Abstract
Purpose
The purpose of this study was to perform a comprehensive survey of all known Leber congenital amaurosis (LCA) genes and loci in a collection of 37 consanguineous LCA families from Saudi Arabia.
Methods
Direct PCR and sequencing were used to screen 13 known LCA genes (GUCY2D, CRX, RPE65, TULP1, AIPL1, CRB1, RPGRIP1, LRAT, RDH12, IMPDH1, CEP290, RD3, LCA5). In addition, families without mutations identified were further screened with STR markers around these 13 known LCA genes and two loci.
Results
Disease-causing mutations were identified in nine of the 37 families: five in TULP1, two in CRB1, one in RPE65, and one in GUCY2D. Mutations in known genes only accounted for 24% of the Saudi families—much less than what has been observed in the European population (65%). Phenotype-genotype analysis was carried out to investigate the LCA disease penetrance for all families whose mutations identified. All identified mutations were found to segregate perfectly with the disease phenotype. On the other hand, severity of the disease varies for different patients carrying the same mutation and even within the same family. Furthermore, based on homozygosity mapping with both STR and SNP markers, one family is likely to map to the LCA3 locus.
Conclusions
These results underscore the importance of studying LCA disease families from different ethnic backgrounds to identify additional novel LCA disease genes. Furthermore, perfect segregation between mutation and disease indicates that LCA is fully penetrant. However, phenotypic variations among patients carrying the same mutation suggest that at least some of the variations in the clinical phenotype is due to modification from the genetic background, environment, or other factors.
Leber congenital amaurosis (LCA; Mendelian Inheritance in Man [MIM] #204,000) was first described in 1869 by Theodor Leber as an “intrauterine” form of retinitis pigmentosa.1 Among the most common genetic causes of visual impairment in infants and children, LCA accounts for more than 5% of all retinal dystrophies.2,3 The clinical phenotype of LCA is usually severe and is characterized by several visual perturbations identifiable at birth or within the first year of life, including infantile nystagmus, various fundus changes, and minimal or absent responses on the electroretinogram, each of which occurs with an autosomal recessive mode of inheritance.4 In addition, numerous variations in retinal pathology have been defined in LCA patients, including heterogeneity in retinal appearance, refractive errors, photoaversion, nyctalopia, and the oculodigital sign.3-5 These phenotypic variations suggest a heterogeneous genetic nature of the clinical disorder.
To date, at least 14 genes associated with LCA have been identified (Table 1). These genes are involved in various genetic pathways, including retina development (CRB16 and CRX7-9), phototransduction (GUCY2D2 and AIPL110), vitamin A metabolism (RPE65,11,12 LRAT,13 and RDH1214,15), protein transport (TULP1,16 RPGRIP1,17,18 CEP290,19 and LCA520), and RPE phagocytosis (MERTK21). In addition, the function of RD3 remains unclear.20,22 Furthermore, in rare cases, certain mutations in CRX and IMPDH1, which is involved in guanine nucleotide synthesis, have been shown to cause a dominant form of LCA.23,24 Finally, two loci (LCA3 at 14q24 and LCA9 at 1p36) have been linked to LCA, but the underlying mutations remain unknown. Several mutation survey studies of subsets of these genes from LCA patients have been reported.5,19,25-27 Cumulatively, mutations in these genes account for ~65% of LCA patients, with CEP290 accounting for the most (21%), followed by GUCY2D (11.8%), CRB1 (8.6%), AIPL1 (5.6%), RPE65 (4.4%), RPGRIP1 (4.1%), RDH12 (4%), TULP1 (1.7%), CRX (1.5%), LRAT (1.1%), and RD3 ([lt]1%), respectively. The frequency of mutations in the recently cloned lebercilin gene (LCA5) has not been studied in detail but probably accounts for less than 3% of all cases.20 However, since most of the samples in these surveys are of European descent, the distribution of mutations in LCA patients from other ethnic populations is not necessarily represented.
Table 1.
Human LCA Gene and Locus Summary
| Gene | Gene | Chr | Genetics | Cloned |
|---|---|---|---|---|
| GUCY2D | Photoreceptor-specific guanylate cyclase | 17p13.1 | Recessive | 1996 |
| CRX | Photoreceptor-specific homeobox gene | 19q13.32 | Rec/Dom | 1997 |
| RPE65 | Retinal pigment epithelium 65 Kd protein | 1p31.2 | Recessive | 1997 |
| TULP1 | Tubby-like gene 1 | 6q21.31 | Recessive | 1998 |
| AIPL1 | Aryl hydrocarbon receptor-interacting protein like 1 | 17p13.2 | Recessive | 2000 |
| CRB1 | Crumbs homologue 1 | 1q31.3 | Recessive | 2001 |
| RPGRIP1 | Retinitis pigmentosa GTPase regulator interacting protein 1 | 14q11.2 | Recessive | 2001 |
| LRAT | Lecithin retinol acyltransferase | 4q32.1 | Recessive | 2001 |
| RDH12 | Retinal dehydrogenase 12 | 14q24.1 | Recessive | 2004 |
| IMPDH1 | Inosine monophosphate dehydrogenase 1 | 7q32.1 | Dominant | 2006 |
| CEP290 | Centrosome protein 290 Kd | 12q21.32 | Recessive | 2006 |
| RD3 | Retina degeneration 3 | 1q32.3 | Recessive | 2006 |
| LCA5 | Lebercilin | 6q11-16 | Recessive | 2007 |
| Locus | Supporting Information | Chr | Genetics | Mapped |
|---|---|---|---|---|
| LCA3 | Mapped with two Saudi Arabian families, one from this report | 14q24 | Recessive | 1998 |
| LCA9 | Mapped with one Pakistani family | 1p36 | Recessive | 2003 |
In contrast to the heterogeneity of a given retinal degenerative disorder, different retinal dystrophies share many common clinical features. For example, although many persons with LCA may have normal or near normal fundi as infants, retinal examinations of some LCA patients evolve over time to exhibit pigmentary retinopathy similar to retinitis pigmentosa (RP). Consistent with this observation, four known LCA disease genes, including CRX, CRB1, RPE65, and TULP1, have also been linked to the clinical appearance of RP in other families.28 Interestingly, different mutant alleles of CRX lead to different clinical presentations, including RP, LCA, and cone rod dystrophy, possibly due to differences in severity between alleles and modification by other factors in the genome.23 Similar phenomena have also been observed with most of the known LCA genes. Mutations in some LCA disease genes also lead to cone rod dystrophy (CRX, AIPL1, GUCY2D, RPE65, and RPGRIP1), retinal dystrophy (RDH12), and Bardet-Biedl syndrome (BBS; CEP290). Taken together, these results suggest that the molecular mechanisms underlying different retinal dystrophies may often be shared. Therefore, the study of LCA provides valuable insights and understanding of other retinal dystrophies.
To assess the mutation distribution of LCA in a non-European population, we studied 37 consanguineous LCA families from Saudi Arabia. These families were screened for mutations in 13 known LCA genes (GUCY2D, CRX, RPE65, TULP1, AIPL1, CRB1, RPGRIP1, LRAT, RDH12, IMPDH1, CEP290, RD3, and LCA5, with the exception of MERTK, which is rare) by both direct sequencing and analysis of polymorphic simple tandem repeat markers (STRs). Surprisingly, only 24% (9/37) of the families were found to contain mutations in these 13 known LCA genes, much less than the 65% observed in populations of European descent. Among them, five families carry one identical mutation in TULP1, two families have the same mutation in CRB1, one family has a homozygous mutation in RPE65, and one family has a homozygous mutation in GUCY2D. Most of the mutant alleles identified (3/5) are novel, further underscoring the utility of studying multiple ethnic populations. In addition, STR markers have been used to potentially map these families to known LCA loci. A new LCA3 family has been identified and has a different haplotype than the original LCA3 family, which was also from Saudi Arabia.29 Furthermore, phenotype-genotype correlation studies with the nine Saudi families whose mutations have been identified in our studies have been conducted. Given the large number of patients and unaffected individuals in these pedigrees (a total of 180 individuals, with 53 affected and 127 unaffected), we were able to assess the penetrance of LCA. By direct sequencing all members of these nine families, we found that mutations segregate with disease perfectly, suggesting that these LCA associated mutations are fully penetrant. Finally, we have found that the clinical phenotype of families with different mutations exhibit variable expressivity, supporting the idea that accurate molecular diagnosis is essential for developing effective treatments of these disorders.
Materials and Methods
Patient Sample Collection
This research adhered to the tenets of the Declaration of Helsinki. LCA patient families were ascertained through the King Khaled Eye Specialist Hospital (KKESH) in Riyadh, Saudi Arabia. Ophthalmologic information for each individual was scored as LCA or unaffected by at least one ophthalmologist at the time of referral to the hospital for evaluation, and then reviewed independently and examined independently by one of us (RAL). Detailed family histories and pedigrees of these families were obtained and confirmed through interviews with appropriate family members. Each subject, or a responsible adult on behalf of minors or wards, signed a Consent Statement for Participation approved by the Research Council at KKESH for these investigations and part of a larger program approved by the Baylor College of Medicine Affiliates Review Board for Human Subject Research. We obtained blood samples from subjects where appropriate. A total of 37 multigenerational consanguineous families with recessive LCA were collected; of 417 individuals from these families, 117 were affected. Pedigrees of the families with mutation identified in this study are shown in Figure 1. Once recruited, blood samples were collected from all available family members, and DNA was extracted with a blood genomic DNA extraction kit (Qiagen, Valencia, CA), following the protocol provided by the manufacturer.
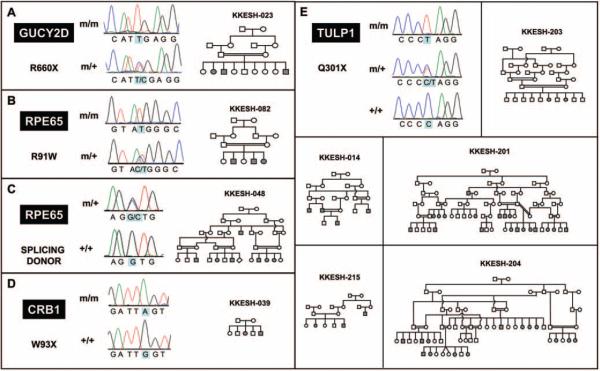
Figure 1.
Pedigrees of nine families with mutations in known LCA genes. Pedigrees are drawn using the HaploPainter program.35 Solid shapes represent affected members. DNA from patients whose identifiers start with n is not available.
Screening for Mutations
Direct PCR and sequencing were used to screen the exons and splicing sites of the 13 known LCA genes, along with the immediately flanking intronic sequences, for point mutations and other small-scale sequence changes. DNA from one affected individual from each family was sequenced for both strands of all exons, including splicing sites, of all known LCA genes. In addition, the intronic region that contains a mutation that results in splicing of a cryptic exon downstream of exon 26 in CEP290 was also sequenced.19 Each exon or intron was individually amplified from leukocyte DNA samples by PCR with primer pairs (primer sequences available on request). Exons longer than 500 base pairs were split into multiple amplicons. Each amplicon is 300 to 500 base pairs in length and was sequenced directly in both forward and reverse directions (ABI3730; AME Bioscience Ltd., Sharnbrook, Bedfordshire, UK). Each read was aligned to the reference sequence and base changes identified with a sequencing program (Sequencher; Gene Codes Corporation, Ann Arbor, MI).
In addition to direct sequencing, families without mutations identified were further screened with STR markers around the 13 LCA genes and two loci. At least one marker near each gene and three markers near each locus were chosen from the GDB human genome database,30 as listed in Table 2. Locations of these markers on the genome were determined by mapping the primer sequences to the human genome using bioinformatic software (BLAT; open source software). A universal primer was added to the end of each primer for fluorescent detection as described previously.31 The size of each PCR fragment was determined with a machine (ABI3730; AME Bioscience Ltd.) and the data were analyzed with genotyping software (GeneMapper; Applied Biosystems, Inc., Foster City, CA) by standard procedures.
Table 2.
Microsatellite Markers for Known LCA Loci and Genes
| Marker | Max Heterozygosity | Type | Range (bp) |
|---|---|---|---|
| LCA3 Locus | |||
| D14S61 | 0.81 | Dinucleotide Repeat | 197-227 |
| D14S1022 | 0.73 | Dinucleotide Repeat | 236-246 |
| D14S68 | 0.91 | Dinucleotide Repeat | 148-172 |
| LCA9 Locus | |||
| D1S450 | 0.82 | Dinucleotide Repeat | 243-267 |
| D1S244 | 0.82 | Dinucleotide Repeat | 285-296 |
| D1S2667 | 0.83 | Dinucleotide Repeat | 224-272 |
| Marker | Gene | Type | Range (bp) |
|---|---|---|---|
| Known LCA Genes | |||
| D19S879L | CRX | Dinucleotide Repeat | 217-265 |
| GATA158H04L | AIPL1 | Tetrinucleotide Repeat | 174-194 |
| D1S1660L | CRB1 | Tetrinucleotide Repeat | 226-250 |
| D14S72L | RPGRIP1 | Dinucleotide Repeat | 257-271 |
| D14S1069L | RDH12 | Dinucleotide Repeat | 200-220 |
| D6S1645L | TULP1 | Tetrinucleotide Repeat | 226-252 |
| D17S1844L | GUCY2D | Dinucleotide Repeat | 227-249 |
| D4S3049L | LRAT | Dinucleotide Repeat | 210-236 |
| D12S1719L | CEP290 | Dinucleotide Repeat | 223-239 |
| D1S219L | RPE65 | Dinucleotide Repeat | 154-176 |
| D7S530L | IMPDH1 | Dinucleotide Repeat | 106-118 |
| D1S414L | RD3 | Dinucleotide Repeat | 185-205 |
| D1S425L | RD3 | Dinucleotide Repeat | 92-108 |
| D6S300L | LCA5 | Dinucleotide Repeat | 188-207 |
| D6S1619L | LCA5 | Dinucleotide Repeat | 189-211 |
| D6S286L | LCA5 | Dinucleotide Repeat | 206-232 |
Results
Identification of Families Who Carry Mutations in Known LCA Genes
To screen for mutations associated with an LCA phenotype in the 37 families in our collection, we selected one affected patient from each family for direct DNA sequencing. By comparison with the reference sequence, five different mutations were identified in nine of the 37 families (Table 3 and Fig. 1), all of which are homozygous for the mutation. Of these nine families, five families (KKESH-014, -201, -203, -204, and -215) carry the same novel nonsense mutation at codon position 301 (Q301X) in TULP1. Two families, KKESH-048 and KKESH-082, carry mutations in RPE65. KKESH-082 has an RPE65 mutation reported previously (R91W), while KKESH-048 carries a novel mutation at the splice donor site of exon 6 (GGT3→GCT). In addition, KKESH-039 carries a novel nonsense mutation (W93X) in CRB1. Finally, a previously reported disease-causing mutation (R660X) in GUCY2D was found in family KKESH-023. In addition to the two previously known mutations, all three novel mutations identified in this study are also likely to be pathogenic. First, the nonsense mutations in TULP1 and CRB1 both occur early in the coding region and are predicted by conceptual translation to result in severe protein truncations. Similarly, the mutation at the splice donor site of exon 6 in RPE65 causes inclusion of intron 6, leading to a frameshift and early termination. Second, clinical phenotypes observed in these subjects are consistent with the premise that these novel mutations are pathogenic. For example, patients from the KKESH-048 family that carry the mutation at the splicing site of exon 6 in RPE65 exhibit a RP-like phenotype, a previously reported feature for patients with mutations in RPE65 (Table 4). Finally, all mutations we have identified are likely to be disease-causative, since each identified mutant allele cosegre-gates with LCA and no unaffected person in any pedigree has two mutant alleles. All available affected and unaffected family members have been genotyped for the identified mutation by direct sequencing. Consistent with the pattern of consanguinity, all LCA subjects are homozygous for the identified mutant allele while unaffected individuals are either heterozygous for the mutant allele or homozygous wild type.
Table 3.
Summary of Mutations and Variations Identified in Known LCA Genes
| Family ID | Gene | Exon | Codon | Base Change | AA Change | Status |
|---|---|---|---|---|---|---|
| Mutations Identified | ||||||
| KKESH-023 | GUCY2D | 10 | 660 | C→T | R→Ter | Known |
| KKESH-082 | RPE65 | 4 | 91 | C→T | R→W | Known |
| KKESH-014 | TULP1 | 10 | 301 | C→T | Q→Ter | Novel |
| KKESH-201 | TULP1 | 10 | 301 | C→T | Q→Ter | Novel |
| KKESH-203 | TULP1 | 10 | 301 | C→T | Q→Ter | Novel |
| KKESH-204 | TULP1 | 10 | 301 | C→T | Q→Ter | Novel |
| KKESH-215 | TULP1 | 10 | 301 | C→T | Q→Ter | Novel |
| KKESH-039 | CRB1 | 6 | 93 | G→A | W→Ter | Novel |
| KKESH-048 | RPE65 | 6 | NA | GGT→GCT | Splice donor | Novel |
| Benign Variation Identified | ||||||
| KKESH-087/-023 | GUCY2D | 12 | 782 | T→A | L→H | rs8069344 |
| KKESH-031 | RPGRIP1 | 13 | 560 | G→T | A→S | rs34263042 |
| KKESH-001/-089 | RPGRIP1 | 18 | 1033 | G→C | E→Q | rs3748361 |
| KKESH-041 | CRB1 | 2 | 27 | G→T | C→F | Novel |
| KKESH-087 | GUCY2D | 3 | 248 | C→T | S→L | Novel |
Table 4.
Clinical Phenotype of Patients with Mutations in Known LCA Genes
| Family | Patient | Gene | Phenotype |
|---|---|---|---|
| KKESH-014 | 04 | TULP1 | Poor vision, nystagmus, hyperopic, vessel attenuation; diminished foveal and ILM macular reflexivities; fundus shows a gray depigmentation with pigment clumping. |
| 01 | Atrophic macula with fovea spared; bone spicules in the mid periphery; Unstable nystagmus; peripheral cortical cuneiform spokes. | ||
| 10 | Irregular coarse nystagmus; hyperopic astigmatism; nystagmus; vertically oval discs; 2/4 pallor; 2/4 vascular attenuation; coarse RP-like dense bone spicules beyond the arcade. | ||
| KKESH-201 | 11 | TULP1 | “RP” with macula spared. |
| 10 | Disc elevation; macula spared; minimal vascular attenuation; sandy peripheral depigmentation; vision CF; horizontal nystagmus; small posterior subcapsular cataracts; vitreous syneresis and cells; midperipheral bone spicules. | ||
| 24 | Disc elevation; macula spared; minimal vascular attenuation; sandy peripheral depigmentation; vision CF; horizontal nystagmus; small posterior subcapsular cataracts; vitreous syneresis and cells; midperipheral bone spicules. | ||
| 20 | Nystagmus; minimal disc pallor; full discs; sandy granular RPE periphery. | ||
| 04 | Pale discs; 3/4 vessel attenuation; full “pseudo-papilledema” discs; fine bone spicules. | ||
| 12 | Small round pale discs; bone spicules in mid-periphery; vessel attenuation. | ||
| 06 | +2 pallor of discs; +2 vessel attenuation; fine bone spicule in mid periphery; PVD OU; ERG nonrecordable. | ||
| 05 | +5 D hyperopic; vertically oval discs; +2 pink pallor; slight elevation; +2 vessel attenuation; fine mid-peripheral bone spicules beyond the arcade. | ||
| 07 | Nystagmus; poor vision to CF OU; disc hyperemia and temporal pallor; vessels mildly attenuated; bone spicules in midperiphery; CT normal; EKG normal; ERG nonrecordable. | ||
| 08 | Jerk nystagmus; vision “follows objects poorly”; pseudopapilledema; CT normal; EKG normal; ERG nonrecordable; attenuated vessels; slightly pale discs. | ||
| 11 | Fundus shows “RP”. | ||
| 12 | No features of deafness, neurologic impairment or mental deficiency; normal EKG; CT scan normal; ERG nonrecordable; fundus shows pigmentary retinopathy with attenuated vessels. | ||
| KKESH-203 | 08 | TULP1 | ERG nonrecordable; compound hyperopic astigmatism; diffuse optic nerve pallor attenuated vessels and RPE granularity; pendular nystagmus. |
| 10 | Irregular horizontal nystagmus; mild horizontal head titubation; onset at age 6 months; hyperopia ≈ +4.00D; ERG nonrecordable; +2 disc pallor; mild vascular attenuation; atrophic foveal light reflexes; minimal RPE degeneration. | ||
| KKESH-204 | 07 | TULP1 | High hyperopia OU: +7.00 + 2.25 × 90; fundus periphery shows confluent outer retinal peripheral whitening and patchy dots at the posterior edge distantly similar to fundus albipunctatus. |
| 20 | Rotatory nystagmus; vertically oval discs; rosy pale discs; vascular attenuation; diffuse pigmentary degeneration. | ||
| 19 | Microrotatory nystagmus; discs, vessels, and pigment same as sister. | ||
| 18 | Microrotatory nystagmus; rosy orange discs; 2-3+ vessel attenuation; diffuse pigmentary changes without bone spicules. | ||
| 04 | Rotatory nystagmus; high hyperopia +5.50 + 1.50 × 90 OU; full elevated discs; +1-2 vessel attenuation; periphery brown grainy RPE; ERG nonrecordable. | ||
| 06 | +8.25 hyperopic; minimal end-gaze nystagmus; macula normal; anterior to equator confluent outer retinal whitening. | ||
| 42 | Rotatory nystagmus; rosy flat vertically oval discs; +1 vessel attenuation; fine midperipheral bone spicules. | ||
| 46 | Rotatory nystagmus; flat rosy discs; +1 vessel attenuation; absent fovea and umbo; greybrown peripheral RPE. | ||
| 48 | Rotatory nystagmus; +1 elevated discs; +1 vessel attenuation; dirty brown peripheral granular RPE. | ||
| 26 | Rotatory nystagmus; minimal hyperopia; rosy pale discs; typical RP in periphery. | ||
| 27 | 30 diopter XT; rotatory nystagmus; +4.00 D hyperopia; vascular attenuation, modest bone spicule midperiphery OU; +6.00 + 0,74 × 180 OU. | ||
| 21 | Rotatory nystagmus; flat discs; vascular attenuation; midperipheral bone spicules. | ||
| 15 | Rotatory nystagmus; 4-prism diopter XT; sluggish pupils; 2/4 vascular attenuation; rate bone spicules in periphery; +3.50 sphere hyperopia. | ||
| KKESH-215 | 04 | TULP1 | Microrotatory nystagmus; +3.50 sph hyperopia; PVD and vitreous condensation OU; vascular attenuation; equatorial and peripheral dense pigmentary RP-like degeneration OU. |
| 07 | Microrotatory nystagmus; full elevated discs with grey corona; far peripheral retina shows confluent grey-white belt similar to family 204. | ||
| KKESH-082 | 04 | RPE65 | Clear lenses; hypermetropic +3.00 + 1.00 × 90 OU; Syneresis of vitreous; Microrotatory infantile nystagmus; Vertically oval discs, mild optic atrophy and vascular attenuation; peripheral depigmentation and NO bone spicules; ERG nonrecordable. |
| 06 | Mild enophthalmos; clear lenses; clear vitreous; vertically oval discs, slight hyperopic optic nerve fullness; decreased retinal light reflexes; minimal peripheral pigment changes; infantile nystagmus +1.50 +1.00 × 90 OU; ERG nonrecordable. | ||
| 07 | Enophthalmos; nystagmus; clear lenses; minimal peripheral pigmentary changes; decreased foveal light reflexes; hyperopic full discs and mild hyperemia; Minimal astigmatism, +0.25 + 1.50 × 90 OU; ERG nonrecordable. | ||
| KKESH-048 | 21 | RPE65 | Micronystagmus; pigmentary granular dystrophy; foveal atrophy; disc atrophy; widespread vascular attenuation and grainy midperiphery. |
| 22 | Micronystagmus; spared macula; +2/4 disc atrophy; +2/4 vascular attenuation; sandy RPE degeneration from inside the arcade to the periphery. | ||
| 15 | No nystagmus; +2/4 vascular attenuation; +1/4 disc pallor; midperiphery sandy depigmentation; “nightblind” according to his father. | ||
| 04 | Disc pallor' vessels attenuated; ERG nonrecordable. | ||
| 06 | Non-recordable ERG; disc pallor, neuroepithelial atrophy; nystagmus. | ||
| KKESH-023 | 05 | GUCY2D | Navigational vision; spectacles do not help; reads Braille; fundus exam (by others) “normal” at age 13 years. |
| 04 | Poor vision from birth; ERG at age 10 years nonrecordable; vision 10/200 OU; pendular nystagmus; pupils sluggish to light; exotropia ≈30 diopters; hyperopic +4.00 D OU. Fundus “normal optic nerve and no pigmentary changes”. Later examiner felt that there were minimally attenuated vessels and mild Optic Nerve pallor OU. | ||
| 11 | Poor vision from birth; vision “fix and follow only”; refraction +6.00 sph OU; ERG nonrecordable; fundus normal to minimal vascular attenuation and diffuse RPE atrophy OU. | ||
| KKESH-039 | 05 | CRB1 | Bilateral enophthalmos; irregular infantile nystagmus; +3/4 optic atrophy; macular geographic atrophy without excavation; mild peripheral pigment clumping; far periphery normal. |
| 07 | Bilateral enophthalmos; irregular conjugate nystagmus; hypermetropic hyperemic discs' + 1-2 vascular attenuation; granular pigmentary dispersion in macula and salt + pepper mid-periphery; ERG nonrecordable. |
In addition to the five pathogenic mutations, five missense variations have also been identified during the initial sequencing of affected individuals (Table 3, section 2). These variations are likely to represent benign polymorphisms. First, three of these variations (marked with rs number) occur frequently in normal populations as common SNPs as documented in the dbSNP database.32 Second, although relatively rare, the other two novel variants do not segregate with LCA. Heterozygous affected and/or homozygous unaffected members have been found in the families, suggesting that these changes are not disease-causing.
LCA as an Early-Onset Genetic Disease with High Penetrance
LCA is considered an early-onset genetic disease with high penetrance. To assess the degree of penetrance of the disease phenotype, we conducted phenotype-genotype correlation studies with nine Saudi Arabian families with the mutations identified above. The pedigree of each family is shown as Figure 1. DNA from a total of 180 individuals (53 affected and 127 unaffected) from these nine families were genotyped for mutations identified above by direct sequencing of the causative mutations. Strikingly, all affected individuals are homozygous for the mutations while unaffected siblings are either heterozygous for the mutations or completely wild type. Therefore, a perfect correlation has been observed between the diagnosis and the underlying genotype. Furthermore, the ages of affected individuals range from 1 year to 27 years old, while the unaffected individuals range from 1 year to 65 years old at the time of evaluation, suggesting that LCA is an early-onset genetic disease with complete penetrance in these families. Detailed clinical phenotypic data for each affected in these nine families are listed in Table 4.
Although patients with LCA all show early loss of vision, the clinical phenotype among them varies. Recent studies have suggested that LCA can be classified into multiple categories and that each category may be caused by mutation in different sets of genes.3-5,25 Consistent with this idea, examination of the clinical phenotype of 53 affected individuals in these nine LCA families revealed that patients with mutations in different genes exhibit distinct phenotypes (Table 4). For example, patients carrying mutations in TULP1 often also show a sandy granular RPE periphery phenotype. In contrast, patients carrying mutations in GUCY2D show relatively normal retinal appearance with no or minimal vascular attenuation. Among the three GUCY2D patients, one also shows navigational vision and another has “fix and follow only” vision. At the same time, the clinical phenotype among patients with the same mutation often varies. For example, we have identified five LCA families carrying the same novel nonsense mutation at codon position 301 (Q301X) in TULP1 (Table 3). Not all patients show an RP-like phenotype, whereas several patients from the family KKESH-204 show a novel phenotype of retinal peripheral whitening and patchy dots at the posterior edge distantly resembling fundus albipunctatus (Table 4).
Excluding Families from Mutations in Known LCA Genes Using STR Markers
It is possible that at least some of the remaining 28 families whose mutations have not been identified by direct sequencing of coding regions of known LCA genes actually carry disease-causative mutations in other functionally important parts of these genes, such as promoters or enhancers. To exclude all 13 known LCA disease genes from these families, we conducted further screening by STR markers (Table 2). As all 28 Saudi families are consanguineous, one can exclude a family from carrying a mutation in a known gene if an affected individual is heterozygous for an STR marker linked to that gene. At least one marker near each LCA disease gene was selected to genotype one or more affected individual from each family. Segregation of these markers with disease was examined for each pedigree and in each case at least one marker was found to be heterozygous in the affected individuals, therefore allowing us to exclude all 28 families from the 13 known LCA genes.
KKESH-060 Maps to the LCA3 Locus
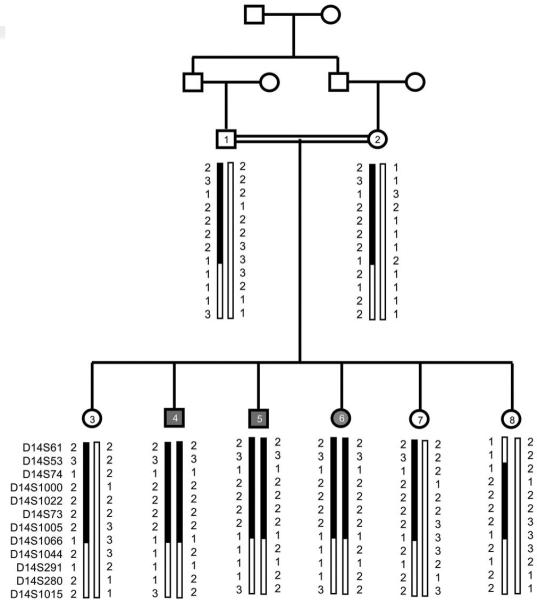
To test whether any of these 28 families who had been excluded from the 13 LCA genes' maps to either of the two known LCA loci (LCA3 and LCA9) for which a specific gene has not been identified, three STR markers from each locus were tested with DNA from the affected individuals from these families (Table 2). At least one subject of each family was found to be heterozygous for one or more STR marker(s) for the LCA9 locus, suggesting that none of the 28 families carry mutations in LCA9. In contrast, an affected individual from one family, KKESH-060, is homozygous for all markers from the LCA3 locus (Fig. 2). To determine whether this family is indeed linked to the LCA3 locus, further genotyping of the other two affected and six unaffected members of the family was performed. We found that all three affected individuals are homozygous for all three STR markers in the LCA3 locus, whereas all unaffected individuals are either heterozygous or completely wild type. In addition to poor vision, all three affected individuals have non-recordable ERGs, hypermetropic astigmatism, and infantile nystagmus. Two patients (KKESH-060-04 and KKESH-060-06) also showed neuroretinal atrophy and markedly attenuated vessels of the fundi at the time (age) of examination.
Figure 2.
Pedigree and haplotype of KKESH-060 that mapped to the LCA3 locus.
Discussion
As one of the most severe causes of congenital blindness, both the clinical phenotype and the underlying molecular bases of the LCA disorders are very complex. To date, 14 genes and two loci have been linked to this disease, but these are estimated to account for only approximately 65% of all families of LCA.5,19,25-27 Therefore, identification of all genes underlying LCA and the development of accurate molecular diagnoses are essential for understanding disease mechanisms and designing proper intervention.
We found that the distribution of mutations in known LCA genes in subjects varies between populations. Previous mutation surveys of LCA patients have estimated that mutations in known LCA genes account for approximately 65% of all families.5,19,25-27 Since most surveys were conducted in Caucasian populations, the fraction of patients carrying mutations in the currently known LCA genes could differ in other populations. To assess this issue, we conducted a comprehensive survey for mutations in 13 known LCA genes (GUCY2D, CRX, RPE65, TULP1, AIPL1, CRB1, RPGRIP1, LRAT, RDH12, IMPDH1, CEP290, RD3, and LCA5) and two loci (LCA3 and LCA9) inour LCA patient collection from Saudi Arabia. Interestingly, mutations in these 13 LCA genes were identified in only 24% of these families (9/37). This difference more likely reflects a difference in the populations rather than that a large portion of the Saudi families in our collection are related and share the same mutation. First, mutations in five known genes have been identified in nine families in our collection, and in most cases, only one or two families carry mutations in the same gene. Similarly, only two families have been mapped to the LCA3 locus. Second, three of the five mutations are novel mutations, indicating that the allele distribution is different from previous studies. For example, despite the fact that 8.6% of all LCA cases are due to mutation in CRB1 in previous surveys and more than 25 disease alleles have been reported, the 660R→Stop mutation identified in our study has not been identified previously.33 Third, none of these 37 families carry mutations in CEP290, the most frequently mutated gene in LCA patients in the European population. While it has been reported that mutations in CEP290 are involved in 21% of all cases in American families, no mutation has been found in this gene in our collection, despite both comprehensive sequencing and STR marker analysis. Interestingly, a recent survey of Italian LCA patients found that only 4% of patients have mutations in CEP290, further underscoring distinct mutation distributions even among different sub-populations of the same ethnicity. Fourth, based on our preliminary genome linkage scans, multiple novel LCA loci may be identified in the remaining 28 families who do not have mutations in the known LCA disease genes or loci, each of which is shared among only a few families (Chen R, unpublished data, 2008). Finally, historical information suggests that these Saudi families have widely divergent tribal origins (data not shown). Taken together, these results suggest that to uncover the entire complement of LCA disease genes, it is essential to examine diverse populations some of which may harbor different genetic variations and diverse mutant allele distributions.
We have confirmed that LCA, as selected here, is indeed an early-onset genetic disease with complete penetrance, consistent with previous reports. In our study, we have identified nine multi-generation, consanguineous families, including a total of 180 individuals with 53 affected and 127 unaffected members, carrying mutations in known LCA genes. All members of these families have been genotyped for the causative mutations, and complete cosegregation between a homozygous mutant genotype and the disease phenotype has been observed (Fig. 1). Such a feature of LCA makes it amenable to identifying mutations with homozygosity mapping with consanguineous families such as the ones that we have reported here. Together with the large sequencing capacity offered by the next generation sequencing technologies, such as the 454 and Solexa platforms, it becomes possible to conduct positional cloning starting with even small consanguineous families.
With STR markers, we have identified another family in our collection, KKESH-060, whose disease maps to the LCA3 locus at 14q24. One LCA3 family, also from Saudi Arabia, has been reported previously.29 A LOD score of 3 was obtained with this new family, and as in the previously reported LCA3 family, all three affected members of this family are homozygous between STR markers D14S61 to D14S1066, a 13.4 Mb interval. In addition, both families present a very similar clinical pheno-type. Interestingly, this new LCA3 family carries a different haplotype than the first LCA3 family based on STR markers, suggesting different origin of the mutant alleles. The identification of this second LCA3 family further supports the linkage between the LCA3 locus and the disease and will provide valuable reagents for identifying the underlying causative mutant gene.
Complex genetic inheritance exists for many human disorders, including retinal diseases.34 For example, digenic triallelic inheritance has been observed in families segregating the Bardet-Biedl syndrome phenotype where inheritance of a mutation in a second gene is required for an individual who has two mutations in the first gene to exhibit a clinical phenotype or to modify the severity of the primary phenotype.34 However, we have not found evidence of tri-allelic inheritance in our inbred Saudi Arabian LCA families. No affected individuals from our cohort carry any coding or splicing mutations in other known LCA disease genes while all unaffected individuals are either wild-type or heterozygous for the mutation. Due to the relatively small sample size and homogenous genetic background, this result does not completely eliminate the possibility of complex inheritance for LCA in every instance.
Acknowledgments
The authors thank Donna Munzy, Jerry Fowler, David Wheeler, and other staff members at the Human Genome Sequencing Center for technical assistance; John Cavender, MD, the Research Director of King Khalid Eye Specialist Hospital at the time of these studies; the Research Council of KKESH for its financial support and the staff of its Research Department for their diligent commitment to this program. In addition, we thank the families reported here for their willing cooperation with these studies.
Supported by the Retinal Research Foundation and National Eye Institute Grant RO1EY018571 (RC). Richard A. Lewis is a Senior Scientific Investigator of Research to Prevent Blindness, New York.
References
- 1.Leber T. Ueber retinitis pigmentosa und angeborene Amaurose. Albrecht von Graefes Arch Ophthal. 1869;15(3):1–25. [Google Scholar]
- 2.Perrault I, Rozet JM, Calvas P, et al. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nat Genet. 1996;14(4):461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 3.Koenekoop RK. An overview of Leber congenital amaurosis: a model to understand human retinal development. Surv Ophthalmol. 2004;49(4):379–398. doi: 10.1016/j.survophthal.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Lewis RA. Retinal Dystrophies and Degenerations. Raven Press; New York: 1988. Juvenile Hereditary Macular Dystrophies; pp. 115–134. [Google Scholar]
- 5.Dharmaraj SR, Silva ER, Pina AL, et al. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet. 2000;21(3):135–150. [PubMed] [Google Scholar]
- 6.Gerber S, Perrault I, Hanein S, et al. A novel mutation disrupting the cytoplasmic domain of CRB1 in a large consanguineous family of Palestinian origin affected with Leber congenital amaurosis. Ophthalmic Genet. 2002;23(4):225–235. doi: 10.1076/opge.23.4.225.13879. [DOI] [PubMed] [Google Scholar]
- 7.Freund CL, Wang QL, Chen S, et al. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18(4):311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- 8.Swaroop A, Wang QL, Wu W, et al. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet. 1999;8(2):299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson SG, Cideciyan AV, Huang Y, et al. Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest Ophthalmol Vis Sci. 1998;39(12):2417–2426. [PubMed] [Google Scholar]
- 10.Sohocki MM, Bowne SJ, Sullivan LS, et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat Genet. 2000;24(1):79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marlhens F, Bareil C, Griffoin JM, et al. Mutations in RPE65 cause Leber's congenital amaurosis. Nat Genet. 1997;17(2):139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 12.Perrault I, Rozet JM, Ghazi I, et al. Different functional outcome of RetGC1 and RPE65 gene mutations in Leber congenital amaurosis. Am J Hum Genet. 1999;64(4):1225–1228. doi: 10.1086/302335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson DA, Li Y, McHenry CL, et al. Mutations in the gene encoding lecithin retinol acyltransferase are associated with earlyonset severe retinal dystrophy. Nat Genet. 2001;28(2):123–124. doi: 10.1038/88828. [DOI] [PubMed] [Google Scholar]
- 14.Janecke AR, Thompson DA, Utermann G, et al. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet. 2004;36(8):850–854. doi: 10.1038/ng1394. [DOI] [PubMed] [Google Scholar]
- 15.Perrault I, Hanein S, Gerber S, et al. Retinal dehydrogenase 12 (RDH12) mutations in leber congenital amaurosis. Am J Hum Genet. 2004;75(4):639–646. doi: 10.1086/424889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagstrom SA, North MA, Nishina PL, et al. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18(2):174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 17.Gerber S, Perrault I, Hanein S, et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur J Hum Genet. 2001;9(8):561–571. doi: 10.1038/sj.ejhg.5200689. [DOI] [PubMed] [Google Scholar]
- 18.Dryja TP, Adams SM, Grimsby JL, et al. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet. 2001;68(5):1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79(3):556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Hollander AI, Koenekoop RK, Mohamed MD, et al. Mutations in LCA5, encoding the ciliary protein lebercilin, cause Leber congenital amaurosis. Nat Genet. 2007;39(7):889–895. doi: 10.1038/ng2066. [DOI] [PubMed] [Google Scholar]
- 21.Gal A, Li Y, Thompson DA, et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26(3):270–271. doi: 10.1038/81555. [DOI] [PubMed] [Google Scholar]
- 22.Friedman JS, Chang B, Kannabiran C, et al. Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am J Hum Genet. 2006;79(6):1059–1070. doi: 10.1086/510021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sohocki MM, Sullivan LS, Mintz-Hittner HA, et al. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63(5):1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowne SJ, Sullivan LS, Mortimer SE, et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2006;47(1):34–42. doi: 10.1167/iovs.05-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanein S, Perrault I, Gerber S, et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23(4):306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 26.Lotery AJ, Namperumalsamy P, Jacobson SG, et al. Mutation analysis of 3 genes in patients with Leber congenital amaurosis. Arch Ophthalmol. 2000;118(4):538–543. doi: 10.1001/archopht.118.4.538. [DOI] [PubMed] [Google Scholar]
- 27.Yzer S, Leroy BP, De Baere E, et al. Microarray-based mutation detection and phenotypic characterization of patients with Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2006;47(3):1167–1176. doi: 10.1167/iovs.05-0848. [DOI] [PubMed] [Google Scholar]
- 28.The University of Texas School of Public Health Retinal Information Network. Available at: http://www.sph.uth.tmc.edu/Retnet. Accessed May 1, 2008.
- 29.Stockton DW, Lewis RA, Abboud EB, et al. A novel locus for Leber congenital amaurosis on chromosome 14q24. Hum Genet. 1998;103(3):328–333. doi: 10.1007/s004390050825. [DOI] [PubMed] [Google Scholar]
- 30.UCSC Genome Browser Web Site. Available at: http://genome. ucsc.edu/. Accessed May 1, 2008.
- 31.Guo DC, Milewicz DM. Methodology for using a universal primer to label amplified DNA segments for molecular analysis. Biotechnol Lett. 2003;25(24):2079–2083. doi: 10.1023/b:bile.0000007075.24434.5e. [DOI] [PubMed] [Google Scholar]
- 32.National Center for Biotechnology Information (NCBI) Single Nucleotide Polymorphism Web site. Available at: http://www.ncbi. nlm.nih.gov/projects/SNP/. Accessed May 1, 2008.
- 33.The Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff. Available at: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=CRX. Accessed May 1, 2008.
- 34.Katsanis N, Ansley SJ, Badano JL, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293(5538):2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 35.Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21(8):1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]