Abstract
A number of abundant mobile genetic elements called retrotransposons reverse transcribe RNA to generate DNA for insertion into eukaryotic genomes. Four major classes of retrotransposons are described here. First, the long-terminal-repeat (LTR) retrotransposons have similar structures and mechanisms to those of the vertebrate retroviruses. Genes that may enable these retrotransposons to leave a cell have been acquired by these elements in a number of animal and plant lineages. Second, the tyrosine recombinase retrotransposons are similar to the LTR retrotransposons except that they have substituted a recombinase for the integrase and recombine into the host chromosomes. Third, the non-LTR retrotransposons use a cleaved chromosomal target site generated by an encoded endonuclease to prime reverse transcription. Finally, the Penelope-like retrotransposons are not well understood but appear to also use cleaved DNA or the ends of chromosomes as primer for reverse transcription. Described in the second part of this review are the enzymatic properties of the reverse transcriptases (RTs) encoded by retrotransposons. The RTs of the LTR retrotransposons are highly divergent in sequence but have similar enzymatic activities to those of retroviruses. The RTs of the non-LTR retrotransposons have several unique properties reflecting their adaptation to a different mechanism of retrotransposition.
Keywords: retrotransposon, reverse transcriptase, phylogenetic relationship, integration mechanism
1. Introduction
Vertebrate retroviruses represent but one lineage of an ever-growing family of mobile genetic elements that utilize reverse transcriptase to generate a DNA copy from their RNA transcript. While a few of these other lineages, such as hepadnaviruses and caulimoviruses, are true viruses the largest number of lineages are classified as retrotransposable elements, or retrotransposons. The first retrotransposons to be identified were discovered because they caused mutations in two favorite model organisms: yeast, Saccharomyces cerevisiae, and the fruitfly, Drosophila melanogaster. The sequences of these elements revealed long-terminal repeats (LTRs) and open reading frames that encoded reverse transcriptase, RNase H, integrase, proteinase and gag-like proteins in an organization that was suggestive of retroviruses (Mount and Rubin 1985; Clare and Farabaugh 1985). Elegant experiments demonstrated that the yeast element made new copies by reverse transcription of their RNA transcripts (Boeke et al., 1985). These retrotransposons, however, did not encode a protein similar to retroviral envelope (env) genes and did not spread between individuals in a population. The retrotransposons were viewed as possible progenitors of the retroviruses, or alternatively as descendants of the retroviruses by loss of their envelope gene. Today the number of characterized retrotransposons has expanded dramatically and many new examples of elements with different putative env genes have been found. The wide diversity of retrotransposons compared to the limited diversity of vertebrate retroviruses suggests the ancestral forms were retrotransposons.
Without an env-like gene retrotransposons are unable to leave the environment of one cell for another cell, thus they must insert into the chromosomes of the germ cells to insure passage to the next generation. The inability to leave an organism also means that retrotransposons must be more circumspect than a virus in how often they replicate due to the potential damage caused by their insertion into the host genome. Any insertion that significantly reduces the fitness of the host will be lost from the population. Given this constraint, it is remarkable that large numbers of retrotransposon families using a variety of mechanisms to reverse transcribe and insert their genetic information into a genome have become highly successful in every lineage of eukaryotic organisms.
Even more remarkable than their diversity is the abundance of retrotransposons in most organisms. Indeed, the reason why many eukaryotic genomes are so enormous in size is because of the accumulation of retrotransposable elements. For example, retrotransposons constitute 42% of the human genome (Lander et al., 2001) and 75% of the maize genome (SanMiguel et al., 1998). Even these percentages are underestimates because the scrambling of DNA sequences by mutation, recombination and continued retrotransposon insertions make the oldest insertions impossible to recognize. Only those organisms that need to replicate their DNA quickly, or have found recombinational mechanisms to remove insertions, appear to be able to prevent the accumulation of elements over time (Charlesworth et al., 1994).
In the following sections we describe the major classes of retrotransposons that are known today emphasizing their structure, their phylogenetic relationship to each other and their mechanism of retrotransposition. Finally, for those retrotransposons where the reverse transcriptase have been studied, we compare the properties of their reverse transcriptases (RTs) with that of retroviral RTs.
2. The use of RT sequences to evaluate the relationship of retrotransposons
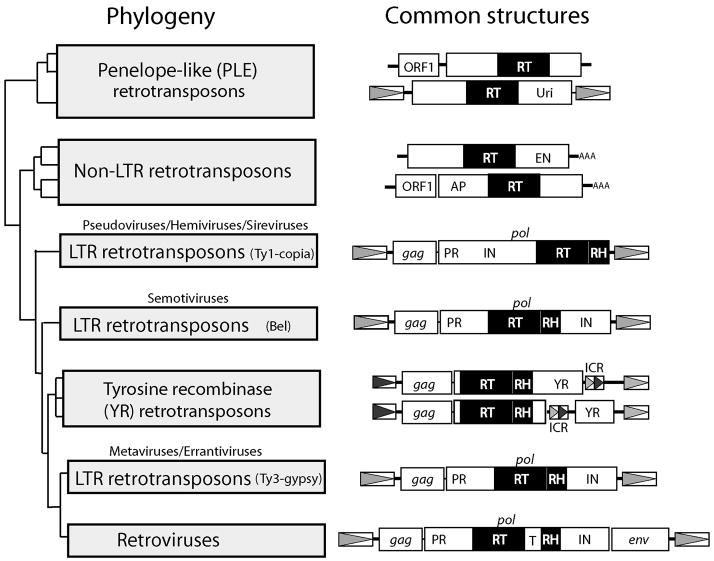
Determining the relationships between the different classes of retrotransposons has been challenging. Grouping elements by their common structural features and mechanism of insertion works well for those groups that have uniform structures and well-defined mechanisms of integration. However, as will be described below there are few shared features for some groups of retrotransposons and our knowledge of their mechanism of integration is limited. A second approach classifies elements based on the level of sequence identity of genes common to all elements. Eukaryotic retrotransposons have such different coding capacities that the only protein sequence shared by all classes of elements is the RT domain (Figure 1). For 20 years various attempts have been made to use these RT sequences to determine the phylogenetic relationship of retrotransposons (Xiong and Eickbush, 1988; Doolittle et al., 1989). Only the seven regions that define the catalytic regions of the enzyme have evolved slowly enough to enable the alignment of sequences in all retrotransposons (Poch et al., 1989; Xiong and Eickbush, 1990, Kohlstaedt et al., 1992). Using sequence similarity within these regions has served as a simple reliable approach to separate known elements into major groups as well classify even partially characterized elements.
Figure 1.
Structure and phylogenetic relationship of the various groups of retrotransposons. Left side: phylogenetic relationship of the retrotransposons based on the sequence of their reverse transcriptase domains. The figure is not intended to represent a specific phylogenetic analysis, rather it represents a summary of the generally accepted relationships based on a number of different studies (Eickbush, 1994; Goodwin and Poulter, 2004; Arkhipova et al. 2003) and rooted on the sequences of telomerase and bacterial reverse transcriptases. All elements within a specific lineage are represented by a box labeled with the commonly used name to describe the elements. Above the three lineages of LTR-retrotransposons are names suggested by the International Committee on the Taxonomy of Viruses to indicate some members of the lineage may be viruses (Boeke et al., 2005; Eickbush et al., 2005). Right side: common structures for the elements in each group. If significant structural variation occurs within a group, then two structures are diagrammed to represent the types of variation most commonly seen. The open reading frames (ORFs) of each element are shown as horizontal boxes. Multiple ORFs in the same element may be in different reading frames or separated by termination codons. ORFs with similarity to the gag, pol and env genes of vertebrate retroviruses are labeled as such. Abbreviations for protein encoding domains: RT, reverse transcriptase DNA polymerase domain; RH, reverse transcriptase RNase H domain; PR, proteinase, IN; integrase; T, tether; APE, apurinic endonuclease; EN, endonuclease; Uri, domain similar to the endonuclease of some mobile group I introns; YR, domain with similarity to tyrosine recombinanses. Shaded arrowheads in boxes, long terminal repeats (LTRs) or internal complimentary repeats (ICR) with sequence identity to the LTRs; thin lines, non-translated regions of the elements; AAA, poly(A) tails.
When using sequence similarity to determine the phylogeny of retrotransposons one can make the assumption that the most divergent sequences represent the most ancient lineages. However, it has been estimated that after a few hundred million years even the seven highly conserved segments of the RT domain are as divergent as the selective constraints to retain function on the encoded protein will allow (Malik et al., 1999). Therefore, the sequence divergence between the major groups of retrotransposons represent equilibrium levels based on the selective constraints on the RT protein, rather than the divergence time between groups. As a consequence, it should be emphasized that the relative ages of the different retrotransposon groups are the least reliable property estimated by this approach. Unfortunately, there is no other means to estimate their ages.
The phylogenetic relationships of the different groups of retrotransposons based on their RT sequences are summarized in Figure 1. This figure does not represent a specific phylogenetic analysis but is intended to represent the relationships between elements that has the greatest level of support from the various attempts that have been made. For more detailed comparisons the reader can turn to a number of studies (Eickbush and Malik, 2002; Arkhipova et al., 2003; Goodwin and Poulter, 2004; Lorenzi et al., 2006). Various attempts have also been made to extend the phylogeny of retrotransposons to include bacterial and mitochondrial genetic elements that encode RT sequences (e.g. Group II introns, retrons) as well as eukaryotic telomerases, which like RTs catalyze the formation of DNA from RNA template. The phylogenies obtained depend upon the extent of the RT sequences and the algorithms used, and there is at present no commonly held view. While it is fascinating to consider the possible origins of retrotransposons from other cellular components, that is not the subject of this chapter, and interested readers are referred to other discussions of this topic (Nakamura and Cech, 1998; Eickbush, 1997; Eickbush and Malik, 2002; Arkhipova et al., 2003; Gladyshev and Arkhipova, 2007).
A consensus structure for each group of retrotransposon is also shown in Figure 1. Within most groups there can be significant variation involving the loss of coding domains, the rearrangement of coding domains, and the structure of terminal repeats. Some of this variation may be artifactual resulting from the recovery of elements from genomic sequencing initiatives. Frequently the structure of the complete (functional) element has not been confirmed and only a consensus structure can be proposed based on the available sequences. The summary structures shown in Figure 1 and the discussions in this report are attempts to emphasize only those characteristic that are shared by multiple elements that have directly been shown to be active (in vivo retrotransposition assays), or are inferred to be recently active (insertions whose locations differ between individuals of a population). When significant differences in structure occur in elements from the same group, two examples are presented to show the range of variation within the group.
3. The major families of retrotransposons
3.1. LTR retrotransposons
Based on the phylogeny of their RT domains (Figure 1) the LTR retrotransposons can be divided into major lineages that are historically referred to as the Ty1/copia group, the Bel group and the Ty3/gypsy group. Ty1 and Ty3 are well-characterized elements from S. cerevisiae, while Copia, Bel and Gypsy are elements from D. melanogaster. These lineages have recently been classified by the International Committee on the Taxonomy of Viruses into two major groups: the Pseudoviridae with three genera, the Pseudoviruses, the Hemiviruses and the Sireviruses (Boeke et al., 2005), and the Metaviridae also with three genera, the Metaviruses, the Errantiviruses and the Semotiviruses (Eickbush et al., 2005). These new classifications are still not in common use today, thus for simplicity the original names will be used throughout this report. The Ty1/copia and Ty3/gypsy groups of elements have extremely broad distributions in animals, plants and fungi, while the Bel class of elements have to date only been reported in animals. The abundance of these elements is usually low in fungi, highly variable in animals, and high in plants. For example the 75% increase in size of the maize genome in the last 5 million years is a result of the proliferation of 11 families of these elements (SanMiguel et al., 1998). On the other hand, LTR retrotransposons represent less than 8% of the human genome, and no elements appear to have been active in this lineage for the past 50 million years (Lander et al., 2001).
Structure and mechanism of retrotransposition
The consensus structures of the elements from each group of LTR retrotransposons are similar to that of retroviruses except for the absence of the env gene in most elements (Figure 1). All LTR retrotransposons contain apparent gag and pol genes that overlap in different reading frames, or be separated by one or more termination codons. There are numerous examples in all lineages however where the gag and pol genes have fused into a single ORF. The gag gene is the most variable but typically encodes major structural and nucleic acid binding domains which may be involved in reverse transcription. The pol gene encodes the various enzymatic domains: the proteinase (PR), RT DNA polymerase domain, RT RNase H domain, and integrase (IN). Functional equivalence of these domains to that of the retroviruses has in most cases only been directly shown with the yeast elements where overexpression and sensitive in vivo retrotransposition assays have been developed to monitor the mutagenesis of donor elements (Boeke, 1989; Sandmeyer et al., 1990; Boeke and Stoye, 1997). The conservation of critical residues in each of these protein domains by the retrotransposons identified in higher animals and plants suggest similar equivalency for all elements. Thus with few exceptions the mechanism of retrotransposition for the LTR retrotransposons is believed to be similar to that of retroviruses.
There are two ways in which the arrangement of protein domains within the pol genes of the LTR retrotransposons has shifted relative to the retroviruses. In the Ty1/copia classes the IN domain is located amino-terminal to the RT DNA polymerase and RNase H domains, while in the Bel and Ty3/gypsy classes the IN domain is in most cases located as in retroviruses at the carboxyl-terminal end of the pol gene. The exception is the Gmr1 elements which based on its RT DNA polymerase sequence is clearly a member of the Ty3/gypsy group but its IN domain is located amino terminal of the RT domain (Goodwin and Poulter, 2002). Based on their RT divergence the Ty1-copia group is the oldest lineage, thus the arrangement with the IN domain at the N-terminal end of the pol gene appears to be ancestral. Sequence similarity of the IN domain is too low to determine if the relocation of the IN domain downstream of the RT domains represents a rearrangement in the order of the pol domains or the addition of a new integrase domain and the loss of the ancestral IN domain (Capy et al., 1998). The other change in the organization of the pol domain involves the RT RNase H domain. In retroviruses the RT DNA polymerase and RNase H domains are separated by a tether (or connection) domain. This tether domain has a three-dimensional structure similar to that of an RT RNase H domain even though it no longer shows significant sequence similarity. Phylogenetic analysis of these retroviral RNase H domains with those of the LTR retrotransposons revealed that the retroviral RNase H sequences are highly divergent from those of all groups of LTR retrotransposons (Malik and Eickbush, 2001). This suggests that the retroviruses have acquired a new RNase H domain downstream of the ancestral domain with the ancestral domain degenerating to become the tether. It has been suggested that the maintenance of the tether domain in retroviruses helps to control the activity of the new RNase H domain (Malik, 2005).
The long-terminal repeats (LTR) of the retrotransposon are functionally similar to those of retroviruses and are involved in the intricate template jumps of the RT from one end of the transcript to the other (Boeke, 1989). Each LTR has a central R region found repeated at both ends of the RNA transcripts, a upstream U3 region found only at the 3′ terminus of the transcript, and a U5 region found only at the 5′ end of the transcript. For most LTR retrotransposons first strand DNA synthesis is primed by the annealing of the 3′ end of a tRNA to a primer binding site near the left LTR, while second strand DNA synthesis is primed from a polypurine tract near the right LTR.
The only known exception to RT priming by tRNA is found in a subgroup of elements within the Ty3/gyspy group. A novel mechanism to prime RT has been identified for Tf1 of Schizosaccharomyces pombe (Levin, 1995; 1996; Lin and Levin, 1997). In Tf1 elements, the first 11 bases of the primary RNA transcript anneal to a sequence downstream of the left LTR at the typical tRNA primer binding site. Cleavage of the looped RNA by the RNase H domain of the Tf1 protein occurs after base 11 enabling the first 11 bases to serve as the primer. Several LTR retrotransposons in fungi and plants that are most related to Tf1 based on their RT sequences also retain this internal complementarity to their 5′ RNA ends suggesting this RT priming mechanism has a long history (Levin, 1997).
Acquisition of env genes
The phylogenetic location of the vertebrate retroviruses well within the various lineages of LTR retrotransposons as shown in Figure 1 strongly suggests that vertebrate retroviruses evolved from the LTR retrotransposons by the acquisition of an env domain. It is becoming increasingly apparent that each of the major lineages of LTR retrotransposons have undergone additional instances in which an env-like gene was acquired downstream of their pol genes. These events occurred in various groups of animals and plants, and in several cases the acquisition was recent enough that the possible origin of the gene can be identified.
The best-studied example of an env-like gene acquisition is in the gypsy element from D. melanogaster. Gypsy has been shown to be able to infect oocytes and evidence is consistent with the env-like gene being responsible for this infection ability (Kim et al., 1994; Song et al., 1994). Multiple other gypsy-like elements (eg. TED, Zam) have been detected in other Drosophila species as well as in other insects. Consistent with their ability to function as viruses, Gypsy-like elements in diverse species are nearly identical in sequence suggesting that transfer (infection) between species has occurred frequently (Heredia et al., 2004). Comparison of the env-like ORF of these gypsy-like elements revealed sequence similarity to a gene encoded by a number of baculoviruses (Malik et al., 2000). This baculoviral gene has been shown to be responsible for the infectious ability of the virus (Kuzio et al., 1999). The N-terminal signal peptide and a C-terminal transmembrane domain of the baculoviral protein are strictly maintained in the gypsy-like elements. Because baculoviruses are double-stranded DNA viruses that infect insects, the transfer of one of its genes to an LTR retrotransposon represents the most likely origin of the gypsy viruses.
The origin of two env-like ORFs can be traced in nematodes for elements in the Bel group (the Semotiviruses). Sequence similarity was found between the env-like genes of certain Caenorhabditis elegans elements (Bowen and McDonald, 1999) and the G2 glycoproteins from Phleoboviruses (Bateman et al., 1999; Malik et al., 2000). The similarity extended throughout the length of the protein, and included proteolytic cleavage sites and a transmembrane domain at the C-terminal end. In the second nematode case, the env-like gene of the TAS element of Ascaris lumbricoides (Felder et al., 1994) was shown to have sequence similarity to the gB glycoproteins of herpesviruses (Malik et al., 2000). The gB protein is an envelope protein suggested to be involved in the attachment and fusion of the virus with the cell membrane (Britt and Mach, 1996).
The most likely example of a Ty1/copia class element acquiring a env-like gene is the SIRE1 element originally identified in the soybean, Glycine max (Laten et al., 1998). Elements related to SIRE1 (the Sireviruses) have been identified in a wide range of plant species, including rice, maize, tomato, lotus and Arabidopsis (Havecker et al., 2005). The likely origin or function of the env-like third ORF has not been identified, but the conservation of its sequence including a transmembrane domain suggests these elements may also be able to leave a cell.
Finally, two additional instances have been suggested where the RT and RH domains of the LTR retrotransposons may have fused with other cellular genes or other viruses to form new types of viruses. The RT DNA polymerase and RNase H domains of caulimoviruses and the hepadnaviruses are most closely related to those of the LTR retrotransposons (Doolittle et al., 1989; Xiong and Eickbush, 1990). Caulimoviruses and the hepadnaviruses, however, differ significantly in structure and mode of replication from that of the LTR retrotransposons (Rothnie et al., 1994) and are only mentioned here as additional examples of how the LTR retrotransposons are likely to have contributed to the evolution of viruses, and vice versa. Indeed, the many examples of env-like gene acquisition by retrotransposons in insects, nematodes and plants suggest the classification between the LTR retrotransposons and viruses is no longer a distinct one.
3.2. Tyrosine recombinase-encoding LTR retrotransposons
As early as 1985 a mobile element was identified in the slime mold, Dictyostelium discoideum, which encoded a RT DNA polymerase domain (Cappello et al., 1985). The element, called DIRS, had a number of properties that differed from LTR retrotransposons and retroviruses. For example it did not encode an integrase domain, and while it had LTRs, they were inverted in orientation and a segment of the LTR sequences was repeated within the element giving rise to the internal complementary repeats (ICR). The authors proposed a model for replication that had many features of the standard retroviral mechanism, with the ICR playing a critical role in the reverse transcription of an RNA transcript into a DNA intermediate. Because many DIRS elements appeared to insert into pre-existing copies of DIRS, a circular DNA intermediate was proposed to recombine into the chromosome. Since this first discovery, retrotransposons with RT domains most similar in sequence to DIRS have continually been identified. A model for the insertion of these elements by recombination rather than by integration was greatly strengthen when it was found that the elements encoded a domain with sequence similarity to tyrosine recombinases (Goodwin and Poulter, 2001). The analysis of the insertion sites of DIRS-like elements pre- and post-insertion was consistent with the recombination of a circular DNA intermediate (Duncan et al., 2002).
Today these tyrosine recombinase encoding LTR retrotransposons (YR retrotransposons) have been discovered in many organisms, including highly primitive organisms such as volvox and trypanosomes (Duncan et al., 2002; Goodwin and Poulter, 2004; Lorenzi et al., 2006). The orientation of the LTRs and of the ICRs varies between the different elements as does the location of the tyrosine recombinase (YR). The RT sequences of these YR-encoding retrotransposons are nearly as divergent as the LTR retrotransposons, with at least three distinct ancient lineages known at present (Lorenzi et al., 2006). Most phylogenetic analyses of the RT domain place the YR retrotransposons within the LTR retrotransposon diversity as shown in Figure 1, suggesting that the original IN domain was replaced with YR. However, the age and phylogenetic position of the YR-encoding elements are unclear. Thus one can not exclude the possibility that the YR elements were the original LTR elements, and their YR domain replaced with an IN domain to form the present day LTR retrotransposons. Many questions remain as to the mechanism of generating a DNA intermediate for the insertion of YR retrotransposons, including the means by which the reverse transcripton is initiated (Goodwin and Poulter, 2004). Biological assays to directly address the mechanism of YR retrotransposon reverse transcription and insertion have not been reported, nor have the individual protein domains been tested in vitro for enzymatic activity.
3.3. non-LTR retrotransposons
This class of retrotransposons is highly abundant in eukaryotes but many copies of these elements in a number of organisms were sequenced before they were recognized as a distinct, autonomous class of retrotransposons. These elements have neither inverted nor tandem terminal repeats, instead ending most frequently with a poly(A) tail at their 3′ ends, while their 5′ ends often contained variable deletions (5′ truncations). The elements were found to encode ORFs, but these ORFs were usually disrupted by mutations. The highly abundant insertions identified in mammals were termed LINEs (long interspersed nucleotide elements), to differentiate them from SINEs (short interspersed nucleotide elements). The insertion of the LINEs appeared to be by reverse transcription in a manner similar to that of processed pseudogenes and SINEs. Thus it was initially proposed that their insertion was catalyzed by the retrotransposition machinery of the LTR retrotransposons or retroviruses (Weiner et al., 1986). The rapid accumulation of more sequences eventually lead to the recovery of elements from different animals and plants with ORFs that encoded intact RT domains. Phylogenetic comparison of these RT sequences with that of all other RT sequences revealed that they represented a distinct class of retrotransposons (Xiong and Eickbush, 1988; Doolittle et al., 1989). The RT domains of several elements were soon shown to encode authentic RT DNA polymerase activity (Ivanov et al. 1991; Gabriel and Boeke, 1991; Mathias et al., 1991) Because of this unusual history, these elements have been referred to by a variety of names including the poly(A) retrotransposons, the nonviral retroposons, or simply retroposons. Generally today these elements are called either the LINE-like elements, to emphasize their similarity to the highly abundant sequences in mammals, or as used here the non-LTR retrotransposons to emphasize their different structure and mechanism of retrotransposition from that of the LTR elements. The non-LTR integration machinery is thought to be the mechanism used for the insertion of SINEs and processed pseudogenes (Esnault et al., 2000; Ostertag and Kazazian, 2001; Kajikawa and Okada, 2002; Dewannieux et al., 2003). SINE elements, also referred to as non-autonomous retrotransposons or retroposons, can represent a large fraction of eukaryotic genomes. For example, there are over 1.4 million such insertions in humans representing 13% of our genome (Lander et al. 2001).
There are a variety of distinct lineages of non-LTR retrotransposons (Malik et al., 1999) and new lineages of elements continue to be identified. These lineages have somewhat different coding capacities but generally there appear to be two major structures for the non-LTR elements (see Figure 1). The first class encodes a single ORF with a centrally located RT domain. The most extensively studied members of this class are the R2 elements (Eickbush, 2002). The N-terminal domain of various ORFs show little similarity to each other or to known proteins except that some elements appear to encode DNA-binding motifs (Christensen et al., 2006). C-terminal to the RT domain is another conserved domain that appears to be the endonuclease for the element (EN). Not much is known about this EN domain except that it has conserved residues that are similar to the active sites of various type II and type IIs restriction enzymes. In only one instance has mutagenesis of this restriction-like domain directly demonstrated the role of this domain in the cleavage of the target site (Yang et al., 1999). The key residues of this C-terminal endonuclease domain are conserved in many other lineages of non-LTR elements suggesting that this domain functions as the endonuclease in many lineages. An unusual feature shared by many of the non-LTR elements with this C-terminal endonuclease domain is that they insert in a sequence-specific manner into highly conserved host genes such as the rRNA genes of various animals, or the leader exons of nematodes (Eickbush, 2002).
The second major class of non-LTR retrotransposons usually encodes two ORFs. The most extensively studied members of this group are the L1 elements of mammals (Moran and Gilbert, 2002). The first ORF may have functional similarity to the gag gene of retroviruses since conserved zinc-finger domains are found in many lineages, and the protein has been shown to bind RNA (Martin and Bushman, 2001). The second ORF encodes the RT domain as well as an endonuclease domain at the N-terminal end. This endonuclease has been termed APE because of sequence similarity to apurinic-apyrimidinic endonucleases involved in DNA repair (Martin et al., 1995). The APE domain from a number of different non-LTR elements has been separately expressed and shown to be directly involved in recognition and cleavage of the target site of the element (Olivares et al., 1997; Cost and Boeke, 1998; Christensen et al., 2000; Anzai et al., 2001). The phylogeny of the APE domain agrees with the phylogeny of the RT domain suggesting that a single acquisition of the former by a non-LTR retrotransposon has given rise to the many lineages that now encode this domain. There is considerable flexibility in other coding features of this class of non-LTR elements. Some lineages contain a C-terminal domain of unknown function, and a few lineages encode an RNase H domain downstream of the RT domain. The structural variability found in the two major classes of the non-LTR retrotransposons means the only feature held in common by all elements is an RT domain and either a downstream EN or an upstream APE domain (Eickbush and Malik, 2002).
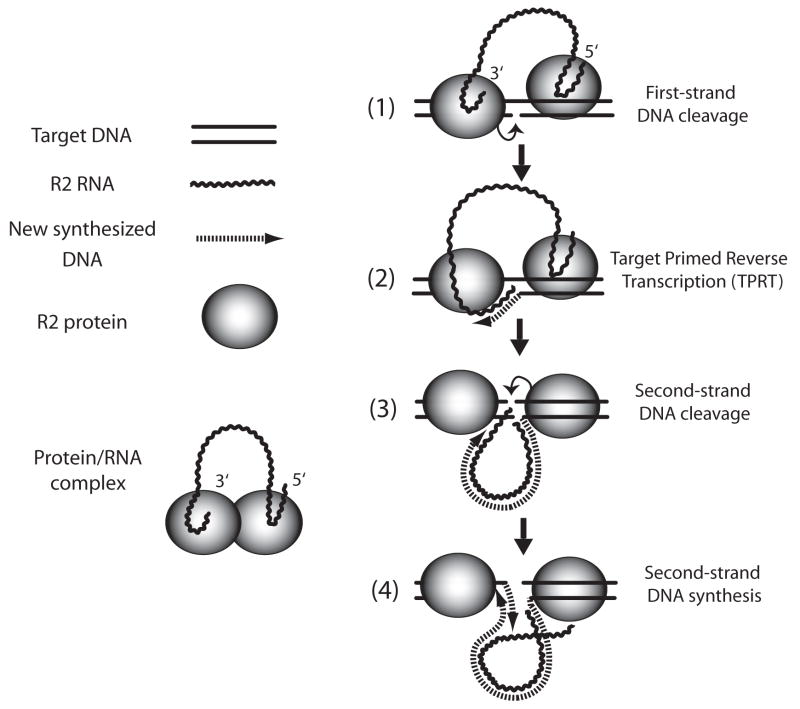
The mechanism of retrotransposition for the non-LTR retrotransposons has only been determined in detail for the R2 element. R2 elements contain a C-terminal EN domain and insert in a sequence specific manner in the 28S rRNA genes in at least five animal phyla. The phylogeny of R2 elements from these species suggests that they have been inserting in this location for most of the evolution of animals (Burke et al., 1998; Kojima et al. 2006). The single ORF of an R2 element was expressed in E. coli, purified and shown to be able to conduct most of the steps of a complete retrotransposition reaction (Luan et al., 1993). The current model for this reaction is diagramed in Figure 2.
Figure 2.
Model for non-LTR retrotransposition based solely on studies of the R2 element. The single ORF of R2 is translated into one protein which contains both RT and endonuclease domains (see Figure 1, top non-LTR retrotransposon structure). R2 protein subunits bind either the 3′ or 5′ end of the R2 transcript to make the RNP complex used for retrotransposition. The RNA end bound determines whether the subunit binds upstream or downstream of the insertion site on the target DNA. The protein subunit bound upstream of the insertion site cleaves the lower DNA strand and use the released 3′ end to prime reverse transcription of the RNA (steps 1 and 2). The protein subunit bound downstream of the insertion site cleaves the upper DNA strand and uses the released 3′ end to prime second-strand DNA synthesis. RNA still bound to the first DNA strand is displaced during this synthesis. For further details of the model see Luan et al., 1993; Christensen et al., 2006; Kurzynska-Kokorniak et al., 2007. While the precise details of protein binding to the RNA transcript and the DNA target site may only be relevant for the R2 elements, the general steps of the reaction, including the use of the first and second strand cleavages to prime the two DNA strands is likely to be common for many non-LTR elements (see review by Ostertag and Kazazian, 2001).
A key feature of the R2 retrotransposition reaction is the ability of the R2 protein to bind RNA sequences near the 5′ and 3′ ends of a full-length R2 transcript (Christensen et al., 2006). When the R2 protein binds the 3′ end of the R2 transcript, it adopts a conformation that binds the 28S gene DNA a short distance upstream of the insertion site. Alternatively, when the R2 protein binds the 5′ end of the R2 transcript, it adopts a conformation that promotes binding of the 28S gene a short distance downstream of the insertion site. The stoichiometry of the reaction suggests a single subunit is involved in either upstream or downstream binding. Binding of the R2 protein to DNA sequences downstream of the insertion site is brought about by DNA-binding motifs at the N-terminal end of the R2 ORF (Christensen et al., 2005). The region of the R2 protein responsible for upstream DNA binding has not been determined.
Current models for a complete R2 retrotransposition reaction involves symmetric reactions first by the upstream and subsequently by the downstream bound subunits. The subunit bound upstream initiates the retrotransposition reaction by cleaving the bottom (first) strand of the DNA target and using the 3′ OH released by this cleavage to prime the reverse transcription reaction. This use of the target site to prime reverse transcription has been termed target-primed reverse transcription (TPRT). When the 5′ RNA sequences are “pulled” from the R2 subunit bound downstream of the insertion site, this subunit initiates the second half of the reaction which involves cleavage of the top (second) DNA strand and again the utilization of the released 3′ end of the DNA to prime second-strand DNA synthesis. As describe below (Sec. 5.1) the R2 protein does not have RNase H activity, thus the R2 protein must displace the annealed RNA as it uses the first DNA strand as template.
This model for R2 retrotransposition can explain two common features of non-LTR retrotransposon insertions. First many of the inserted copies have precise 3′ ends but are variably truncated at their 5′ ends. In the R2 model, if the RNA template is cleaved by cellular RNases or if the RT dissociates before reaching the 5′ end of the complete transcript, then a 5′ truncated copy is likely to arise. Second, unlike retrotransposons or DNA transposons which typically generate a short (<8 bp) target site duplication of defined length, many non-LTR retrotransposons generate variable length target site duplications, and in some cases even deletions of the target site. The separate cleavages of the top and bottom strands of the target site in the R2 model can explain this variability. Cleavage of the top strand downstream of bottom strand site generates target site duplications, while cleavage upstream of the bottom strand site generates a deletion. For those elements with variable duplications, the location of the top strand cleavage may be variable, or the priming of second strand synthesis may involve micro-complementarities with the target site (Ostertag and Kazazian, 2001).
L1 elements are highly abundant non-LTR retrotransposons in mammals with an estimated 800,000 copies representing 17% of the human genome (Lander et al. 2001). L1 elements are representative of the second group of non-LTR elements with two ORFs and a N-terminal AP endonuclease (Ostertag and Kazazian, 2001). The large ORF of the human L1 element has been expressed and directly shown to have RT activity (Mathias et al., 1991). More recently purified protein encoded by L1 was shown to be able to conduct the TPRT reaction by initiating reverse transcription from pre-existing nicks on the DNA target, or from nicks generated by the AP domain (Cost et al., 2002). The protein was also shown capable of synthesizing the second DNA strand primed from the DNA target. Additional studies of L1 integration have been made possible by the development of a powerful in vitro assay to monitor retrotransposition in tissue culture cells (Moran et al., 1996). In this assay, the L1 retrotransposition machinery was shown not to recognize the sequences at the 3′ end of the RNA transcript, requiring only a poly(A) tailed transcript. This lack of specificity would seem to give rise to a highly inefficient process because any polyadenlyated RNA transcript would become a substrate for integration. However it was shown that the L1 machinery predominately uses the RNA transcript from which the L1 proteins were synthesized, greatly increasing the likelihood of reverse transcribing functional L1 transcripts (Wei et al. 2001). This cis-preference is not absolute because both SINEs (e.g. human Alu sequence) and processed pseudogenes have been shown to insert using the L1 machinery (Esnault et al., 2000; Dewannieux et al., 2003).
In vivo retrotransposition assays have also been developed for several other non-LTR retrotransposons with AP domains. These assays include the TRAS, SART and R1 elements of the silkmoth, Bombyx mori (Feng et al., 1998; Anzai et al., 2005; Takahashi and Fujiwara, 2002), the I element of D. melanogaster (Chaboissier et al., 2000), and the UnaL2 element of an eel, Anguilla japonica (Kajikawa and Okada, 2002). In these cases the retrotransposition machinery does seem to recognize the 3′ untranslated region of the element transcript. In the eel system, several SINE elements appear to have taken advantage of this sequence recognition by having short regions near the 3′ end of their transcripts that mimic this recognition sequence (Kajikawa and Okada, 2002). Interestingly, consistent with the R2 model of integration, there is evidence to suggest that in the I element system the non-LTR retrotransposition machinery also recognizes the 5′ end of the RNA transcript (Chambeyron et al., 2002).
3.4. Penelope-like retrotransposons
The last group of retrotransposons to be identified was discovered only recently in Drosophila virilis (Evgen’ev et al., 1997). This element, named Penelope, could actively insert in the genome of the host but its sequence revealed ORFs that had little sequence similarity to other protein domains. When additional Penelope-like elements were found in other animals, fungi and plants, it was possible to recognize an RT domain that was highly divergent from any previously defined sequence. A second domain was also identified with sequence similarity to the Uri (or GIY-YIG) endonucleases of bacterial mobile group I introns, as well as UvrC bacterial DNA-repair endonucleases (Lyozin et al., 2001). The Penelope-like elements (PLE) have the most diverse structures of any class of retrotransposon. Some elements contain apparent LTRs that may be in either direct or inverted orientations, some contain a first ORF, and some lack the Uri domain (see Figure 1). Remarkably many of the elements retain introns, which is unexpected for an element that makes additional copies of itself by reverse transcription. Given the significant divergence of the RT domains, it was important that the ORF of the original Penelope element was expressed, purified and shown to exhibit authentic RT activity (Pyatkov et al., 2004).
The phylogenetic relationship of the PLE retrotransposons based on the RT domain clearly placed it as the most divergent branch of retrotransposon sequences, grouping them in some cases with the telomerases (Arkhipova et al., 2003). The possible phylogenetic relationship with telomerase is particularly intriguing because lineages of PLE retrotransposons have been identified in bdelloid rotifers, fungi and plants that do not encode the Uri domain (Gladyshev and Archipova, 2007). These elements are found near or at the telomeres of the host organisms in an orientation consistent with the utilization of a free chromosomal end to prime reverse transcription. While the mechanism of integration is not established for PLE elements, their frequent 5′ truncations, variable length target site duplications, and the possibility that some elements use the end of a chromosome to prime reverse transcription, all suggest that these elements utilize a TPRT-like mechanism of retrotransposition.
4. Properties of the reverse transcriptases from LTR retrotransposons
Most studies of the RTs from LTR retrotransposons have involved direct comparisons of their activities to that of retroviral RTs. To date all studies have been conducted with elements from S. cerevisiae and S. pombe. Even though these elements encode proteins that are highly divergent in sequence from the retroviral enzymes, they exhibit remarkably similar properties.
4.1 Saccharomyces cerevisiae Ty1
Studies with retroviral RTs have suggested that interactions between the RT and IN domains play an important role in the reverse transcription reaction. For instance, in avian leucosis virus (ALV) an α/β heterodimer composed of a smaller RT (α) and an incompletely processed RT-IN (β) intermediate is the active form of the reverse transcriptase, while in Human T-cell Leukemia Virus Type-1 (HTLV-1) the active form is an α3/β oligomer (Trentin et al., 1998). In other retroviruses (e.g. MLV, HIV-1), the RT and IN are separated during virion maturation, but mutations or deletions of IN affect the initiation of reverse transcription and the level of cDNA produced (Lai et al., 2001; Padow et al., 2003).
As with retroviruses, interactions between IN and RT are important for the function of yeast Ty1 RT. Purified recombinant Ty1 RT exhibited polymerase activity only when a 115 amino acid C-terminal fragment of the Ty1 integrase was fused to the N-terminus of the RT domain (Wilhelm et al., 2000). Subsequent successive deletion of the IN domain revealed a small acidic tail fused to RT could mimic the IN and give rise to an active recombinant RT (Wilhelm and Wilhelm, 2005). Further studies showed that IN acts in cis to activate RT during reverse transcription and remains associated with RT during the formation of the preintegrative complex (Wilhelm and Wilhelm, 2006). Thus the important interactions between the IN and RT domains of retroviruses are also found in Ty1, even though the IN domain in Ty1 is encoded N-terminal to the RT domain (see Figure 1).
The fidelity of Ty1 reverse transcription has been determined both in vivo and in vitro. The in vivo study involved the complete sequencing of new Ty1 insertions in the S. cerevisiae genome after a single cycle of retrotransposition (Gabriel et al., 1996). All observed changes were base substitutions with the template ends representing hot spots for mutations. The observed mutation rate of 2.5 × 10−5 bp per cycle suggested that Ty1 mutated as rapidly as retroviruses. The in vitro study involved steady state kinetics of misinsertion opposite A, T, G and C residues at defined primer-template sites (Boutabout et al., 2001). Ty1 RT was found to be less error prone than lentiviral RTs such as HIV-1, HIV-2 and EIAV and comparable to that of oncoretroviral RTs such as AMV. The X residue of the highly conserved YXDD motif within the active site of retroviral reverse transcriptases is known to be important for RT fidelity with the low fidelity lentiviral RTs containing a methionine at the X position and the high fidelity oncoretroviral RTs containing a valine at the X position (Kaushik et al., 2000; Poch et al., 1989). Consistent with its greater fidelity, Ty1 RT encodes a valine at position X.
Sequence comparisons of all RTs revealed a triad of conserved aspartic acid residues (Doolittle et al., 1989; Poch et al., 1989; Xiong and Eickbush, 1990). Two of these aspartic acids are part of the just described highly conserved YXDD motif, while the other aspartic acid is found about 75 to 100 amino acids N-terminal to this motif. Mutational studies of these three residues in HIV-1 RT resulted in the loss of both in vitro RT activity and in vivo infectivity (Kaushik et al., 1996). Mutational studies of Ty1 RT showed that while D to N mutations at the first two aspartic acid positions eliminated both in vitro and in vivo activity, D to N mutation of the second D in the YXDD motif allowed in vitro polymerization although it prevented in vivo retrotransposition (Uzun and Gabriel, 2001). More recent pre-steady state kinetic studies showed that this D to N mutation in Ty1 RT had similar dNTP binding affinities (Kd) with that of wild type Ty1 RT but over a 200-fold reduced rate of chemical catalysis (kpol) (Pandey et al., 2004). This slower polymerization rate would have a large cumulative effect during synthesis of a complete Ty1 DNA intermediate and thus can explain the loss of in vivo retrotransposition (Pandey et al., 2004).
4.2 Saccharomyces cerevisiae Ty3
Ty3 virus-like particles (VLPs) of S. cerevisiae contain a 115-kDa RT-IN fusion protein as well as the processed 55-kDa RT and 61-kDa IN proteins (Hansen and Sandmeyer, 1990; Kirchner and Sandmeyer, 1993). The major replication competent form of Ty3 RT appears to be an α/β heterodimer similar to that of ALV (Nymark-McMahon et al., 2002). As in retroviruses and Ty1, reverse transcription in Ty3 is disrupted by IN mutations suggesting that the involvement of IN in the stability of or catalysis by the RT domain is a general property shared by retroviruses and LTR retrotransposons (Nymark-McMahon et al., 2002).
Mutagenesis studies have also been done on the aspartic acid residues in the catalytic aspartic acid triad of the Ty3 RT active site. These studies revealed that like Ty1 RT, the second aspartic acid residue in the YLDD motif of Ty3 RT is not essential for in vitro catalysis by the 55-kDa RT but is required for in vivo retrotransposition (Bibillo et al., 2005a). Thus both Ty3 and Ty1 RTs appear to have more relaxed structural constraints with respect to the catalytic aspartic acid triad compared to the precise geometry required of the HIV RT.
The thumb subdomain of DNA polymerases makes contact with the duplex product of DNA synthesis 3–8 bp behind the catalytic site (Steitz and Yin, 2004). Studies done on HIV-1 RT revealed that a helix-turn-helix motif of the thumb serves as an important modulator of both processivity and fidelity (Latham et al., 2000; Powell et al., 1999). Secondary structure predictions and amino acid sequence alignments were used to identify the putative thumb subdomain of Ty3 RT that is equivalent to the HIV-1 RT subdomain αH (Bibillo et al., 2005b). Biochemical studies of the 55-kDa RT conducted with locked nucleic acid analogs and abasic lesions in either template or primer revealed interactions of the Ty3 thumb subdomain with primer nucleotides -3 and -4 and with template nucleotide -6, suggesting a structure similar to that of HIV RT and DNA polymerases in general. Interestingly, mutations in the Ty3 thumb subdomain also affected RNase H activity, an interaction that was not observed for HIV-1 RT (Bibillo et al., 2005b). This finding is consistent with the separation of the RT DNA polymerase and RNase H domains by the tether domain in retroviruses but not in LTR retrotransposons (see section 3.1).
Finally, the nucleocapsid proteins (NCps) are small basic proteins encoded by retroviruses which are required for virion structure and replication (Thomas and Gorelick, 2008). Specific NC proteins, such as NCp7 of HIV-1, have an important chaperone function during reverse transcription in directing specific tRNA-primed cDNA synthesis (Lapadat-Tapolsky et al. 1997). While the role of NC proteins is not established for most LTR retrotransposons (e.g. Ty1 and Tf1), Ty3 has been shown to encode a nucleocapsid protein, NCp9, which is also important in transposition (Orlinsky and Sandmeyer, 1994; Gabus et al., 1998; Cristofari et al., 1999). Ty3 NCp9 was shown to form nucleoprotein complexes between primer tRNAiMet and Ty3 RNA which in turn induced high levels of cDNA synthesis by Ty3 RT. Thus Ty3 NCp9 chaperones cDNA synthesis and appears functionally equivalent to HIV-1 NCp7.
4.3 Schizosaccharomyces pombe Tf1
Tf1 of S. pombe also belongs to the Ty3/gypsy group of LTR retrotransposons. This element utilizes an unusual mechanism of self-primed reverse transcription as described in Section 3.1 (Levin, 1997). Another unusual property of Tf1 retrotransposition was revealed when studies of isolated VLPs revealed 85% of the cDNAs had 1, 2 or 3 non-templated nucleotides at their 3′ ends (Atwood-Moore et al., 2005; 2006). Retroviruses and other LTR retrotransposons are also known to have cDNA species with non-templated additions but these are usually limited to one nucleotide. Studies of the biochemical properties of Tf1 RT have confirmed these additions are a direct result of the Tf1 RT terminal transferase activity (Kirshenboim et al., 2007). Expression of the RT and RH domains of Tf1 gave rise to a 56-kDa protein possessing typical DNA- and RNA-dependent DNA polymerase activity as well as RNase H activity. Tf1 showed higher terminal transferase activity than HIV-1 in some conditions. It was suggested that the higher terminal transferase activity of Tf1 RT compared to other RTs is because in S. pombe the non-templated extra nucleotides are needed to protect the ends of the cDNA from degradation by non-specific cellular 3′ exonucleases.
5. Properties of the reverse transcriptases from non-LTR retrotransposons
5.1 Bombyx mori R2
Biochemical studies of R2 RT have all been conducted with the entire ORF of the element from Bombyx mori expressed and purified from E. coli (Luan et al., 1993). The purified protein is 120-kDa in size and was found to have the RNA and DNA binding properties and enzymatic activities that gave rise to the retrotransposition mechanism shown in Figure 2. During these studies of R2 retrotransposition, R2 RT was shown to have a number of unusual properties that differentiate it from the RTs encoded by LTR retrotransposons and retroviruses.
During target-primed reverse transcription (TPRT), R2 RT uses the 3′ end of DNA generated by the first strand cleavage of the target site to prime reverse transcription of R2 RNA. No sequence complementarity between the template RNA and the DNA target is needed for this priming (Luan and Eickbush, 1996). The most efficiently used RNA templates are those that end at the precise 3′ end of the element. If the RNA template extends beyond the end of the R2 sequences, reverse transcription still initiates at the first nucleotide of the R2 sequence. With RNA templates that contain short deletions of R2 sequences at their 3′ end, R2 RT adds non-templated nucleotides to the target DNA until the extended DNA is of sufficient length to enable it to prime reverse transcription of the RNA template (Luan and Eickbush, 1995). Remarkably, the non-templated nucleotides added to the target DNA are usually T nucleotides. The addition of Ts to initiate first strand synthesis generates, after a complete integration reaction, a short A-rich stretch on the mRNA synonymous strand. Short A-rich 3′ ends are a common property of non-LTR retrotransposons.
The ability of R2-RT to use the 3′ end of DNA to prime reverse transcription is not limited to the cleaved target site. In the absence of its target site, R2 RT can synthesize cDNA efficiently using either the 3′ end of any RNA or the 3′ end of any DNA as the primer. This priming again occurs in the absence of any complementarity between the template and primer (Bibillo and Eickbush, 2002b).
Processivity studies have also been conducted with R2-RT on both RNA and DNA templates. Processivity refers to the length of the DNA product that can be catalyzed by the enzyme before it dissociates from the template. In single cycle reactions, R2 RT synthesized cDNA over twice the length of that synthesized by AMV RT on complex templates and over four times the length of AMV RT on poly(rA) templates (Bibillo and Eickbush, 2002a). The rate of polymerization of R2 RT on RNA templates was approximately similar to that of AMV RTs. The processivity of R2 RT on DNA templates was about 3-fold higher than that on RNA templates. The processivity of R2 RT on these DNA templates was again about twice that of AMV RT (Kurzynska-Kokorniak et al. 2007). The difference in processivity between the RTs may reflect where in the cell the polymerizations occur. First and second strand DNA synthesis by retroviral or LTR RTs occurs within or associated with a VLP within the cell. When these RTs dissociate from the template the reaction simply stalls until they reassociate, therefore multiple rounds of dissociation and reassociation occur in each cycle. DNA synthesis by R2 RT occurs in the nucleus directly at the target site (Figure 2). If R2 RT dissociates from the RNA template, reassociation is less likely, because DNA repair may take over or second strand DNA synthesis could initiate, resulting in a 5′ truncated copy. Thus there should be strong selective pressure on non-LTR retrotransposons to evolve RTs with high processivity.
Another unusual ability of R2 RT is that it can jump from the 5′ end of one RNA template to the 3′ end of another RNA template. This end-to-end template jumping can generate continuous cDNA products from two or more templates and occurs in the absence of sequence identity between the templates (Bibillo and Eickbush, 2002b; 2004). This activity is related to the ability of R2 RT to use the free 3′ end of any RNA or DNA to prime polymerization. However, end-to-end template jumping is more efficient and is brought about by R2 RT’s ability to add non-templated nucleotides to the cDNA when it reaches the end of the template. The terminal transferase activity of R2 RT can add up to 5 nucleotides, thus is even higher than that observed with Tf1 (Kirshenboim et al., 2007). In these terminal transferase reactions R2 RT preferentially adds purines, rather than the non-templated T’s seen in a TPRT reaction. This activity The overhanging nucleotides generated by R2 RT anneal to the sequences at the 3′ end of the acceptor template promoting higher frequencies of the template jumps (Bibillo and Eickbush, 2004). End-to-end template jumps are similar to the template jumps that have been seen for viral RNA directed RNA polymerases (Arnold and Cameron, 1999). However, they differ from the template switching reaction associated with retroviral cDNA synthesis because they do not require initial sequence identity between the donor and acceptor RNA templates (Peliska and Benkovic, 1992).
The high processivity of R2 RT on both RNA and single-stranded DNA templates is consistent with the ability of this enzyme to make both DNA strands during a retrotransposition reaction. However, the R2 ORF has no RNase H domain (Malik et al., 1999), and no RNase H activity has been detected in vitro (Luan et al. 1993; Kurzynska-Kokorniak et al. 2007). Thus the template for second strand DNA synthesis is an RNA:DNA duplex (see Figure 2). How is the RNA removed from the first strand of synthesized DNA to allow second strand synthesis? We have shown that R2 RT can efficiently displace an annealed RNA or DNA strand as it uses an RNA or DNA strand as template (Bibillo and Eickbush, 2002a; Kurzynska-Kokorniak et al. 2007). Indeed, the processivity of R2 RT on DNA templates is not significantly reduced by the presence of an annealed RNA or DNA strand. Retroviral RT, on the other hand, shows limited ability to displace RNA annealed to DNA and greatly reduced processivity when displacing DNA from a DNA strand. We have postulated that the more extensive finger subdomains of non-LTR retrotransposon RTs enable additional binding of the RT to the template upstream of the active site, permitting more extensive displacement synthesis. Because most non-LTR retrotransposons do not have RNase H domains, it seems likely that this displacement ability might be a common property of their RTs.
Finally, recent studies conducted with R2 RT have shown that this polymerase has a relatively low fidelity, comparable with that of HIV-1 RT (Jamburuthugoda and Eickbush, unpublished data). The low fidelity was found in assays that monitored either misincorporation or mismatch extension. The proficiency at which R2 RT could extend mismatched base pairs could be related to its ability to use the 3′ end of any nucleic acid to prime reverse transcription in the absence of sequence homology. Interestingly, consistent with the low fidelity of R2 RT, this enzyme has an alanine in position X in the conserved YXDD motif. As described above, the amino acid in this position has a crucial affect on the fidelity of RTs from retroviruses. Replacement of the hydrophobic residue at this position in retroviral RT with an alanine gave rise to a 4–8 fold reduction in fidelity (Kaushik et al., 2000).
5.2 Human L1
Studies conducted with the human L1 element were among the first to express the protein of a non-LTR retrotransposon and show that it had authentic reverse transcriptase activity (Mathias et al. 1991). Unfortunately, the L1 protein has proven very difficult to express in an active form that would enable more detailed studies. The entire ORF containing the RT has been expressed in a baculoviral system and several of the critical steps of retrotransposition have been documented (Cost et al., 2002). The L1 RT can utilize the 3′ hydroxyl of nicks generated in DNA by its APE domain in a TPRT reaction (Figure 2). L1 RT can also use pre-existing nicks to initiate reverse transcription on double-stranded DNA ends with either 5′ or 3′ overhangs. As in the case of R2, the junctions between the target DNA and the L1 sequences often contained non-templated residues. Finally, products were generated that were primed by the DNA target that appeared to correspond to second strand DNA synthesis. The efficiency of these reactions were extremely low, however, in that PCR amplification was needed to monitor the DNA products.
In a more recent study, the human L1 RT ORF was expressed in a manner that enabled greater activity (Piskareva et al., 2003; Piskareva and Schmatchenko, 2006). Primer extensions by L1 RT directly demonstrated both DNA- and RNA-directed DNA polymerase activities. Again, as with R2 RT the processivity of L1 RT was found to be significantly higher than that of MLV RT on RNA templates. Finally, RNP complexes containing L1 RT have also been isolated from mammalian tissue culture cells (Kulpa and Moran, 2006). The addition of DNA primers that could anneal to the 3′ ends of L1 transcripts gave rise to reverse transcription indicating that the complexes were active. Interestingly, many of the products had been extended from primer-template complexes that contained terminally mismatched bases. These combined studies suggest that many of the basic enzymatic properties of R2 RT are also characteristic of L1 RT.
5.3 Trypanosome cruzi L1
Studies of this non-LTR retrotransposons L1Tc were the first to identify the APE domain (Martin et al., 1995) and the enzymatic activities of this endonuclease were among the first to be characterized in vitro (Olivares et al. 1997; 1999). L1Tc represents a distinct lineage of non-LTR retrotransposons that is somewhat unusual in that the element does not contain a first gag-like ORF typical of most non-LTR elements with APE domains. However the C-terminal region of L1Tc’s single ORF has been shown to contain nucleic acid chaperone activity similar to the L1 ORF1 (Heras et al. 2005). This chaperone activity has been suggested to be involved in the TPRT reaction because it can promote the annealing of complimentary oligonucleotides and can facility strand exchange between DNAs to form the most stable duplexes. L1Tc is among the small fraction of non-LTR retrotransposons that contain RNase H domains, and represents the only element to date where this activity has been characterized in vitro (Olivares et al. 2002). The RNase H activity could be monitored as a separate 25-kDa protein, and showed many similarities to that of retroviral and E.coli RNase H domains. Finally the protein encoded by L1Tc was shown to have both RNA-directed and DNA-directed DNA polymerase activity (Garcia-Perez et al. 2003). Interestingly this RT activity has the ability to jump between oligonucleotide templates in a manner reminiscent of the R2 element (section 5.1). Template jumping was extremely efficient when the oligonucleotides complementary sequences of their terminal 2 nts. However, L1Tc does not appear to have the extensive terminal transferase activity of R2 to allow template jumps in the absence of short terminal complementarity between templates (Garcia-Perez et al. 2003).
6. Concluding remarks
It is likely that we have only scratched the surface in documenting the variety and the distribution of retrotransposons within eukaryotic genomes. New elements belonging to each of the four groups of retrotransposons as well as completely new groups of elements will no doubt be discovered in the massive amounts of repetitive DNA present in most genomes. These elements have played a major role in determining the size and composition of eukaryotic genomes, and are responsible for much of their instability. Yet we still know surprisingly little about the nature of the proteins these elements encode and their mechanism of insertion. At present the Penelope-like elements are especially interesting since the analysis of their abundance in eukaryotes has only started, and their different structures suggest a variety of retrotransposition mechanisms. Continued characterization of the enzymatic activity of the RTs associated with all retrotransposons will also be important as it will help us to understand in general how polymerases function. For example, the RTs of non-LTR retrotransposons and Penelope-like elements have a variety of activities that resemble the enzymatic activity of telomerase. Indeed, retrotransposons of both classes may even serve as the telomeres in some species. The era of “genomics” is indeed an exciting time for the retrotransposon field.
Acknowledgments
We are very appreciative of the insightful comments made by Danna Eickbush on various drafts of this manuscript, as well as the suggestions of several reviewers. Our work on R2 specifically and of retrotransposon evolution in general has been with support from the National Institutes of Health (GM42790) and from the National Science Foundation (MCB0544071).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anzai T, Takahashi H, Fujiwara H. Sequence specific recognition and cleavage of telomeric repeats by the endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol Cell Biol. 2001;21:100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai T, Osanai M, Hamada M, Fujiwara H. Functional roles of 3′-terminal structures of template RNA during in vivo retrotransposition on non-LTR retrotransposon, R1Bm. Nucleic Acids Res. 2005;33:1993–2002. doi: 10.1093/nar/gki347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J Biol Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- Atwood-Moore A, Ejebe K, Levin HL. Specific recognition and cleavage of the plus-strand primer by reverse transcriptase. J Virol. 2005;79:14863–75. doi: 10.1128/JVI.79.23.14863-14875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood-Moore A, Yan K, Judson RL, Levin HL. The self primer of the long terminal repeat retrotransposon Tf1 is not removed during reverse transcription. J Virol. 2006;80:8267–70. doi: 10.1128/JVI.01915-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J Biol Chem. 2002a;277:34836–34845. doi: 10.1074/jbc.M204345200. [DOI] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J Mol Biol. 2002b;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- Bibillo A, Lener D, Klarmann GJ, Le Grice SF. Functional roles of carboxylate residues comprising the DNA polymerase active site triad of Ty3 reverse transcriptase. Nucleic Acids Res. 2005a;33:171–181. doi: 10.1093/nar/gki150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibillo A, Lener D, Tewari A, Le Grice SF. Interaction of the Ty3 reverse transcriptase thumb subdomain with template-primer. J Biol Chem. 2005b;280:30282–30290. doi: 10.1074/jbc.M502457200. [DOI] [PubMed] [Google Scholar]
- Boeke JD. Transposable elements in Saccharomyces cerevisiae. In: Berg DE, Howe MM, editors. Mobile DNA. American Society for Microbiology; Washington D.C.: 1989. pp. 335–374. [Google Scholar]
- Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF. Pseudoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press; London: 2005. pp. 397–407. [Google Scholar]
- Boeke JD, Garfinkel CA, Styles A, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1997. pp. 343–435. [PubMed] [Google Scholar]
- Burke WD, Malik HS, Lathe WC, III, Eickbush TH. Are retrotransposons long-term hitchhikers. Nature. 1998;392:141–142. doi: 10.1038/32330. [DOI] [PubMed] [Google Scholar]
- Boutabout M, Wilhelm M, Wilhelm FX. DNA synthesis fidelity by the reverse transcriptase of the yeast retrotransposon Ty1. Nucleic Acids Res. 2001;29:2217–2222. doi: 10.1093/nar/29.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, McDonald JF. Genomic analysis of Caenorhabditis elegans reveals ancient families of retroviral-like elements. Genome Res. 1999;9:924–935. doi: 10.1101/gr.9.10.924. [DOI] [PubMed] [Google Scholar]
- Britt WJ, Mach M. Human cytomegalovirus glycoproteins. Intervirology. 1996;39:401–412. doi: 10.1159/000150510. [DOI] [PubMed] [Google Scholar]
- Cappello J, Handelsman K, Lodish H. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 1985;43:105–115. doi: 10.1016/0092-8674(85)90016-9. [DOI] [PubMed] [Google Scholar]
- Capy P, Basin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Springer-Verlag; New York: 1998. [Google Scholar]
- Chaboissier MC, Finnegan D, Bucheton A. Retrotransposition of the I factor, a non-long terminal repeat retrotransposon of Drosophila, generates tandem repeats at the 3′ end. Nucleic Acids Res. 2000;28:2467–2472. doi: 10.1093/nar/28.13.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bucheton A, Busseau I. Tandem UAA repeats at the 3′-end of the transcript are essential for the precise initiation of reverse transcription of the I factor in Drosophila melanogaster. J Biol Chem. 2002;277:17877–17882. doi: 10.1074/jbc.M200996200. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Christensen S, Pont-Kingdom G, Carroll D. Target specificity of the endonuclease from Xenopus laevis non-long terminal repeat retrotransposons, Tx1L. Mol Cell Biol. 2000;20:1219–1226. doi: 10.1128/mcb.20.4.1219-1226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SM, Bibillo A, Eickbush TH. Role of the Bombyx mori R2 element N-terminal domain in the target-primed reverse transcription (TPRT) reaction. Nucleic Acids Res. 2005;33:6461–6468. doi: 10.1093/nar/gki957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SM, Ye J, Eickbush TH. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci USA. 2006;103:17602–17607. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J, Farabaugh P. Nucleotide sequence ofa yeast Ty element: evidence for an unusal mechanism of gene expression. Proc Natl Acad Sci USA. 1985;82:2829–2933. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD. Targeting of human retrotransposons integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Gabus C, Ficheux D, Bona M, Le Grice SF, Darlix JL. Characterization of active reverse transcriptase and nucleoprotein complexes of the yeast retrotransposon Ty3 in vitro. J Biol Chem. 1999;274:36643–36648. doi: 10.1074/jbc.274.51.36643. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Doolittle RF, Feng DF, Johnson MS, McClure MA. Origins and evolutionary relationships of retroviruses. Quart Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Duncan L, Bouckaert K, Yeh F, Kirk DL. kangaroo, a mobile element from Volvox carteri, is a member of a newly recognized third class of retrotransposons. Genetics. 2002;162:1617–1630. doi: 10.1093/genetics/162.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH. Origin and evolutionary relationships of retroelements. In: Morse SS, editor. The Evolutionary Biology of Viruses. Raven Press; New York: 1994. pp. 121–157. [Google Scholar]
- Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- Eickbush TH. R2 and Related Site-specific non-LTR Retrotransposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. American Society of Microbiology Press; Washington D.C.: 2002. pp. 813–835. [Google Scholar]
- Eickbush TH, Malik HS. Evolution of retrotransposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. American Society of Microbiology Press; Washington D.C.: 2002. pp. 1111–1144. [Google Scholar]
- Eickbush TH, Boeke JD, Sandmeyer SB, Voytas DF. Metaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press; London: 2005. pp. 409–420. [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Evgen’ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, Lankenau DH, Corces VG. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci USA. 1997;94:196–201. doi: 10.1073/pnas.94.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder H, Herzceq A, de Chastonay Y, Aeby P, Tobler H, Muller F. TAS, a retrotransposon from the parasitic nematode Ascaris lumbricoides. Gene. 1994;149:219–225. doi: 10.1016/0378-1119(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Feng Q, Schumann G, Boeke JD. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc Natl Acad Sci USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Heyman T, Wilhelm ML, Wilhelm FX. Extended interactions between the primer tRNA(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res. 1996;24:441–449. doi: 10.1093/nar/24.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel A, Boeke JD. Reverse transcriptase encoded by a retrotransposon from the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci USA. 1991;88:9794–9798. doi: 10.1073/pnas.88.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel A, Willems M, Mules EH, Boeke JD. Replication infidelity during a single cycle of Ty1 retrotransposition. Proc Natl Acad Sci USA. 1996;93:7767–7771. doi: 10.1073/pnas.93.15.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C, Ficheux D, Rau M, Keith G, Sandmeyer S, Darlix JL. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Gonzalez CI, Thomas MC, Olivaries M, Lopez MC. Characterization of reverse transcriptase activity of the L1Tc retroelement from Trypanosoma cruzi. Cell Mol Life Sci. 2003;60:2692–2701. doi: 10.1007/s00018-003-3342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. The DIRS1 group of retrotransposons. Mol Biol Evol. 2001;18:2067–2082. doi: 10.1093/oxfordjournals.molbev.a003748. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. A group of deuterostome Ty3/gypsy-like retrotransposons with the Ty1/copia-like pol-domain order. Mol Genet Genomics. 2002;267:481–491. doi: 10.1007/s00438-002-0679-0. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. A new group of tyrosine recombinase-encoding retrotransposons. Mol Biol Evol. 2004;21:746–59. doi: 10.1093/molbev/msh072. [DOI] [PubMed] [Google Scholar]
- Hansen LJ, Sandmeyer SB. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J Virol. 1990;64:2599–2607. doi: 10.1128/jvi.64.6.2599-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Gao X, Voytas DF. The sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain 8. Plant Physiology. 2005;139:857–868. doi: 10.1104/pp.105.065680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Lopez MC, Garcia-Perez JL, Martin SL, Thomas MC. The L1Tc C-terminal domain from Trypanosoma cruzi non-long terminal repeat retrotransposon codes for a protein that bears two C2H2 zinc finger motifs and is endowed with nucleic acid chaperone activity. Mol Cell Biol. 2005;25:9209–9220. doi: 10.1128/MCB.25.21.9209-9220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia F, Loreto ELS, Valent VL. Complex evolution of gypsy in Drosophilid species. Mol Biol Evol. 2004;21:1831–1842. doi: 10.1093/molbev/msh183. [DOI] [PubMed] [Google Scholar]
- Ivanov VA, Melnikov AA, Siunov AV, Fodor II, Ilyin YV. Authentic reverse transcriptase is coded by jockey, a mobile Drosophila element related to mammalian lines. EMBO J. 1991;10:2489–2495. doi: 10.1002/j.1460-2075.1991.tb07788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- Kaushik N, Chowdhury K, Pandey VN, Modak MJ. Valine of the YVDD motif of moloney murine leukemia virus reverse transcriptase: role in the fidelity of DNA synthesis. Biochemistry. 2000;39(17):5155–5165. doi: 10.1021/bi992223b. [DOI] [PubMed] [Google Scholar]
- Kaushik N, Rege N, Yadav PN, Sarafianos SG, Modak MJ, Pandey VN. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- Ke N, Gao X, Keeney JB, Boeke JD, Voytas DF. The yeast retrotransposon Ty5 uses the anticodon stem-loop of the initiator methionine tRNA as a primer for reverse transcription. RNA. 1999;5:929–938. doi: 10.1017/s1355838299990015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Terzian C, Santamaria P, Pelisson A, Prudhomme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshenboim N, Hayouka Z, Friedler A, Hizi A. Expression and characterization of a novel reverse transcriptase of the LTR retrotransposon Tf1. Virology. 2007;366:263–276. doi: 10.1016/j.virol.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 angstrom resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Kuma K, Toh H, Fujiwara H. Identification of rDNA-specific nonLTR retrotransposons in Cnidaria. Mol Biol Evol. 2006;23:1984–1993. doi: 10.1093/molbev/msl067. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzio J, Pearson MN, Harwood SH, Funk CJ, Evans JT, Slavicek JM, Rohrmann GF. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- Lai L, Liu H, Wu X, Kappes JC. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J Virol. 2001;75:11365–11372. doi: 10.1128/JVI.75.23.11365-11372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, Gabus C, Rau M, Darlix JL. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- Larder BA, Purifoy DJ, Powell KL, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Laten HM, Majumdar A, Gaucher EA. SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci USA. 1998;95:6897–6902. doi: 10.1073/pnas.95.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham GJ, Forgacs E, Beard WA, Prasad R, Bebenek K, Kunkel TA, Wilson SH, Lloyd RS. Vertical-scanning mutagenesis of a critical tryptophan in the “minor groove binding track” of HIV-1 reverse transcriptase. Major groove DNA adducts identify specific protein interactions in the minor groove. J Biol Chem. 2000;275:15025–15033. doi: 10.1074/jbc.M000279200. [DOI] [PubMed] [Google Scholar]
- Levin HL. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol Cell Biol. 1996;16:5645–5654. doi: 10.1128/mcb.16.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL. It’s prime time for reverse transcriptase. Cell. 1997;88:5–8. doi: 10.1016/s0092-8674(00)81851-6. [DOI] [PubMed] [Google Scholar]
- Lin JH, Levin HL. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes & Develp. 1997;11:270–285. doi: 10.1101/gad.11.2.270. [DOI] [PubMed] [Google Scholar]
- Lorenzi HA, Robledo G, Levin MJ. The VIPER elements of trypanosomes constitute a novel group of tyrosine recombinase-enconding retrotransposons. Mol Biochem Parasitol. 2006;145:184–194. doi: 10.1016/j.molbiopara.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol Cell Biol. 1996;16:4726–4734. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Lyozin GT, Makarova KS, Velikodvorskaja VV, Zelentsova HS, Khechumian RR, Kidwell MG, Koonin EV, Evgen’ev MB. The structure and evolution of Penelope in the virilis species group of Drosophila: an ancient lineage of retroelements. J Mol Evol. 2001;52:445–456. doi: 10.1007/s002390010174. [DOI] [PubMed] [Google Scholar]
- Malik HS. Ribonuclease H evolution in retrotransposable elements. Cytogenet Genome Res. 2005;110:392–401. doi: 10.1159/000084971. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH. Phylogenetic analysis of Ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- Martin FC, Maranon Olivares M, Alonso C, Lopez MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the APE family of DNA repair enzymes. J Mol Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HHJ, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Moran JV, Gilbert N. Mammalian LINE-1 retrotransposons and related elements. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. American Society of Microbiology Press; Washington D.C.: 2002. pp. 836–869. [Google Scholar]
- Mount SM, Rubin GM. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985;5:1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Nymark-McMahon MH, Beliakova-Bethell NS, Darlix JL, Le Grice SF, Sandmeyer SB. Ty3 integrase is required for initiation of reverse transcription. J Virol. 2002;76:2804–2816. doi: 10.1128/JVI.76.6.2804-2816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M, Alonso C, Lopez MC. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J Biol Chem. 1997;272:25224–25228. doi: 10.1074/jbc.272.40.25224. [DOI] [PubMed] [Google Scholar]
- Olivares M, Garcia-Perez JL, Thomas MC, Heras SR, Lopez MC. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J Biol Chem. 2002;277:28025–28030. doi: 10.1074/jbc.M202896200. [DOI] [PubMed] [Google Scholar]
- Olivares M, Thomas MC, Alonso C, Lopez MC. The L1Tc, long interspersed nucleotide element from Trypanosoma cruzi, encodes a protein with 3′-phosphatase and 3′-phosphodiesterase enzymatic activities. J Biol Chem. 1999;274:23883–23886. doi: 10.1074/jbc.274.34.23883. [DOI] [PubMed] [Google Scholar]
- Orlinsky KJ, Sandmeyer SB. The cys-his motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J Virol. 1994;68:4152–4166. doi: 10.1128/jvi.68.7.4152-4166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Padow M, Lai L, Deivanayagam C, DeLucas LJ, Weiss RB, Dunn DM, Wu X, Kappes JC. Replication of chimeric human immunodeficiency virus type 1 (HIV-1) containing HIV-2 integrase (IN): naturally selected mutations in IN augment DNA synthesis. J Virol. 2003;77:11050–11059. doi: 10.1128/JVI.77.20.11050-11059.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M, Patel S, Gabriel A. Insights into the role of an active site aspartate in Ty1 reverse transcriptase polymerization. J Biol Chem. 2004;279:47840–47848. doi: 10.1074/jbc.M406019200. [DOI] [PubMed] [Google Scholar]
- Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Piskareva O, Denmukhametova S, Schmatchenko V. Functional reverse transcriptase encoded by the human LINE-1 from baculovirus-infected insect cells. Protein Expr Purif. 2003;28:125–130. doi: 10.1016/s1046-5928(02)00655-1. [DOI] [PubMed] [Google Scholar]
- Piskareva O, Schmatchenko V. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett. 2006;580:661–668. doi: 10.1016/j.febslet.2005.12.077. [DOI] [PubMed] [Google Scholar]
- Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MD, Beard WA, Bebenek K, Howard KJ, Le Grice SF, Darden TA, Kunkel TA, Wilson SH, Levin JG. Residues in the αH and αI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J Biol Chem. 1999;274:19885–19893. doi: 10.1074/jbc.274.28.19885. [DOI] [PubMed] [Google Scholar]
- Pyatkov KI, Arkhipova IR, Malkova NV, Finnegan DJ, Evgen’ev MB. Reverse transcriptase and endonuclease activities encoded by Penelope-like retroelements. Proc Natl Acad Sci USA. 2004;101:14719–14724. doi: 10.1073/pnas.0406281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie HM, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- Sandmeyer SB, Hansen LJ, Chalker DL. Integration specificity of retrotransposons and Retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tiknonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nature Genetics. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Steitz TA, Yin YW. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2004;359:17–23. doi: 10.1098/rstb.2003.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Fujiwara H. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 2002;21:408–417. doi: 10.1093/emboj/21.3.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Gorelick R. Nucleocapsid protein function in early infection processes. Virus Res. 2008 doi: 10.1016/j.virusres.2007.12.006. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentin B, Rebeyrotte N, Mamoun RZ. Human T-cell leukemia virus type 1 reverse transcriptase (RT) originates from the pro and pol open reading frames and requires the presence of RT-RNase H (RH) and RT-RH-integrase proteins for its activity. J Virol. 1998;72:6504–6510. doi: 10.1128/jvi.72.8.6504-6510.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzun O, Gabriel A. A Ty1 reverse transcriptase active-site aspartate mutation blocks transposition but not polymerization. J Virol. 2001;75:6337–6347. doi: 10.1128/JVI.75.14.6337-6347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Boutabout M, Wilhelm FX. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem J. 2000;38:337–342. [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Wilhelm FX. Role of integrase in reverse transcription of the Saccharomyces cerevisiae retrotransposon Ty1. Eukaryot Cell. 2005;4:1057–1065. doi: 10.1128/EC.4.6.1057-1065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Wilhelm FX. Cooperation between reverse transcriptase and integrase during reverse transcription and formation of the preintegrative complex of Ty1. Eukaryot Cell. 2006;5:1760–1769. doi: 10.1128/EC.00159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988;5:675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Malik HS, Eickbush TH. Identification of the endonuclease domain encoded by R2 and other site-specific non-LTR retrotransposable elements. Proc Natl Acad Sci USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]