Summary
Many bacteria assemble hair-like fibers termed pili or fimbriae on their cell surface. These fibers mediate adhesion to various surfaces, including host cells, and play critical roles in pathogenesis. Pili are polymers composed of thousands of individual subunit proteins. Understanding how these subunit proteins cross the bacterial envelope and correctly assemble at the cell surface is important not only for basic biology but also for the development of novel antimicrobial agents. The chaperone/usher pilus biogenesis pathway is one of the best-understood protein secretion systems, thanks in large part to innovative efforts in biophysical techniques such as X-ray crystallography and cryo-electron microscopy. Such a combined approach holds promise for further elucidating remaining questions regarding the multi-step and highly dynamic pilus assembly process, as well as for studying other protein secretion and organelle biogenesis systems.
Introduction to the combined approach of X-ray crystallography and cryo-electron microscopy
Single particle cryo-electron microscopy (SPEM) is a biophysical method for visualizing the structures of macromolecular assemblies [1]. SPEM starts with purified protein complexes in solution. Individual molecules in solution at random orientation are pipetted onto an EM grid and rapidly frozen in liquid ethane to achieve buffer vitrification. Frozen samples are kept at low temperature during electron imaging in a transmission electron microscope. A large number of particle images are recorded, computationally selected, aligned, averaged, and eventually the three-dimensional structure of the particle is reconstructed. Traditionally, X-ray crystallography and cryo-EM have focused on relatively different biological systems: crystallography favored smaller proteins and cryo-EM focused on extremely large biological oligomers and polymers, such as the ribosome, microtubules, and viruses. Advances in methodology have steadily increased the protein size that X-ray crystallography can deal with, and decreased the size for which cryo-EM is applicable, which is currently around 200 kDa. Hence, there is a trend toward gradual overlapping of molecular targets for which both methods are applicable. Synergy between X-ray crystallography and SPEM exists in two ways. In cases where the entire macromolecular structure can be determined by both methods, cryo-EM is usually at lower resolution, but more likely to provide flexible or biologically relevant conformations, due to the absence of crystal lattice constraints. More commonly, however, a large complex is difficult to crystallize and instead structures of individual protein components are determined to atomic resolution by X-ray crystallography. These structures are then docked into a cryo-EM map to assemble a pseudo-atomic resolution structural model of the whole complex and to detect protein contact interfaces.
The power of the combined approach of cryo-EM and X-ray crystallography has been amply illustrated by many highly insightful works, such as the racheting ribosome [2], the spinning bacterial flagella [3,4], the type III secretion injectisome [5,6], and the molecular motors tracking along F-actin or microtubules [7,8], to name a few. In the following sections, we briefly review how the combined structural approaches of SPEM and X-ray crystallography have yielded mechanistic insights into the highly dynamic and multi-step process of pilus assembly by the chaperone/usher (CU) pathway.
1. Bacterial pilus assembly by the usher and chaperone pathway
Pili (fimbriae) are non-flagellar, hair-like surface appendages. Pili perform a variety of functions, including adhesion to surfaces, motility, biofilm formation, colonization of host tissues, and invasion of host cells. This review focuses on P and type 1 pili expressed by uropathogenic E. coli (UPEC), which mediate adhesion in the urinary tract and are critical virulence determinants. P and type 1 pili are encoded by the chromosomal pap and fim gene clusters, respectively. Both pili are composite fibers composed of a short, linear tip fibrillum followed by a long (∼1-3 μm), helical rod that is attached to the bacterial surface (Fig. 1) [9,10]. The pilus rod contains over 1,000 copies of the major pilin subunit (PapA for P pili, FimA for type 1 pili). The P pilus rod is terminated by the PapH minor pilin [11,12]. The P pilus tip contains the PapG adhesin at the distal end, followed by the PapF, PapE, and PapK minor subunits [10]. PapE is present in multiple copies (∼5-10) in the tip. PapK functions as an adaptor subunit, linking the tip to the PapA rod. Similarly, PapF links the PapG adhesin to PapE. The type 1 pilus tip is shorter, containing the FimH adhesin at the distal end, followed by single copies of the FimG and FimF minor pilins/adaptors (Fig. 1) [9,13,14].
Fig. 1. Model for pilus biogenesis by the CU pathway.
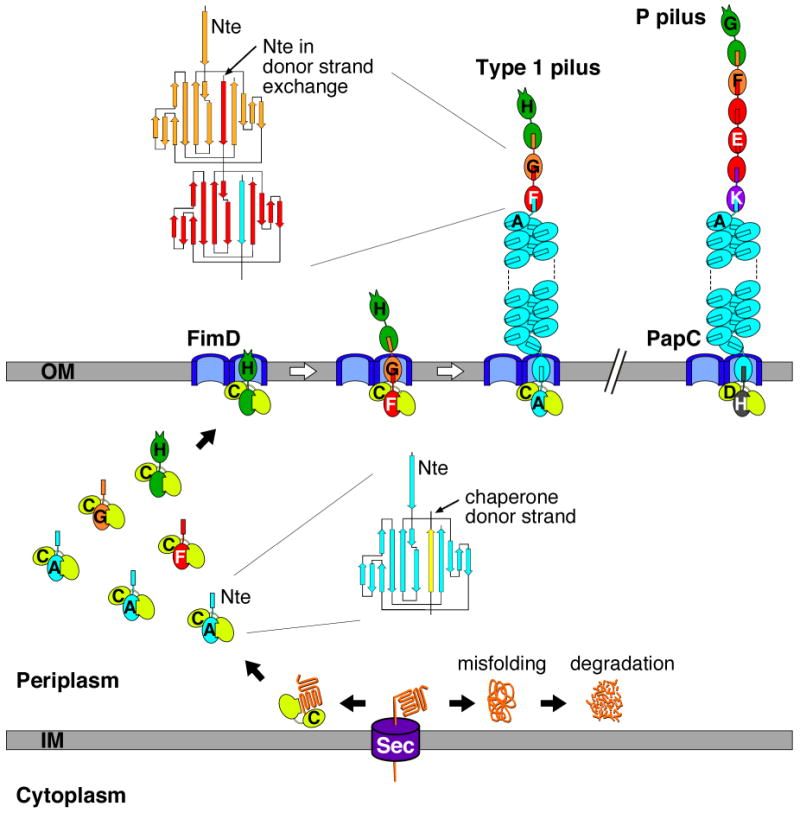
The assembly steps for type 1 pili are shown, with each of the Fim proteins indicated by a single letter (C, FimC; A, FimA; etc.). Please see the main text for details. A model of an assembled P pilus is shown in the upper right. The topology diagrams depict the donor strand complementation and exchange reactions occurring in the periplasm and assembled pilus fiber, respectively.
Biogenesis of P and type 1 pili requires a dedicated periplasmic chaperone (PapD for P pili, FimC for type 1 pili) and an integral outer membrane protein termed the usher (PapC for P pili, FimD for type 1 pili), which together form the secretory machinery of the chaperone/usher (CU) pathway (Fig. 1). Pilus subunits enter the periplasm as unfolded polypeptides via the Sec general secretory pathway [15], and then must interact with the periplasmic chaperone for proper folding and stabilization. In the absence of the chaperone, subunits misfold and form aggregates that are degraded by the DegP periplasmic protease [16]. Periplasmic chaperone-subunit complexes must next target to the outer membrane usher for subunit assembly into the pilus fiber and secretion through the usher to the cell surface (Fig. 1). Pili are assembled in a top down fashion, with the adhesin incorporated first, followed by assembly of the tip fibrillum and finally the rod. Each subunit specifically interacts with its appropriate neighbor subunit in the pilus [17,18*] and the usher facilitates this organization by differentially recognizing chaperone-subunit complexes according to their final position in the pilus [19,20].
2. The pilus structure as revealed by cryo-EM
The nascently assembled pilus fiber is constrained to a linear form during translocation through the usher channel, and the rod must undergo a quaternary structural transition to adopt its final helical form on the bacterial surface (Fig. 1). The rod is extensible and can unravel back into a linear fiber under stress [21]. Such structural flexibility, though important for maintenance of bacterial adhesion [22,23], makes it difficult to determine pilus structures by cryo-EM using the well-established helical image reconstruction technique. To tackle structures of this nature, a real space single particle-based technique was developed [24]. Application of the new method yielded several cryo-EM maps of pilus rods, and models have been built by docking crystal structures of pilus subunits into these maps [14,25,26]. A similar single particle cryo-EM approach was applied recently to reveal the structure of the Neisseria gonorrhoeae Type IV pilus [27].
The P pilus rod measures 82 Å in diameter and contains a 25 Å axial channel (Fig. 2C) [25]. There are 3.28 subunits per helical turn. The structure of the type 1 pilus rod is very similar, with an external diameter of 69 Å, a 2.1-2.5 Å axial channel, and 3.375 subunits per turn [14]. The rod is stabilized by horizontal interactions between neighboring subunits and by lateral interactions between n and n+3 subunits. For P pili, the lateral interactions involve a loop region in PapA (residues 106-109). Disruption of this loop region by mutagenesis abolished the helical rod structure, resulting in formation of a linear fiber [26]. In addition, residues at the N terminus of PapA form a protruding hinge region that appears to provide the flexibility required for rotation of the PapA monomers to coil into the helix [25]. Mutations of residues within this region affected PapA-PapA interactions and also prevented formation of the native pilus helix [25,28].
Fig. 2. Donor strand complementation (DSC) and donor strand exchange (DSE) as observed in crystal structures and cryo-EM map of the P pilus rod.
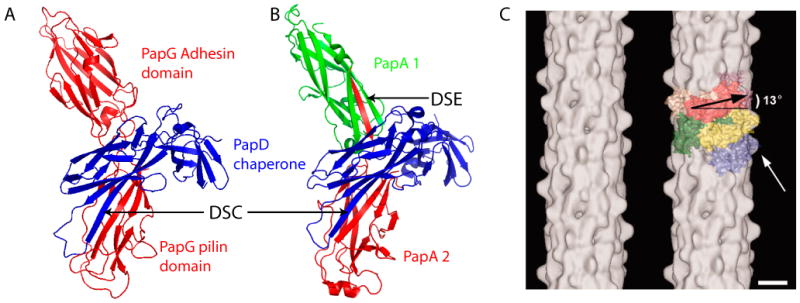
(A) Crystal structure of the PapD-PapG chaperone-adhesin complex (PDB ID: 1QUN). (B) Crystal structure of a PapD-PapA1-PapA2 complex (PDB ID: 2UY6). The arrows labeled DSC indicate the chaperone G1 β-strand engaged in DSC with the pilin domain of PapG (A) or the PapA2 subunit (B). The arrow labeled DSE indicates the Nte of the PapA2 subunit engaged in DSE with the PapA1 subunit (B). (C) Cryo-EM map (left) and modeling with the crystal structure of PapK (right) of the P pilus rod at an estimated resolution of 10 Å. The major pilin PapA appears to sit 13° tilted from horizontal in the helical rod. The white arrow points to a surface protrusion that is presumably the hinge loop formed by the PapA N terminus. Scale bar: 25 Å. Panel C is adapted from reference [25].
3. Donor strand complementation and donor strand exchange mechanisms govern pilus biogenesis
A series of groundbreaking X-ray crystal structures revealed the molecular mechanisms governing chaperone-subunit and subunit-subunit interactions during pilus biogenesis [29-32]. All pilus subunits consist of a single immunoglobulin-like (Ig) fold termed the pilin domain (Figs. 1 and 2A). (The exception is the adhesin, which contains an additional, N-terminal adhesin domain). Notably, the pilin domain lacks the seventh β-strand (the G strand) present in canonical Ig folds. This results in a deep groove on the pilin surface, exposing the hydrophobic core. In the periplasm, the chaperone, which consists of two complete Ig domains oriented in an L shape [33], completes the Ig fold of the subunit by a mechanism termed donor strand complementation (DSC) [29,30]. In DSC, the chaperone inserts a motif of alternating hydrophobic residues from its G1 β-strand into the subunit groove, forming a β-zipper interaction (Figs. 1 and 2A). Interestingly, the chaperone G1 β-strand inserts parallel, rather than anti-parallel, to the F strand of the subunit, resulting in a non-canonical Ig fold. This maintains subunits in an “activated” state that is competent for subsequent pilus assembly [31,32].
Subunit-subunit interactions occur by a mechanism similar to DSC, termed donor strand exchange (DSE) [31,32]. All pilus subunits (with the exception of the adhesin) have a conserved N-terminal extension (Nte), which contains a motif of alternating hydrophobic residues similar to the chaperone G1 β-strand. In DSE, the Nte from one subunit inserts into the groove of the preceding subunit to complete that subunit's Ig fold (Figs. 1 and 2B) [31,32]. The DSE reaction occurs via a concerted strand displacement mechanism, in which an incoming chaperone-subunit complex forms a transient intermediate with the preceding chaperone-subunit complex. The chaperone is displaced from the transient intermediate via a ‘zip-in–zip-out’ mechanism, in which the G1 β-strand of the preceding chaperone zips out, residue by residue, as the Nte of the incoming subunit simultaneously zips in [34*,35*,36]. In DSE, the Nte inserts anti-parallel to the F strand, forming a canonical Ig fold. Thus, DSE allows subunits to adopt a more compact, lower energy state compared to DSC, and it is this topological transition that appears to provide the driving force for pilus assembly [31,32,37].
4. The versatile usher is a dimeric structure
Pilus fibers assemble on the order of minutes in vivo [38,39], but on the order of hours in vitro in the absence of the usher [34*,35*,40**]. Using an in vitro reconstituted type 1 pilus system, the FimD usher was recently shown to speed up the formation of FimA polymers by a factor of > 1000 [40**]. Therefore, the usher is a highly effective pilus assembly catalyst.
The usher contains four distinct domains: a central, transmembrane β-barrel domain that forms the secretion channel, a middle domain located within the β-barrel region that forms a channel gate or plug, and soluble N- and C-terminal domains that are located in the periplasm (see Fig. 4A) [41**-44]. The usher N-terminal domain provides the initial binding site for periplasmic chaperone-subunit complexes [42,45,46], whereas the C-terminal domain is required for subsequent pilus assembly steps. The C-terminal domain may stabilize the binding of chaperone-subunit complexes on the usher and may also participate in controlling access to the usher channel [43,47].
Fig. 4. Model for type 1 pilus biogenesis at the outer membrane usher.
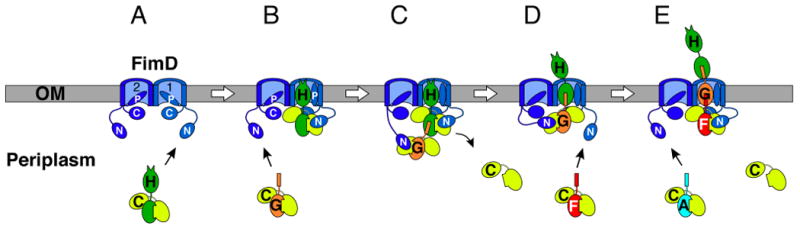
(A) The two ushers in the FimD dimer are labeled 1 and 2. Each usher monomer contains periplasmic N- and C-terminal domains (labeled N and C, respectively) and the translocation channel is gated shut by a plug domain (labeled P). (B) Interaction of a FimCH complex with usher 1 opens the plug domain of usher 1, shown as rotating up within the channel. The plug domain could also rotate out of the channel into the periplasm. (B-E) The N-terminal domains of ushers 1 and 2 alternate in recruiting periplasmic chaperone-subunit complexes to the usher for donor strand exchange, subunit incorporation into the pilus fiber, and secretion of the fiber through the usher to the cell surface.
A medium resolution (18 Å) projection structure of the usher was obtained by reconstituting purified PapC into a lipid bilayer to generate two-dimensional crystals [48]. Cryo-EM analysis of these crystals showed that the usher formed a dimeric complex in the membrane, with each usher monomer containing an apparent central channel of ∼2 nm diameter (Fig. 3A, left). A 2 nm channel is consistent with the pore-forming activity of the usher [49], and is sufficiently large to accommodate secretion of a linear fiber of folded pilus subunits. Subsequent biochemical and genetic evidence further supported the notion that the usher functions as a dimeric complex in vivo [43].
Fig. 3. The usher caught in action.
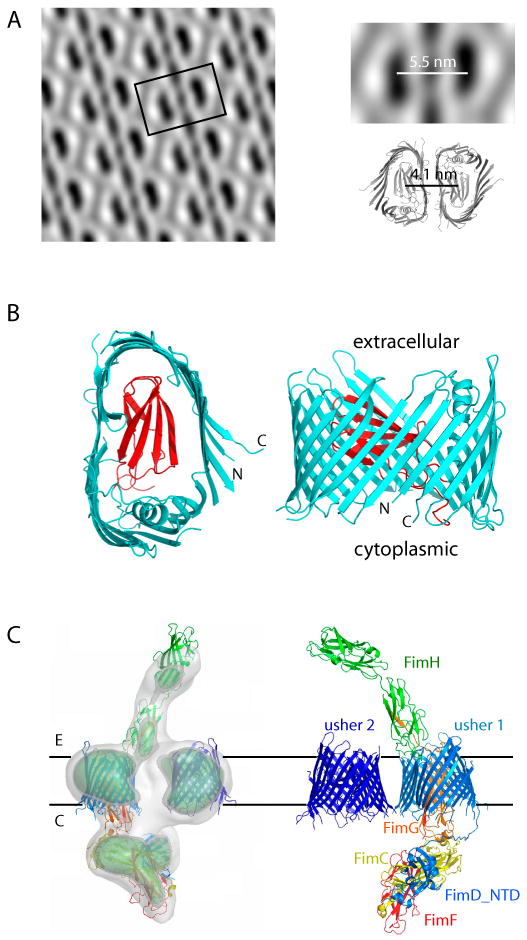
(A) The P pilus usher PapC forms a dimer when reconstituted in a lipid bilayer as a two-dimensional crystalline sheet. The center-to-center distance between the two PapC monomers is 14 Å larger than that observed in the crystal structure of the PapC dimer, suggesting a lipid mediated dimerization interface in vivo. (B) The crystal structure of the PapC translocation domain shown in extracellular (left) and side (right) views (PDB ID 2VQI). The 24-stranded β-barrel is shown in cyan and the plug domain in red. (C) A surface rendered side view of the cryo-EM map of the FimDCFGH tip complex (left) and rigid-body docking of the crystal structures of the components (right). The structures used are the FimD translocation domain (PDB ID 2VQI), FimH (PDB ID 1QUN), FimDNCHP (PDB ID 1ZE3), and the PapDAA ternary complex (PB ID 2UY6) as a guide for positioning FimCFG.
The recent high-resolution X-ray crystal structure of the translocation domain of the PapC usher provided a quantum leap forward in understanding fiber assembly and secretion by the CU pathway (Fig. 3B) [41**]. The structure revealed the usher to be the largest single-protein β-barrel channel to date, comprised of 24 transmembrane β-strands and, unexpectedly, an Ig-like middle domain that plugged the channel of the β-barrel. In hindsight, both features are perfectly reasonable and necessary: translocation of fully folded pilins requires a sufficiently large channel and such a large channel requires a plug to prevent leakage of periplasmic contents. In agreement with the cryo-EM structure, the PapC translocation domain formed a dimer in the crystal structure, with a similar usher:usher interface (Fig. 3A, right). There are two detergent molecules at the interface in the crystal structure, suggesting a lipid-mediated dimer interface in vivo. This is supported by the observation that the center-to-center distance between the two monomers in the EM structure is 14 Å larger than in the crystal structure [41**].
5. The usher assembles pili asymmetrically
The best approach to understand a process as dynamic as pilus biogenesis is to take a series of snapshots of the assembly process. This requires capturing critical assembly intermediates [13] and revealing their structures in sufficient detail. One such assembly intermediate was recently captured for type 1 pili using cryo-EM [41**]. An usher-pilus tip complex comprised of FimD:FimC:FimF:FimG:FimH was isolated from bacteria by expressing these proteins in the absence of the FimA rod subunit. The stoichiometry of this complex was 2:1:1:1:1, corresponding to a single tip fiber bound to a dimeric FimD usher and a single periplasmic chaperone. Cryo-EM analysis was then used to generate a 3D reconstruction of the FimD-tip complex at 23 Å resolution (Fig. 3C) [41**]. Since the crystal structures of all components or their homologs were available, these structures were docked into the 3D cryo-EM envelope as rigid bodies (Fig. 3C) [41**]. The resulting model of the tip complex presented fresh insights into the assembly mechanism as described below.
The tip complex model suggests that the action of the ushers within the dimeric complex is coordinated to assemble only one pilus at a time, as depicted in Fig. 4 for type 1 pili. Pilus assembly initiates when a chaperone-adhesin complex (FimCH) binds to the N-terminal domain of one of the usher monomers (usher 1 in Fig. 4A). The chaperone-adhesin complex then makes stable interactions with the usher C terminus and induces a conformational change in the usher to gate open the usher by rotation or displacement of the plug domain, activating the usher for pilus biogenesis (Fig. 4B). The N-terminal domain of the second usher (usher 2) remains available to bind the next chaperone-subunit complex (FimCG). The channel within usher 2 will remain closed, as only chaperone-adhesin complexes cause a conformational change to activate the usher [20,40**]. The two chaperone-subunit complexes are now positioned for the Nte of FimG to displace the G1 β-strand of the FimC chaperone from FimH (Fig. 4C), resulting in DSE between FimG and FimH and formation of the first link in the pilus fiber (Fig. 4D). Formation of the FimGH complex also results in dissociation of FimC from FimH and the release of usher 1's N-terminal domain. The cycle then repeats with binding of the next chaperone-subunit complex, FimCF, to the liberated N-terminal domain of usher 1 (Fig. 4D). DSE between FimF and FimG will generate the next link in the pilus fiber, displacing the chaperone that had been bound to FimG and liberating the N-terminal domain of usher 2 (Fig. 4E). This assembly intermediate corresponds to the tip complex visualized by cryo-EM (Fig. 3C). Multiple rounds of binding and DSE by FimCA subunits will then drive assembly and secretion of the pilus rod through the usher to the cell surface. Note that the N-terminal domains of the two ushers alternate in recruiting chaperone-subunit complexes from the periplasmic pool. Thus, although only one usher channel is used for secretion of the pilus fiber, two ushers are required for pilus elongation. In addition, the positioning of the chaperone-subunit complexes by the two ushers (see Fig. 4C) is likely to be key for the catalytic activity of the usher in pilus assembly.
6. Remaining questions
The work described above shows the powerful synergy and mechanistic insights that can result from a combined X-ray crystallography and cryo-EM approach. Several outstanding questions regarding the pilus assembly mechanism remain to be answered. (i) What is the mechanism for gating open the usher, how does the plug domain move to accommodate the incoming subunit, and what is the exact role of C-terminal domain of the usher? These questions will likely be answered by solving the crystal structure of the full-length FimDCH ternary complex. (ii) The FimD tip complex (FimDCFGH) represents only one of the several tip assembly steps. What are the structures, by cryo-EM and/or X-ray crystallography, of several other intermediates, such as FimDCFH and FimDCGH? These structures will provide insights into the step-by-step assembly process, elucidate the catalytic mechanism of the usher, and reveal if the usher dimer functions as proposed in the model presented in Fig. 4. (iii) How is the pilus anchored on the cell surface? The terminator subunit (PapH for P pili) alone might not be sufficient, since interaction between chaperone-subunit is weaker than that between subunit-subunit. It is possible that the extracellular surface of the usher, enlarged by its dimeric configuration, contributes to attachment of the helical rod on the cell. Cryo-EM visualization of the pilus rod emerging from the usher might help answer this question. (iv) What does the asymmetrical translocation mechanism observed in the CU system tell us about other protein secretion pathways? There is intriguing evidence that asymmetric mechanisms are also utilized in systems ranging from the Sec general secretory pathway to protein import into mitochondria [50,51]. Elucidation of all these questions will undoubtedly require a full range of structural approaches as part of integrated, multidisciplinary efforts.
Acknowledgments
Work in the Li laboratory is supported by NIH grants AI70285 and GM74985, and LDRD grant 06-60 at the Brookhaven National Laboratory under contract with the U.S. Department of Energy. Work in the Thanassi laboratory is supported by NIH grants GM62987 and AI55621.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State. New Ed. New York, USA: Oxford University Press; 2006. [Google Scholar]
- 2.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 3.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 5.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 6.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2008.12.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volkmann N, Liu H, Hazelwood L, Krementsova EB, Lowey S, Trybus KM, Hanein D. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Jones CH, Pinkner JS, Roth R, Heuser J, Nicholoes AV, Abraham SN, Hultgren SJ. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehn MJ, Heuser J, Normark S, Hultgren SJ. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature. 1992;356:252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- 11.Baga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- 12.Verger D, Miller E, Remaut H, Waksman G, Hultgren S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 2006;7:1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci USA. 2000;97:9240–9245. doi: 10.1073/pnas.160070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Haner M, Taschner N, Burkhard P, Aebi U, Muller SA. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J Mol Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- 15.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 16.Jones CH, Dexter P, Evans AK, Liu C, Hultgren SJ, Hruby DE. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J Bacteriol. 2002;184:5762–5771. doi: 10.1128/JB.184.20.5762-5771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YM, Dodson KW, Hultgren SJ. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J Bacteriol. 2007;189:5276–5283. doi: 10.1128/JB.01648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose RJ, Verger D, Daviter T, Remaut H, Paci E, Waksman G, Ashcroft AE, Radford SE. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc Natl Acad Sci U S A. 2008;105:12873–12878. doi: 10.1073/pnas.0802177105. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The new technique of noncovalent mass spectrometry was employeed to reveal a correlation between the assembly order of subunits in pili and their propensity to undergo DSE in vitro. This is a physical demonstration that subunit ordering is dominated by the Nte sequence and the acceptor binding groove.
- 19.Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ. Outer membrane PapC usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bullitt E, Makowski L. Structural polymorphism of bacterial adhesion pili. Nature. 1995;373:164–167. doi: 10.1038/373164a0. [DOI] [PubMed] [Google Scholar]
- 22.Forero M, Yakovenko O, Sokurenko EV, Thomas WE, Vogel V. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 2006;4:e298. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller E, Garcia T, Hultgren S, Oberhauser AF. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 25.Mu XQ, Bullitt E. Structure and assembly of P-pili: a protruding hinge region used for assembly of a bacterial adhesion filament. Proc Natl Acad Sci U S A. 2006;103:9861–9866. doi: 10.1073/pnas.0509620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu XQ, Jiang ZG, Bullitt E. Localization of a critical interface for helical rod formation of bacterial adhesion P-pili. J Mol Biol. 2005;346:13–20. doi: 10.1016/j.jmb.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 27.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Verger D, Bullitt E, Hultgren SJ, Waksman G. Crystal structure of the P pilus rod subunit PapA. PLoS Pathog. 2007;3:e73. doi: 10.1371/journal.ppat.0030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 30.Sauer FG, Fütterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 31.Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 32.Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 33.Holmgren A, Brändén C. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature. 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 34.Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol Cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PubMed] [Google Scholar]; * With the combined physical techniques of X-ray crystallography and real-time electrospray ionization mass spectrometry, the authors captured a transient ternary complex between the chaperone-subunit complex and the Nte of the incoming subunit. This work demonstrates that DSE proceeds in a zip-in-zip-out mechanism that requires an initiation site (the P5 pocket) in the sununit.
- 35.Vetsch M, Erilov D, Moliere N, Nishiyama M, Ignatov O, Glockshuber R. Mechanism of fibre assembly through the chaperone-usher pathway. EMBO Rep. 2006;7:734–738. doi: 10.1038/sj.embor.7400722. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Complementing the above physical approaches, these authors performed biochemical measurements of kinetic and thermodynamic interactions bewteen chaperone-subunit complexes and the Nte, demonstrating the same concerted reaction mechanism of DSE in which a chaperone-bound acceptor subunit is attacked by the incoming chaperoned subunit.
- 36.Rose RJ, Welsh TS, Waksman G, Ashcroft AE, Radford SE, Paci E. Donor-strand exchange in chaperone-assisted pilus assembly revealed in atomic detail by molecular dynamics. J Mol Biol. 2008;375:908–919. doi: 10.1016/j.jmb.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 37.Zavialov AV, Tischenko VM, Fooks LJ, Brandsdal BO, Aqvist J, Zav'yalov VP, Macintyre S, Knight SD. Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem J. 2005;389:685–694. doi: 10.1042/BJ20050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob-Dubuisson F, Striker R, Hultgren SJ. Chaperone-assisted self-assembly of pili independent of cellular energy. J Biol Chem. 1994;269:12447–12455. [PubMed] [Google Scholar]
- 39.Dodd DC, Eisenstein BI. Kinetic analysis of the synthesis and assembly of type 1 fimbriae of Escherichia coli. J Bacteriol. 1984;160:227–232. doi: 10.1128/jb.160.1.227-232.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of Pilus Assembly Reveals a Bacterial Outer Membrane Catalyst. Science. 2008;320:376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]; ** As the first in vitro reconstitution of the usher catalyzed subunit polymerization, this work demonstrates the high efficiency of the usher as a catalyst, and provides an effective in vitro assay for identifying compounds that hinder pilus assembly.
- 41.Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, Hultgren SJ, Thanassi DG, Waksman G, Li H. Fiber Formation across the Bacterial Outer Membrane by the Chaperone/Usher Pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** In this two-pronged study, the structure of the translocation domain of the PapC usher was determined by X-ray crystallography, and the FimD-tip assembly intermediate was captured and visualized by cryo-EM. This work provides so far the strongest evidence that the usher functions as a dimer in the concerted recruitment and assembly of pilus subunits.
- 42.Nishiyama M, Horst R, Eidam O, Herrmann T, Ignatov O, Vetsch M, Bettendorff P, Jelesarov I, Grutter MG, Wuthrich K, et al. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shu Kin So S, Thanassi DG. Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol Microbiol. 2006;60:364–375. doi: 10.1111/j.1365-2958.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- 44.Capitani G, Eidam O, Grutter MG. Evidence for a novel domain of bacterial outer membrane ushers. Proteins. 2006;65:816–823. doi: 10.1002/prot.21147. [DOI] [PubMed] [Google Scholar]
- 45.Ng TW, Akman L, Osisami M, Thanassi DG. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J Bacteriol. 2004;186:5321–5331. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishiyama M, Vetsch M, Puorger C, Jelesarov I, Glockshuber R. Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J Mol Biol. 2003;330:513–525. doi: 10.1016/s0022-2836(03)00591-6. [DOI] [PubMed] [Google Scholar]
- 47.Thanassi DG, Stathopoulos C, Dodson KW, Geiger D, Hultgren SJ. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J Bacteriol. 2002;184:6260–6269. doi: 10.1128/JB.184.22.6260-6269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Qian L, Chen Z, Thahbot D, Liu G, Liu T, Thanassi DG. The outer membrane usher forms a twin-pore secretion complex. J Mol Biol. 2004;344:1397–1407. doi: 10.1016/j.jmb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Thanassi DG, Saulino ET, Lombardo MJ, Roth R, Heuser J, Hultgren SJ. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]


