Abstract
The two major pathways for repair of DNA double-strand breaks (DSBs) are homologous recombination (HR) and nonhomologous end joining (NHEJ). HR leads to accurate repair, while NHEJ is intrinsically mutagenic. To understand human somatic mutation it is essential to know the relationship between these pathways in human cells. Here we provide a comparison of the kinetics and relative contributions of HR and NHEJ in normal human cells. We used chromosomally integrated fluorescent reporter substrates for real-time in vivo monitoring of the NHEJ and HR. By examining multiple integrated clones we show that the efficiency of NHEJ and HR is strongly influenced by chromosomal location. Furthermore, we show that NHEJ of compatible ends (NHEJ-C) and NHEJ of incompatible ends (NHEJ-I) are fast processes, which can be completed in approximately 30 min, while HR is much slower and takes 7h or longer to complete. In actively cycling cells NHEJ-C is twice as efficient as NHEJ-I, and NHEJ-I is three times more efficient than HR. Our results suggest that NHEJ is a faster and more efficient DSB repair pathway than HR.
Keywords: DNA repair, Normal human fibroblasts, Nonhomologous end joining, Homologous recombination
1. Introduction
Double-stranded DNA break (DSB) is a dangerous DNA lesion. If left unrepaired DSBs result in massive loss of genetic information, chromosomal aberrations, or cell death. The two major pathways for the repair of DSBs, which differ in the fidelity and template requirements, are nonhomologous end joining (NHEJ) and homologous recombination (HR) [1]. NHEJ modifies the broken DNA ends, and ligates them together with no regard for homology, generating deletions or insertions [2]. In contrast, HR uses an undamaged DNA template to repair the break, leading to the reconstitution of the original sequence [3].
Both DSB repair pathways play important roles in mammalian DSB repair [4,5]. The exact mechanism by which NHEJ and HR interact, and how the choice is made between the two pathways remains unclear. NHEJ and HR may compete for a DSB, or the choice may be determined by the structure of the broken ends [6]. To understand human somatic mutation it is essential to know the relationship between these pathways in human cells. Surprisingly, the efficiency and kinetics of HR and NHEJ have not been directly compared.
Here we employed sensitive fluorescent reporter assays to examine NHEJ and HR in human cells. Reporter cassettes were chromosomally integrated in normal human fibroblasts immortalized by ectopic expression of telomerase, which retain all characteristics of untransformed primary cells [7]. The assays are based on the reconstitution of a functional GFP gene, where the completion of NHEJ or HR is monitored in real-time by the appearance of green cells. Using these assays we compared the kinetics of NHEJ and HR, and determined the ratio between the two processes in multiple genomic locations. We show that NHEJ is a faster and more efficient pathway than HR in human cells.
2. Materials and methods
2.1. Cell culture
Cells were cultured at 37 °C in a 5% CO2, 3% O2 incubator, in EMEM media supplemented with 15% fetal bovine serum, 100 units/ml penicillin and 100µg/ml streptomycin.
2.2. Construction of cell lines for detecting NHEJ and HR efficiency
To generate reporter cell lines HCA2-hTERT cells were transfected with 0.5µg of linearized NHEJ-I, NHEJ-C, or HR reporter constructs. G418, at 1mg/ml, was added to the media 1 day post-transfection. Colonies were picked after 8–10 days on selection. Genomic DNA was then extracted and analyzed by Southern blotting with 3′ and 5′ probes to confirm that the cell lines contained a single integrated copy of the reporter cassettes. The numbers of cell lines analyzed with Southern blotting were 47 for NHEJ-I, 38 for NHEJ-C, and 28 for HR. Of those cell lines, 14 NHEJ-I cell lines, 14 NHEJ-S cell lines, and 7 HR cell lines had single reporter cassettes. Seven cell lines of each type were randomly picked for further research.
2.3. Transfections
The transfections were performed using Amaxa Nucleofector; program U-20. In each transfection 2 million cells were transfected.
2.4. FACS analysis
For analysis of NHEJ and HR cells were harvested, resuspended in ~1 ml 1× PBS, put on ice, and run on a FACS machine. GFP and DsRed fluorescence was analyzed using the red-versus-green plot as described previously [8]. FACS analysis was performed on a FACS Calibur instrument, and data were analyzed using CellQuest software.
2.5. Antibodies
I-SceI expression was analyzed with anti-HA 2367 antibody (Cell Signaling). Actin expression was detected with sc-8432 antibody (Santa Cruz).
3. Results
3.1. Reporter cell lines for analysis of NHEJ and HR
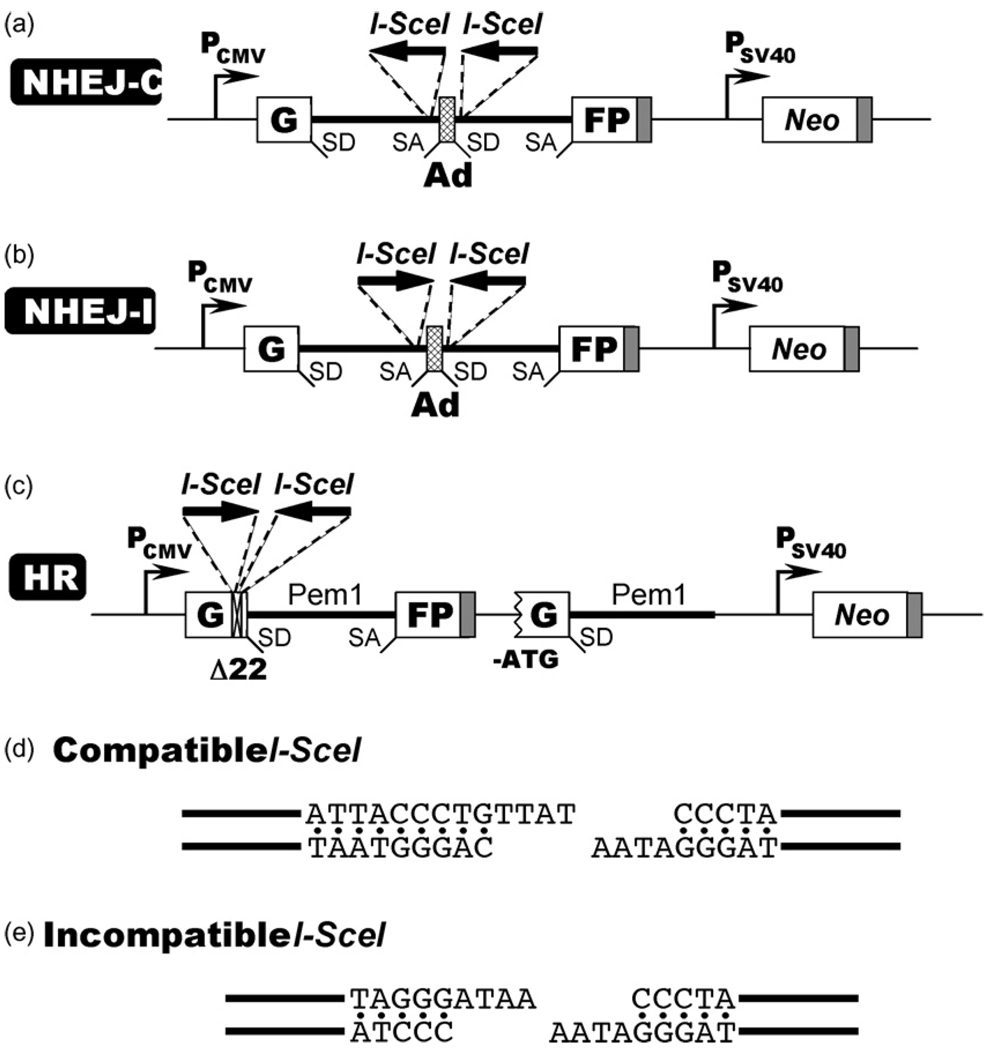
To investigate the roles of NHEJ and HR in DSB repair in normal human cells we generated a series of reporter cell lines containing chromosomally integrated GFP-based reporter constructs. The reporter cassette for detecting NHEJ [8] contains a GFP gene with an artificially engineered 3 kb intron from the Pem1 gene (GFP-Pem1). The Pem1 intron contains an adenoviral exon flanked by recognition sequences for I-SceI endonuclease in direct (Fig. 1a) or inverted (Fig. 1b) orientation, which generate compatible (Fig. 1d) or incompatible ends (Fig. 1e) respectively. Incompatible DNA ends best mimic the naturally occurring DSBs. An un-rearranged NHEJ cassette is GFP negative since the adenoviral exon disrupts the GFP ORF. Upon induction of DSBs by the expression of I-SceI, the adenoviral exon is removed and NHEJ restores function of the GFP gene. This reporter can detect a wide spectrum of NHEJ events since the intron can tolerate deletions and insertions. The NHEJ reporters will not detect the events where only one I-SceI site is digested and then repaired without a deletion. Therefore, NHEJ frequencies measured by these constructs may be an underestimate.
Fig. 1.
Reporter constructs for analysis of DSB repair. (a) Reporter cassette for detection of NHEJ of compatible DNA ends (NHEJ-C). The cassette consists of a GFP gene under a CMV promoter with an engineered intron from the rat Pem1 gene, interrupted by an adenoviral exon (Ad). The adenoviral exon is flanked by I-SceI recognition sites in direct orientation for induction of DSBs. In this construct the GFP gene is inactive; however upon induction of a DSB and successful NHEJ the construct becomes GFP+. SD, splice donor; SA, splice acceptor; shaded squares, polyadenylation sites. (b) Reporter cassette for detection of NHEJ of incompatible DNA ends (NHEJ-I). The cassette is similar to the one shown above, but the I-SceI sites are in inverted orientation, therefore I-SceI digestion produces incompatible ends. (c) Reporter cassette for detection of HR. The cassette consists of two mutated copies of GFP-Pem1. In the first copy of GFP-Pem1 the first GFP exon contains a deletion of 22 nt and an insertion of two I-SceI recognition sites in inverted orientation. The 22 nt deletion ensures that GFP cannot be reconstituted by a NHEJ event. The second copy of GFP-Pem1 lacks the ATG and the second exon of GFP. Upon induction of DSBs by I-SceI, gene conversion events reconstitute an active GFP gene. (d) Compatible DNA ends generated by digestion of two I-SceI sites in direct orientation. (e) Incompatible DNA ends generated by digestion of two inverted I-SceI sites.
The HR reporter (Fig. 1c) is built on the same GFP-Pem1 basis as the NHEJ reporter. In the HR reporter, the first exon of GFP-Pem1 contains a 22 bp deletion combined with the insertion of three restriction sites, I-SceI-HindIII-I-SceI, which are used for inducing DSBs. The deletion ensures that GFP cannot be reconstituted by an NHEJ event. The two I-SceI sites are in an inverted orientation, so that I-SceI digestion leaves incompatible ends (Fig. 1c). The first copy of GFP-Pem1 is followed by a promoter-less/ATG-less first exon and intron of GFP-Pem1. The intact construct is GFP-negative. Upon induction of a DSB by I-SceI digestion the functional GFP gene will be reconstituted by intramolecular or intermolecular gene conversion between the two mutated copies of the first GFP-Pem1 exon. Since the second copy of the GFP gene is lacking the first ATG codon and the second exon, crossing over or single-strand annealing will not restore the GFP activity. This design allows for the exclusive detection of gene conversion, which is the predominant HR pathway in mammalian cells [9].
Since the sequences surrounding the I-SceI sites differ between the NHEJ and HR constructs there is a possibility that the cleavage of the HR substrate occurs more slowly or less efficiently. To test this we digested the plasmids containing the NHEJ and HR substrates with I-SceI enzyme in vitro.We did not detect any difference in the efficiency of digestion (data not shown). This experiment does not completely rule out that a difference exists in vivo, but makes it highly unlikely.
The efficiency of DSB repair may be affected by chromosomal position. Therefore, to obtain a representative picture of the roles of NHEJ and HR in DSB repair we generated multiple randomly integrated reporter cell lines with each construct. The NHEJ or HR reporter cassettes were stably integrated on a chromosome of HCA2 normal human fibroblasts immortalized by expression of hTERT (HCA2-hTERT). Clones containing single un-rearranged copies of each reporter cassette were identified by Southern blot (data not shown), and seven independent cell lines of each kind were randomly chosen for further analysis. The cell lines were designated as follows. Cell lines containing a NHEJ cassette to measure NHEJ of compatible I-SceI ends (NHEJ-C) were designated S35c, S28c, S30c, S44c, S15c, S46c, S13a. The cell lines containing the cassette to measure NHEJ of incompatible I-SceI ends (NHEJ-I) were designated I26c, I29c, I13b, If16a, I9a, I7c, I4a. Finally, the cell lines containing HR construct were named H32c, H29c, H33c, H23c, H15c, H4b, H9b. Construction of the I9a and H15c cell lines was described previously, and formation of the expected NHEJ and HR repair products was confirmed by plasmid rescue and sequencing [10].
3.2. The ratio between NHEJ-C/NHEJ-I/HR is 6:3:1
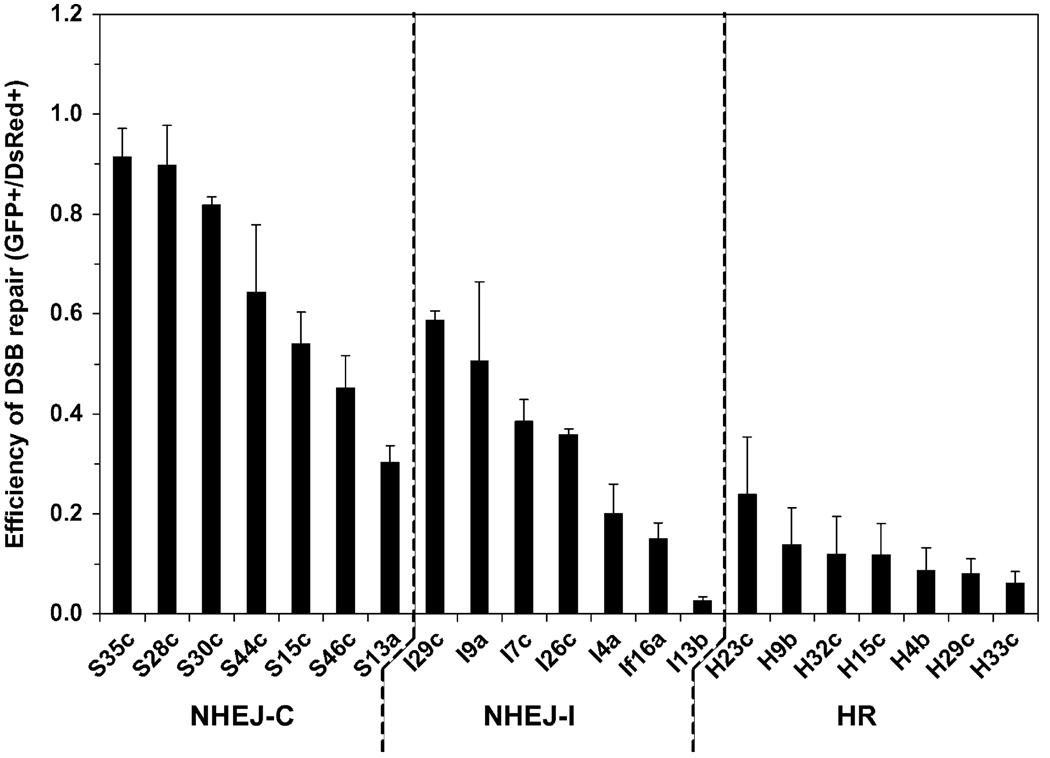
Despite a general belief that NHEJ is a major repair pathway in human cells, the ratio between NHEJ and HR events has not been directly analyzed. To compare the contribution of NHEJ and HR to DSB repair in normal human cells, we determined the efficiencies of NHEJ-C, NHEJ-I and HR in a collection of reporter cell lines with different chromosomal locations of the reporter. For each reporter construct seven cell lines with randomly integrated reporter cassettes were analyzed. Cycling cells were co-transfected with 5µg of plasmid encoding I-SceI endonuclease to induce DSBs and 0.1µg of plasmid encoding DsRed to control for transfection efficiency. Four days after transfection the numbers of GFP+ and DsRed+ cells were determined by flow cytometry as described [8]. The ratio between GFP+ and DsRed+ cells was used as a measure of DSB repair efficiency. Importantly, the gating for flow cytometry was set to detect all GFP+ cells, regardless the intensity, thus differences in expression levels do not affect the results.
The efficiency of repair varied between different cell lines carrying identical reporter constructs, indicating that chromosomal location affects DSB repair (Fig. 2). Nevertheless, NHEJ-C, NHEJ-I, and HR reporter cell lines formed distinct groups in terms of the DSB repair efficiency (Fig. 2). NHEJ-C was the most efficient, NHEJ-I, which requires processing of the ends prior to ligation, was less efficient, and HR was the least efficient pathway. We calculated the averages between the seven cell lines in each group, and the ratio between them. The ratio between NHEJ-C/NHEJ-I/HR was 6:3:1. Since most naturally occurring DSBs have incompatible ends we conclude, that on average NHEJ is three times more efficient than HR in cycling normal human cells.
Fig. 2.
Comparison of the efficiencies of NHEJ and HR across different chromosomal locations. For each DSB repair pathway 7 HCA2-hTERT cell lines, each containing a single randomly integrated copy of the NHEJ-C, NHEJ-I, or HR reporter cassettes were analyzed. Cell lines were co-transfected with the plasmids I-SceI (5µg), and pDsRed2-N1 (0.1µg). I-SceI endonuclease generates DSBs in the GFP-based reporter cassettes (Fig. 1) and successful repair by NHEJ, or HR by gene conversion results in the appearance of GFP+ cells. To quantify NHEJ or HR events the cells were analyzed by flow cytometry 4 days after transfection. Transfection efficiency was normalized by DsRed. The ratio of GFP+ to DsRed+ cells was used as a measure of NHEJ or HR. Typically 20,000 cells were analyzed for each sample; in the treatments where the numbers of GFP+ cells were low, 40,000 cells were scored. The experiments were repeated at least five times and error bars are S.D.
3.3. NHEJ is a faster repair pathway than HR
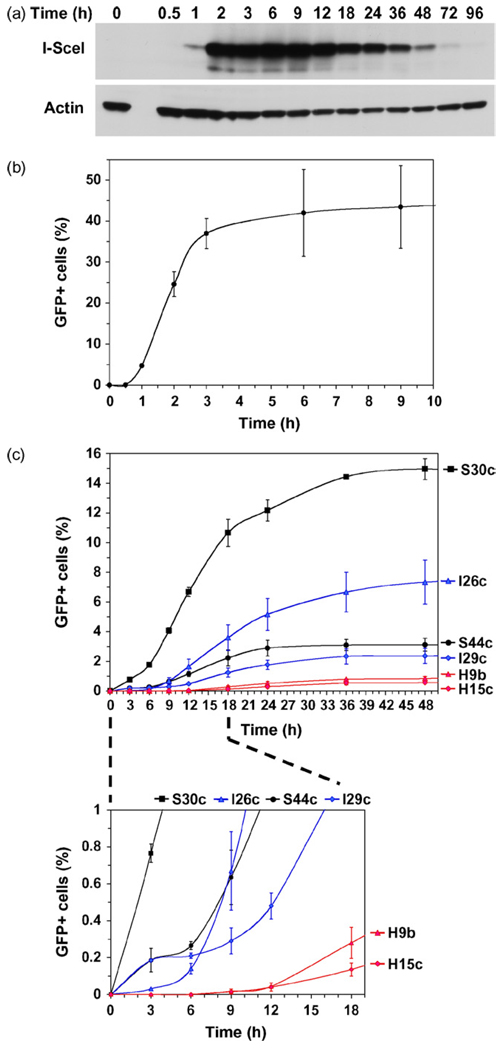
We hypothesized that fluorescent reporter substrates may be utilized for real-time monitoring of DSB repair kinetics via the appearance of green fluorescent cells. This measurement reflects a combination of repair, transcription, splicing and protein synthesis; however, since all three substrates are based on the same gene (GFP-Pem1) transcription, splicing, and protein synthesis are expected to occur with the same kinetics. To determine how soon after DSB induction the first repair events can be detected we examined the kinetics of I-SceI and GFP expression. HCA2-hTERT cells were transfected by Amaxa with 5µg of plasmid encoding I-SceI, or 0.1µg of a control plasmid encoding GFP, and analyzed by Western blot and flow cytometry. I-SceI expression was very rapid and reached high levels at 2h after transfection, then gradually declined after ~12h (Fig. 3a). Thus, Amaxa transfection with I-SceI provides rapid and transient induction of DSBs. The control plasmid encoding GFP was transfected at a low concentration of 0.1µg/2 million cells, which results in transfection of a single plasmid copy per cell [8], to mimic the DSB repair events occurring in chromosomal constructs. GFP expression was also very rapid; the first GFP+ cells were detectable as early as 1h after transfection (Fig. 3b). Thus, assuming that the first I-SceI cuts are introduced 1.5h after transfection, and it takes 1h for GFP to reach detectable levels following DSB repair, the first DSB repair events can be detected as early as 2.5h after I-SceI transfection.
Fig. 3.
NHEJ is a faster process than HR. (a) Western blot showing kinetics of expression of I-SceI after Amaxa transfection. Time indicates hours after transfection. (b) Kinetics of GFP expression following Amaxa transfection. Cells were harvested at indicated time points after transfection and analyzed by flow cytometry. (c) Kinetics of NHEJ and HR. Two representative reporter cell lines containing each type of construct were chosen for this analysis: NHEJ-C, lines S30c and S44c; NHEJ-I, lines I-26c and I-29c; HR, lines H9b and H15c. Cells were transfected with the plasmid encoding I-SceI endonuclease to induce DSBs. For each cell line multiple transfections were pooled, cells were mixed and plated on multiple plates to eliminate variation in transfection efficiency between time points. Cells were harvested at indicated time points post-transfection and analyzed by flow cytometry. GFP+ cells correspond to DSB repair events. The absolute values for DSB repair differ from those reported for the corresponding clones in Fig. 2, because the data in this experiment is not corrected for transfection efficiency with DsRed. Typically, 20,000 cells were analyzed in each sample; in the treatments where the numbers of GFP+ cells were low, 40,000 cells were scored. The experiments were repeated at least four times and error bars are S.D.
To compare the repair rates of NHEJ and HR we chose two representative cell lines from each group: for the analysis of NHEJ-C, S30c and S44c; for the analysis of NHEJ-I, I26c and I29c; and for the analysis of HR, reporter lines H9b and H15c. Exponentially growing cells were transfected by Amaxa with 5µg of plasmid encoding I-SceI to induce DSBs. A single Amaxa electroporation transfects 2 millions cells, therefore multiple transfections were done for each cell line, pooled together, and mixed before plating to eliminate variations in transfection efficiency. Cells were harvested for FACS analysis at various time points after transfection. In this experiment we did not co-transfect with DsRed since DsRed expression has its own kinetics, which may distort the GFP result. The three DSB repair pathways displayed strikingly different kinetics (Fig. 3c and d). NHEJ-C and NHEJ-I were the fastest; 12–308 events per 40,000 cells were detected after 3h. The number of GFP+ cells continued to increase rapidly up to 18h, and then slowed down, which corresponds to the loss of I-SceI expression. From the slope of the curve (Fig. 3c) we conclude that the rate of DSB repair in cell line S30c was the fastest, the I26c and S44c line rates were similar, and I29c was the slowest. Thus, NHEJ of compatible ends may be faster than NHEJ of incompatible ends or they may have similar kinetics. HR was strikingly slower than NHEJ; the first HR events were detected at 9h. As few as 8 HR events were detected for both HR reporter cell lines per 40,000 cells at the 9h time point. After 9h the rate at which new HR events accumulated was much slower than the rate of accumulation of new NHEJ events. From these experiments we conclude that NHEJ is a fast process which takes 0.5h to complete (3h minus 2.5h required for I-SceI and GFP expression), while HR is much slower and takes 7h or more.
4. Discussion
Here we report the first analysis of NHEJ and HR in normal human cells. We employ sensitive fluorescent reporter assays that allow for a direct comparison of the efficiencies of NHEJ and HR events upon induction of chromosomal DSBs with a rare-cutting endonuclease. Fluorescent assays allow for scoring DSB repair events in thousands of cells, and are highly quantitative. Furthermore, rather than relying on a single genomic locus we analyzed DSB repair at multiple chromosomal locations. By doing so we demonstrate that the efficiency of NHEJ and HR is strongly affected by chromosomal position. Nevertheless, when NHEJ and HR are compared across multiple integration sites, NHEJ is more efficient than HR. In unsynchronized proliferating cell populations, NHEJ of compatible DNA ends is twice as efficient as NHEJ of incompatible DNA ends, and NHEJ of incompatible DNA ends is three times more efficient than HR. Since the majority of randomly occurring DSBs have DNA ends that require processing prior to ligation we conclude that in proliferating cells NHEJ repairs 75% of DSBs while HR repairs the remaining 25%. This ratio is similar to the one reported for mouse ES cells [9], however in that study the DSB repair pathways were distinguished by analyzing repair products by Southern blot and only 42 colonies were examined. An overall 3:1 ratio between NHEJ and HR may be a general phenomenon for mammalian cells, although for every individual break it is strongly affected by chromosomal location and the type of DNA ends. It should be noted that the observed frequencies refer to actively proliferating cells. In G1-arrested quiescent or differentiated cells the frequency of HR is likely to be much lower [4,5]. The ratio between NHEJ and HR varies greatly across phylogenetic groups. Yeast rely heavily on HR while in mammals and plants NHEJ is the preferred pathway [11,12]. The choice may be dictated by genome composition [12–14]. In large repetitive genomes of plants and animals overly efficient HR may lead to deleterious genomic rearrangements, such that NHEJ may be a safer choice. The trade-off is the accumulation of small mutations resulting from error-prone NHEJ. Thus, mammalian cells may avoid large genomic rearrangements, but instead accumulate deletions and insertions that contribute to aging and tumorigenesis.
We performed comparison of the kinetics of NHEJ and HR. Our assay allows for rapid and transient expression of I-SceI and an almost instantaneous expression and detection of GFP. We show that NHEJ is much faster than HR. NHEJ can be completed in approximately 30min, while HR takes 7h or more. The kinetics of HR has been analyzed in detail in yeast by directly monitoring recombination products by Southern blot or PCR. The process of recombination was slow(for a fast growing organism like Saccharomyces) and took from 1h for mating type switching [15] to up to 3h for repair of a substrate containing LACZ gene duplication [16]. The kinetics of DSB repair in mammalian cells have previously been examined by pulse field gel electrophoresis and analysis of gamma-H2AX foci [17–20]. Considering that this technique monitors primarily NHEJ events, the kinetics of NHEJ that we observed is consistent with the earlier studies where the majority of DSBs were repaired within 90 min [17]. The in situ studies of Rad51 recruitment to radiation-induced foci showed that Rad51 is recruited within 1h of DSB induction and leaves after 5h [21,22]. It is likely that departure of Rad51 does not manifest the completion of repair, as any remaining gaps, heteroduplexes, and nicks must be filled-in, repaired, and ligated. Therefore, the 7h time course for completion of HR that we observe corresponds well with in situ studies. It is remarkable that HR takes such a long time to complete. What factors determine the choice between NHEJ and HR remain amystery and a subject of intense studies. Our finding that NHEJ is a much faster process than HR offers an explanation of the higher efficiency of NHEJ in mammalian cells.
Acknowledgements
We thank Dara Brown, A’Shantee O’Steen, and Anna Sokolov for help with the construction of the HR reporter cell lines. This work was supported by grants from US National Institute of Health AG027237 (V.G.), American Federation for Aging Research (V.G.), the Komen Foundation (V.G.), and Ellison Medical Foundation (V.G. and A.S.).
Footnotes
Conflict of interest
None.
REFERENCES
- 1.Haber JE. Partners and pathways repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 3.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 4.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst.) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 6.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 7.Morales CP, Holt I, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 8.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double strand break repair in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao Z, Seluanov A, Jiang Y, Gorbunova V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13068–13073. doi: 10.1073/pnas.0702410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sargent RG, Branneman MA, Wilson JH. Repair of site-specific double-strand breaks in mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 2005;56:1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- 13.Gorbunova V, Levy AA. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 1999;4:263–269. doi: 10.1016/s1360-1385(99)01430-2. [DOI] [PubMed] [Google Scholar]
- 14.Lieber MR, Karanjawala ZE. Ageing, repetitive genomes and DNA damage. Nat. Rev. Mol. Cell. Biol. 2004;5:69–75. doi: 10.1038/nrm1281. [DOI] [PubMed] [Google Scholar]
- 15.White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudin N, Sugarman E, Haber JE. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzger L, Iliakis G. Kinetics of DNA double-strand break repair throughout the cell cycle as assayed by pulsed field gel electrophoresis in CHO cells. Int. J. Radiat. Biol. 1991;59:1325–1339. doi: 10.1080/09553009114551201. [DOI] [PubMed] [Google Scholar]
- 18.Iliakis GE, Cicilioni O, Metzger L. Measurement of DNA double-strand breaks in CHO cells at various stages of the cell cycle using pulsed field gel electrophoresis: calibration by means of 125I decay. Int. J. Radiat. Biol. 1991;59:343–357. doi: 10.1080/09553009114550321. [DOI] [PubMed] [Google Scholar]
- 19.Kruger I, Rothkamm K, Lobrich M. Enhanced fidelity for rejoining radiation-induced DNA double-strand breaks in the G2 phase of Chinese hamster ovary cells. Nucleic Acids Res. 2004;32:2677–2684. doi: 10.1093/nar/gkh586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 21.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapp A, Greulich KO. After double-strand break induction by UV-A, homologous recombination and nonhomologous end joining cooperate at the same DSB if both systems are available. J. Cell Sci. 2004;117:4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]