malignancy is complicated. HCC also often presents as a disseminated cancer and safe delivery of RNAi effectors to all malignant cells will require improvement of currently available vectors. Good progress has been made with silencing of HBV and HCV replication using RNAi. It is likely that treatment of these virus infections, as an HCC preventative measure, will be the first RNAi-based therapies to counter the malignancy. Currently, early diagnosis of HCC is critical for its effective treatment. Success of RNAi against HCC is also expected to be dependent on identifying the malignancy in its early stages before tumor bulk becomes excessive. In the near future, it seems that silencing technology may well be used as an adjunct to other liver cancer treatments. Thus, the utility of RNAi-based therapy is also likely to be reliant on improvement of existing treatment and diagnostic modalities. Despite these difficulties, intensive efforts from several quarters have given momentum to development of RNAi-based HCC therapy. It is difficult to anticipate technological advancements, but the field is likely to see considerable progress during the coming years.
Acknowledgments
We thank the Cancer Association of South Africa, the Sixth Research Framework Programme of the European Union Project RIGHT (LSHB-CT-2004-005276), South African National Research Foundation and Poliomyelitis Research Foundation support research conducted in the authors’ laboratory.
Supported by The Cancer Association of South Africa, the Sixth Research Framework Programme of the European Union Project RIGHT, LSHB-CT-2004-005276; South African National Research Foundation and Poliomyelitis Research Foundation support research
Peer reviewer: Anna C Piscaglia, MD, Department of internal medicine and Gastroenterology, Catholic University of Rome, Via Angelo Rocca 19, Roma 00135, Italy
S- Editor Zhong XY L- Editor Alpini GD E- Editor Ma WH
References
- 1.Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77–100. doi: 10.1111/j.1365-2613.2001.iep0082-0077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 4.Trinchet JC, Ganne-Carrie N, Nahon P, N'kontchou G, Beaugrand M. Hepatocellular carcinoma in patients with hepatitis C virus-related chronic liver disease. World J Gastroenterol. 2007;13:2455–2460. doi: 10.3748/wjg.v13.i17.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 6.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 7.Mason WS, Aldrich C, Summers J, Taylor JM. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus-strand DNA. Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 9.Weiser B, Ganem D, Seeger C, Varmus HE. Closed circular viral DNA and asymmetrical heterogeneous forms in livers from animals infected with ground squirrel hepatitis virus. J Virol. 1983;48:1–9. doi: 10.1128/jvi.48.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arbuthnot P, Capovilla A, Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J Gastroenterol Hepatol. 2000;15:357–368. doi: 10.1046/j.1440-1746.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- 11.Bartenschlager R, Frese M, Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res. 2004;63:71–180. doi: 10.1016/S0065-3527(04)63002-8. [DOI] [PubMed] [Google Scholar]
- 12.Boulant S, Becchi M, Penin F, Lavergne JP. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J Biol Chem. 2003;278:45785–45792. doi: 10.1074/jbc.M307174200. [DOI] [PubMed] [Google Scholar]
- 13.Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 2001;7:710–721. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foy E, Li K, Sumpter R Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale MJ Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 17.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 18.Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray RB, Lagging LM, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruggieri A, Harada T, Matsuura Y, Miyamura T. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology. 1997;229:68–76. doi: 10.1006/viro.1996.8420. [DOI] [PubMed] [Google Scholar]
- 21.Prikhod’ko EA, Prikhod’ko GG, Siegel RM, Thompson P, Major ME, Cohen JI. The NS3 protein of hepatitis C virus induces caspase-8-mediated apoptosis independent of its protease or helicase activities. Virology. 2004;329:53–67. doi: 10.1016/j.virol.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai MM. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagoner J, Austin M, Green J, Imaizumi T, Casola A, Brasier A, Khabar KS, Wakita T, Gale M Jr, Polyak SJ. Regulation of CXCL-8 (interleukin-8) induction by double-stranded RNA signaling pathways during hepatitis C virus infection. J Virol. 2007;81:309–318. doi: 10.1128/JVI.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray RB, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176–182. doi: 10.1006/viro.1996.0644. [DOI] [PubMed] [Google Scholar]
- 25.Marusawa H, Hijikata M, Chiba T, Shimotohno K. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappaB activation. J Virol. 1999;73:4713–4720. doi: 10.1128/jvi.73.6.4713-4720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamura H, Govindarajan S, Aswad F, Machida K, Lai MM, Sung VM, Dennert G. HCV core expression in hepatocytes protects against autoimmune liver injury and promotes liver regeneration in mice. Hepatology. 2006;44:936–944. doi: 10.1002/hep.21360. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, Nagano-Fujii M, Deng L, Ishido S, Sada K, Hotta H. Single-point mutations of hepatitis C virus NS3 that impair p53 interaction and anti-apoptotic activity of NS3. Biochem Biophys Res Commun. 2006;340:792–799. doi: 10.1016/j.bbrc.2005.12.076. [DOI] [PubMed] [Google Scholar]
- 28.Ishido S, Muramatsu S, Fujita T, Iwanaga Y, Tong WY, Katayama Y, Itoh M, Hotta H. Wild-type, but not mutant-type, p53 enhances nuclear accumulation of the NS3 protein of hepatitis C virus. Biochem Biophys Res Commun. 1997;230:431–436. doi: 10.1006/bbrc.1996.5980. [DOI] [PubMed] [Google Scholar]
- 29.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 30.Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22:535–545. doi: 10.1093/carcin/22.4.535. [DOI] [PubMed] [Google Scholar]
- 31.Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 32.Shen HM, Ong CN. Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis. Mutat Res. 1996;366:23–44. doi: 10.1016/s0165-1110(96)90005-6. [DOI] [PubMed] [Google Scholar]
- 33.McKillop IH, Moran DM, Jin X, Koniaris LG. Molecular pathogenesis of hepatocellular carcinoma. J Surg Res. 2006;136:125–135. doi: 10.1016/j.jss.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Blum HE. Treatment of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2005;19:129–145. doi: 10.1016/j.bpg.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 37.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 38.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 42.Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- 43.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 46.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 47.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 49.Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27:4238–4247. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Pogribny IP, Tryndyak VP, Boyko A, Rodriguez-Juarez R, Beland FA, Kovalchuk O. Induction of microRNAome deregulation in rat liver by long-term tamoxifen exposure. Mutat Res. 2007;619:30–37. doi: 10.1016/j.mrfmmm.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Zhou F, Xu T, Deng H, Ge YY, Zhang C, Li J, Zhuang SM. Analysis of sequence variations in 59 microRNAs in hepatocellular carcinomas. Mutat Res. 2008;638:205–209. doi: 10.1016/j.mrfmmm.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci USA. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 55.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 56.Turrini P, Sasso R, Germoni S, Marcucci I, Celluci A, Di Marco A, Marra E, Paonessa G, Eutropi A, Laufer R, et al. Development of humanized mice for the study of hepatitis C virus infection. Transplant Proc. 2006;38:1181–1184. doi: 10.1016/j.transproceed.2006.02.149. [DOI] [PubMed] [Google Scholar]
- 57.McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 58.Ren X, Luo G, Xie Z, Zhou L, Kong X, Xu A. Inhibition of multiple gene expression and virus replication of HBV by stable RNA interference in 2.2.15 cells. J Hepatol. 2006;44:663–670. doi: 10.1016/j.jhep.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 59.Ren XR, Zhou LJ, Luo GB, Lin B, Xu A. Inhibition of hepatitis B virus replication in 2.2.15 cells by expressed shRNA. J Viral Hepat. 2005;12:236–242. doi: 10.1111/j.1365-2893.2005.00587.x. [DOI] [PubMed] [Google Scholar]
- 60.Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Du D, Wu J, Chan CP, Tan Y, Kung HF, He ML. Inhibition of hepatitis B virus replication by stably expressed shRNA. Biochem Biophys Res Commun. 2003;311:398–404. doi: 10.1016/j.bbrc.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Carmona S, Ely A, Crowther C, Moolla N, Salazar FH, Marion PL, Ferry N, Weinberg MS, Arbuthnot P. Effective inhibition of HBV replication in vivo by anti-HBx short hairpin RNAs. Mol Ther. 2006;13:411–421. doi: 10.1016/j.ymthe.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 63.Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim YH, Lee JH, Paik NW, Rho HM. RNAi-based knockdown of HBx mRNA in HBx-transformed and HBV-producing human liver cells. DNA Cell Biol. 2006;25:412–417. doi: 10.1089/dna.2006.25.412. [DOI] [PubMed] [Google Scholar]
- 65.Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 66.Xuan B, Qian Z, Hong J, Huang W. EsiRNAs inhibit Hepatitis B virus replication in mice model more efficiently than synthesized siRNAs. Virus Res. 2006;118:150–155. doi: 10.1016/j.virusres.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Chen CC, Ko TM, Ma HI, Wu HL, Xiao X, Li J, Chang CM, Wu PY, Chen CH, Han JM, et al. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 2007;14:11–19. doi: 10.1038/sj.gt.3302846. [DOI] [PubMed] [Google Scholar]
- 68.Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 69.Klein C, Bock CT, Wedemeyer H, Wustefeld T, Locarnini S, Dienes HP, Kubicka S, Manns MP, Trautwein C. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- 70.Konishi M, Wu CH, Wu GY. Inhibition of HBV replication by siRNA in a stable HBV-producing cell line. Hepatology. 2003;38:842–850. doi: 10.1053/jhep.2003.50416. [DOI] [PubMed] [Google Scholar]
- 71.Hamasaki K, Nakao K, Matsumoto K, Ichikawa T, Ishikawa H, Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51–54. doi: 10.1016/s0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- 72.Ying C, De Clercq E, Neyts J. Selective inhibition of hepatitis B virus replication by RNA interference. Biochem Biophys Res Commun. 2003;309:482–484. doi: 10.1016/j.bbrc.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 73.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 74.Kim SI, Shin D, Choi TH, Lee JC, Cheon GJ, Kim KY, Park M, Kim M. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol Ther. 2007;15:1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- 75.Kronke J, Kittler R, Buchholz F, Windisch MP, Pietschmann T, Bartenschlager R, Frese M. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J Virol. 2004;78:3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Randall G, Rice CM. Interfering with hepatitis C virus RNA replication. Virus Res. 2004;102:19–25. doi: 10.1016/j.virusres.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 77.Takigawa Y, Nagano-Fujii M, Deng L, Hidajat R, Tanaka M, Mizuta H, Hotta H. Suppression of hepatitis C virus replicon by RNA interference directed against the NS3 and NS5B regions of the viral genome. Microbiol Immunol. 2004;48:591–598. doi: 10.1111/j.1348-0421.2004.tb03556.x. [DOI] [PubMed] [Google Scholar]
- 78.Wang Q, Contag CH, Ilves H, Johnston BH, Kaspar RL. Small hairpin RNAs efficiently inhibit hepatitis C IRES-mediated gene expression in human tissue culture cells and a mouse model. Mol Ther. 2005;12:562–568. doi: 10.1016/j.ymthe.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 79.Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartenschlager R, Lohmann V. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 2001;52:1–17. doi: 10.1016/s0166-3542(01)00164-4. [DOI] [PubMed] [Google Scholar]
- 81.Kato N, Shimotohno K. Systems to culture hepatitis C virus. Curr Top Microbiol Immunol. 2000;242:261–278. doi: 10.1007/978-3-642-59605-6_12. [DOI] [PubMed] [Google Scholar]
- 82.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 84.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Akashi H, Miyagishi M, Yokota T, Watanabe T, Hino T, Nishina K, Kohara M, Taira K. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol Biosyst. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- 89.Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K, Taira K, Yoshiba M, Kohara M. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- 90.Zhang J, Yamada O, Sakamoto T, Yoshida H, Iwai T, Matsushita Y, Shimamura H, Araki H, Shimotohno K. Down-regulation of viral replication by adenoviral-mediated expression of siRNA against cellular cofactors for hepatitis C virus. Virology. 2004;320:135–143. doi: 10.1016/j.virol.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 91.Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology. 2005;335:72–86. doi: 10.1016/j.virol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 92.Nakagawa M, Sakamoto N, Tanabe Y, Koyama T, Itsui Y, Takeda Y, Chen CH, Kakinuma S, Oooka S, Maekawa S, et al. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology. 2005;129:1031–1041. doi: 10.1053/j.gastro.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 93.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 94.Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166–1174. doi: 10.1053/j.gastro.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 96.Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, Waltemathe M, Gosling T, Flemming P, Malek NP, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci USA. 2003;100:7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cho-Rok J, Yoo J, Jang YJ, Kim S, Chu IS, Yeom YI, Choi JY, Im DS. Adenovirus-mediated transfer of siRNA against PTTG1 inhibits liver cancer cell growth in vitro and in vivo. Hepatology. 2006;43:1042–1052. doi: 10.1002/hep.21137. [DOI] [PubMed] [Google Scholar]
- 98.Tfelt-Hansen J, Kanuparthi D, Chattopadhyay N. The emerging role of pituitary tumor transforming gene in tumorigenesis. Clin Med Res. 2006;4:130–137. doi: 10.3121/cmr.4.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salvi A, Arici B, Alghisi A, Barlati S, De Petro G. RNA interference against urokinase in hepatocellular carcinoma xenografts in nude mice. Tumour Biol. 2007;28:16–26. doi: 10.1159/000097699. [DOI] [PubMed] [Google Scholar]
- 100.Salvi A, Arici B, De Petro G, Barlati S. Small interfering RNA urokinase silencing inhibits invasion and migration of human hepatocellular carcinoma cells. Mol Cancer Ther. 2004;3:671–678. [PubMed] [Google Scholar]
- 101.Duffy MJ. Urokinase-type plasminogen activator: a potent marker of metastatic potential in human cancers. Biochem Soc Trans. 2002;30:207–210. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 102.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 103.Pulukuri SM, Gondi CS, Lakka SS, Jutla A, Estes N, Gujrati M, Rao JS. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J Biol Chem. 2005;280:36529–36540. doi: 10.1074/jbc.M503111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Lakka SS, Gondi CS, Dinh DH, Olivero WC, Gujrati M, Rao VH, Sioka C, Rao JS. Specific interference of urokinase-type plasminogen activator receptor and matrix metalloproteinase-9 gene expression induced by double-stranded RNA results in decreased invasion, tumor growth, and angiogenesis in gliomas. J Biol Chem. 2005;280:21882–21892. doi: 10.1074/jbc.M408520200. [DOI] [PubMed] [Google Scholar]
- 105.Li H, Fu X, Chen Y, Hong Y, Tan Y, Cao H, Wu M, Wang H. Use of adenovirus-delivered siRNA to target oncoprotein p28GANK in hepatocellular carcinoma. Gastroenterology. 2005;128:2029–2041. doi: 10.1053/j.gastro.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 106.Fu XY, Wang HY, Tan L, Liu SQ, Cao HF, Wu MC. Overexpression of p28/gankyrin in human hepatocellular carcinoma and its clinical significance. World J Gastroenterol. 2002;8:638–643. doi: 10.3748/wjg.v8.i4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iwai A, Marusawa H, Kiuchi T, Higashitsuji H, Tanaka K, Fujita J, Chiba T. Role of a novel oncogenic protein, gankyrin, in hepatocyte proliferation. J Gastroenterol. 2003;38:751–758. doi: 10.1007/s00535-003-1141-8. [DOI] [PubMed] [Google Scholar]
- 108.Shan YF, Zhou WP, Fu XY, Yan HX, Yang W, Liu SQ, Cao HF, Kang B, Wu MC, Wang HY. The role of p28GANK in rat oval cells activation and proliferation. Liver Int. 2006;26:240–247. doi: 10.1111/j.1478-3231.2005.01203.x. [DOI] [PubMed] [Google Scholar]
- 109.Tan L, Fu XY, Liu SQ, Li HH, Hong Y, Wu MC, Wang HY. Expression of p28GANK and its correlation with RB in human hepatocellular carcinoma. Liver Int. 2005;25:667–676. doi: 10.1111/j.1478-3231.2005.01003.x. [DOI] [PubMed] [Google Scholar]
- 110.Detwiller KY, Fernando NT, Segal NH, Ryeom SW, D’Amore PA, Yoon SS. Analysis of hypoxia-related gene expression in sarcomas and effect of hypoxia on RNA interference of vascular endothelial cell growth factor A. Cancer Res. 2005;65:5881–5889. doi: 10.1158/0008-5472.CAN-04-4078. [DOI] [PubMed] [Google Scholar]
- 111.Tao J, Tu YT, Huang CZ, Feng AP, Wu Q, Lian YJ, Zhang LX, Zhang XP, Shen GX. Inhibiting the growth of malignant melanoma by blocking the expression of vascular endothelial growth factor using an RNA interference approach. Br J Dermatol. 2005;153:715–724. doi: 10.1111/j.1365-2133.2005.06765.x. [DOI] [PubMed] [Google Scholar]
- 112.Xu WH, Ge YL, Li Q, Zhang X, Duan JH. Inhibitory effect of vascular endothelial growth factors-targeted small interfering RNA on proliferation of gastric cancer cells. World J Gastroenterol. 2007;13:2044–2047. doi: 10.3748/wjg.v13.i14.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li TJ, Song JN, Kang K, Tong SS, Hu ZL, He TC, Zhang BQ, Zhang CQ. RNA interference-mediated gene silencing of vascular endothelial growth factor in colon cancer cells. World J Gastroenterol. 2007;13:5312–5316. doi: 10.3748/wjg.v13.i40.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mulkeen AL, Silva T, Yoo PS, Schmitz JC, Uchio E, Chu E, Cha C. Short interfering RNA-mediated gene silencing of vascular endothelial growth factor: effects on cellular proliferation in colon cancer cells. Arch Surg. 2006;141:367–374; discussion 374. doi: 10.1001/archsurg.141.4.367. [DOI] [PubMed] [Google Scholar]
- 115.Shen HL, Xu W, Wu ZY, Zhou LL, Qin RJ, Tang HR. Vector-based RNAi approach to isoform-specific downregulation of vascular endothelial growth factor (VEGF)165 expression in human leukemia cells. Leuk Res. 2007;31:515–521. doi: 10.1016/j.leukres.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 116.Karpala AJ, Doran TJ, Bean AG. Immune responses to dsRNA: implications for gene silencing technologies. Immunol Cell Biol. 2005;83:211–216. doi: 10.1111/j.1440-1711.2005.01331.x. [DOI] [PubMed] [Google Scholar]
- 117.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 118.Zhou HS, Liu DP, Liang CC. Challenges and strategies: the immune responses in gene therapy. Med Res Rev. 2004;24:748–761. doi: 10.1002/med.20009. [DOI] [PubMed] [Google Scholar]
- 119.Robbins MA, Li M, Leung I, Li H, Boyer DV, Song Y, Behlke MA, Rossi JJ. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat Biotechnol. 2006;24:566–571. doi: 10.1038/nbt1206. [DOI] [PubMed] [Google Scholar]
- 120.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 122.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 123.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 124.Judge A, McClintock K, Phelps JR, Maclachlan I. Hypersensitivity and loss of disease site targeting caused by antibody responses to PEGylated liposomes. Mol Ther. 2006;13:328–337. doi: 10.1016/j.ymthe.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 125.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 126.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arbuthnot P, Bralet MP, Thomassin H, Danan JL, Brechot C, Ferry N. Hepatoma cell-specific expression of a retrovirally transferred gene is achieved by alpha-fetoprotein but not insulinlike growth factor II regulatory sequences. Hepatology. 1995;22:1788–1796. [PubMed] [Google Scholar]
- 128.Arbuthnot PB, Bralet MP, Le Jossic C, Dedieu JF, Perricaudet M, Brechot C, Ferry N. In vitro and in vivo hepatoma cell-specific expression of a gene transferred with an adenoviral vector. Hum Gene Ther. 1996;7:1503–1514. doi: 10.1089/hum.1996.7.13-1503. [DOI] [PubMed] [Google Scholar]
- 129.Huber BE, Richards CA, Krenitsky TA. Retroviral-mediated gene therapy for the treatment of hepatocellular carcinoma: an innovative approach for cancer therapy. Proc Natl Acad Sci USA. 1991;88:8039–8043. doi: 10.1073/pnas.88.18.8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wadman M. Gene therapy might not have caused patient’s death. Nature. 2007;449:270. doi: 10.1038/449270b. [DOI] [PubMed] [Google Scholar]
- 131.Shen X, Storm T, Kay MA. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol Ther. 2007;15:1955–1962. doi: 10.1038/sj.mt.6300293. [DOI] [PubMed] [Google Scholar]
- 132.Alba R, Bosch A, Chillon M. Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther. 2005;12 Suppl 1:S18–S27. doi: 10.1038/sj.gt.3302612. [DOI] [PubMed] [Google Scholar]
- 133.Kreppel F, Kochanek S. Modification of adenovirus gene transfer vectors with synthetic polymers: a scientific review and technical guide. Mol Ther. 2008;16:16–29. doi: 10.1038/sj.mt.6300321. [DOI] [PubMed] [Google Scholar]
- 134.Oka K, Chan L. Construction and characterization of helper-dependent adenoviral vectors for sustained in vivo gene therapy. Methods Mol Med. 2005;108:329–350. doi: 10.1385/1-59259-850-1:329. [DOI] [PubMed] [Google Scholar]
- 135.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iwasaki Y, Ueda M, Yamada T, Kondo A, Seno M, Tanizawa K, Kuroda S, Sakamoto M, Kitajima M. Gene therapy of liver tumors with human liver-specific nanoparticles. Cancer Gene Ther. 2007;14:74–81. doi: 10.1038/sj.cgt.7700990. [DOI] [PubMed] [Google Scholar]
- 137.Wolschek MF, Thallinger C, Kursa M, Rossler V, Allen M, Lichtenberger C, Kircheis R, Lucas T, Willheim M, Reinisch W, et al. Specific systemic nonviral gene delivery to human hepatocellular carcinoma xenografts in SCID mice. Hepatology. 2002;36:1106–1114. doi: 10.1053/jhep.2002.36372. [DOI] [PubMed] [Google Scholar]
- 138.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 139.Watanabe T, Umehara T, Yasui F, Nakagawa S, Yano J, Ohgi T, Sonoke S, Satoh K, Inoue K, Yoshiba M, et al. Liver target delivery of small interfering RNA to the HCV gene by lactosylated cationic liposome. J Hepatol. 2007;47:744–750. doi: 10.1016/j.jhep.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 140.Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.John M, Constien R, Akinc A, Goldberg M, Moon YA, Spranger M, Hadwiger P, Soutschek J, Vornlocher HP, Manoharan M, et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449:745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Melnikova I. RNA-based therapies. Nat Rev Drug Discov. 2007;6:863–864. doi: 10.1038/nrd2314. [DOI] [PubMed] [Google Scholar]