Abstract
Natural products (NPs) are a rich source of novel compound classes and new drugs. In the present study we have used the chemical space navigation tool ChemGPS-NP to evaluate the chemical space occupancy by NPs and bioactive medicinal chemistry compounds from the database WOMBAT. The two sets differ notable in coverage of chemical space, and tangible lead-like NPs were found to cover regions of chemical space that lack representation in WOMBAT. Property based similarity calculations were performed to identify NP neighbours of approved drugs. Several of the NPs revealed by this method, were confirmed to exhibit the same activity as their drug neighbours. The identification of leads from a NP starting point may prove a useful strategy for drug discovery, in the search for novel leads with unique properties.
INTRODUCTION
Space is big. You just won’t believe how vastly, hugely, mind-bogglingly big it is. Even though Douglas Adams in this well known quotation1 relates to astronomy, these words are a striking description of chemical space. It is basically infinite, comprising all possible molecules, which has been estimated to exceed 1060 compounds even when only small (less than 500 Da) carbon-based compounds are considered2. The chemical space of small molecules (CSSM) has recently been mapped with a coarse grained method, namely scaffold topologies, which are mathematical representations of ring structures. The exhaustive enumeration of all 3-node and 4-node topologies for up to eight rings resulted in 1,547,689 distinct scaffolds3. Of these, only 0.6 percent (9,747 unique topologies) are mapped to the known CSSM, sampled by over 52 million compounds from eight different chemical collections representing drugs, natural products, medicinal chemistry, environmental toxicants, and virtual compounds4. As we continue to explore the CSSM, the process of compound selection and prioritization is crucial. It is therefore a challenge for chemical biologists and drug discoverers to identify the limited part of CSSM referred to as biologically relevant chemical space, i.e. the fraction of space where biologically active compounds reside.
A large component of biologically relevant chemical space is occupied by natural products (NPs), i.e. chemical entities produced by living organisms. NPs have been the source of inspiration for chemists and physicians for millennia, and have so far proven to be by far the richest source of novel compound classes, and an essential source of new drugs5–7. NPs can be regarded as pre-validated by Nature. They have a unique and vast chemical diversity and have been optimized for optimal interactions with biological macromolecules through evolutionary selection. Virtually all of the biosynthesized compounds have a biological activity with (from an evolutionary perspective) beneficial purpose for the organism that produces it, thus fulfilling the requirement for biological relevance. Taken together, these facts make them exceptional as design resources in drug discovery, and the interest for NPs remains considerable8, 9. In an earlier study10, we used the concept of chemical space to correlate structural trends among NPs with confirmed cyclo-oxygenase (COX)-1 and COX-2 inhibitory activity. The identification of numerous outliers suggested, what has also been supported by several other authors, e.g.11, that NPs populate unique regions of chemical space.
Pfizer’s Rule of Five (Ro5) provided guidelines to evaluate if a chemical compound has properties that would make it likely orally available in humans12. It was recently established that of the 126,140 unique NPs in The Dictionary of Natural products (DNP), sixty percent had no Ro5 violations13. It should be kept in mind that NPs are often cited as an exception to Pfizer’s Ro5, and even Lipinski himself noted14 that many NPs remain bio-available despite violating the Ro5 – although active mechanisms may be involved. In a recent paper15, a set of NPs, that each led to an approved drug between 1970 and 2006, were analyzed and found to be divided into two equal subsets. One is Ro5 compliant, while the other one violates Ro5 criteria. Interestingly, the two subsets had an identical success rate in delivering an oral drug.
That NPs have properties distinguishing them from other medicinal chemistry compounds has been suggested by several studies, e.g. references10, 11, 16–19. One of the more comprehensive studies was recently reported by Ertl and Schuffenhauer19. They compared the physico-chemical properties and structural features of three classes of compounds: NP structures from DNP, bioactive molecules obtained by combining structures from the World Drug Index20 and the MDDR database21, and an in-house set of organic compounds. They found that the distribution of the octanol-water partition coefficient (logP), polar surface area, and the number of atoms were very similar between the three classes. Additionally, NPs appeared to be less flexible, and to contain fewer aromatic rings. Besides looking at property distributions of these compounds, Ertl and Schuffenhauer also visualized them in a structural chemistry space using principal component analysis (PCA). Instead of using calculated molecular properties, as we have done in the present paper, Ertl and Schuffenhauer used counts of one and two-atomic substructures fragments in the molecules.
High-throughput screening is a hit-finding technique frequently used in pharmaceutical industry where large screening collections are tested against a particular target. These collections generally capture only a fraction of CSSM2 and are occasionally biased such that some areas covered are over-sampled. This is found, in particular, where compounds have been synthesized with focus around targets of current interest, like metabolic enzymes, G-protein-coupled receptors, and kinases. Quite likely, such bias may have resulted, over time, in lack of broad diversity in pharmaceutical screening collections. Extensive compound collection enhancement programs have been described in literature to address this issue and reshape the screening collections22, 23. Recently, available chemical libraries were statistically evaluated, based on a set of commonly used molecular descriptors24. This study found that bioactive collections, which contained compounds with well-characterized biological functions, and NP libraries, came closest to populate the biologically relevant regions of CSSM, albeit with poor density. This observation was also confirmed by comparing scaffold topology coverage of NPs vs. medicinal chemistry collections4.
In this paper we have used the PCA25 based chemical space navigation tool ChemGPS-NP26–28 to analyze large datasets of chemical compounds, thus exploring biologically relevant chemical space. The aim of this paper was four-fold. First, we wanted to compare the coverage of biologically relevant chemical space by bioactive medicinal chemistry compounds, represented by the WOMBAT database, and NPs respectively. Second, we aimed at revealing regions that are sparsely populated by the bioactive medicinal chemistry compounds, here referred to as low density regions, where we could break new grounds in terms of biological activities. Third, we intended to possibly uncover so called lead-like NPs located in any of the low density regions. Fourth and finally, we wanted to compare the chemical space of registered drugs with that of NPs and identify NPs situated close to any of the drugs suggesting possible lead potential.
RESULTS AND DISCUSSION
Differences in coverage of biologically relevant chemical space by medicinal chemistry compounds and NPs
The WOMBAT database29, 30, version 2007.2, was used to estimate the coverage by bioactive medicinal chemistry compounds of the biologically relevant chemical space. WOMBAT is a medicinal chemistry database containing chemical structures and associated experimental biological activity data on 1,820 targets (receptors, enzymes, ion channels, transporters and proteins) for 203,924 records, or 178,210 unique structures30, 31. A data table was constructed, where chemical structures in SMILES32 representation were tagged with demonstrated biological activities, and 35 calculated molecular descriptors. The descriptor array used was the set of 35 previously validated descriptors used in conjunction with the chemical space navigation tool ChemGPS-NP26–28. Briefly, ChemGPS-NP is a PCA based global space map with eight principal components (dimensions) describing physico-chemical properties such as size, shape, polarizability, lipophilicity, polarity, flexibility, rigidity, and hydrogen bond capacity for a reference set of compounds. New compounds are positioned onto this map using interpolation in terms of PCA score prediction25, 27. The properties of the compounds together with trends and clusters can easily be interpreted from the resulting projections. This tool is available as a free web-based resource at http://chemgps.bmc.uu.se/28. The selection of these particular descriptors have been thoroughly described elsewhere26. The bioactive medicinal chemistry compounds from WOMBAT, here referred to as the medicinal chemistry compounds, were then mapped on to these descriptors using ChemGPS-NP.
Coverage of the biologically relevant chemical space by medicinal chemistry compounds reveals several areas that are sparsely populated, a feature discussed in detail below. To investigate the overlap in coverage of biologically relevant chemical space between the medicinal chemistry compounds and NPs, a set of NPs were mapped on to the same chemical space using ChemGPS-NP. DNP33, October 2004 release, was used as the NP dataset. This version of DNP includes entries corresponding to 167,169 compounds (126,140 unique compounds) of natural origin, covering large parts of what has been isolated and published in terms of NPs up until the release date. The difference in coverage of biologically relevant chemical space by these two different sets is noteworthy as can be interpreted from Figures 1 and 2.
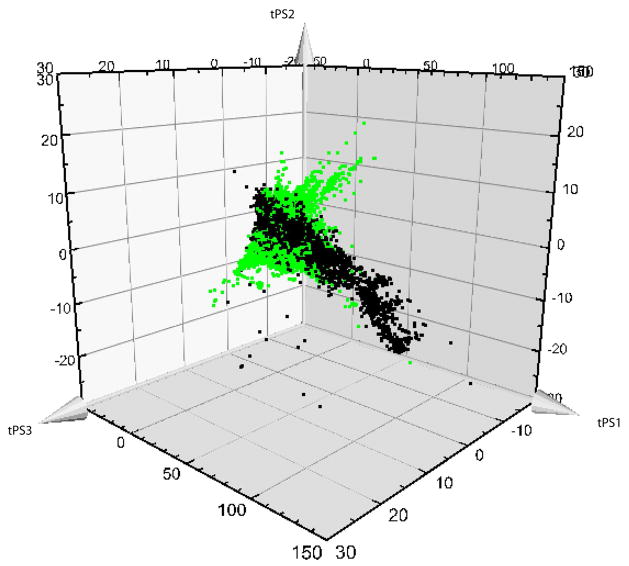
Figure 1.
Predicted score (tPS) plots illustrating the difference in coverage of biologically relevant chemical space by natural products (NPs, in green) and bioactive medicinal chemistry compounds from the database WOMBAT (in black) in the first three principal components. NPs cover parts of chemical space that lack representation in medicinal chemistry compounds, indicating that these areas have yet to be investigated in drug discovery. They appear to contain lead-like NPs that could subsequently be of interest in drug discovery.
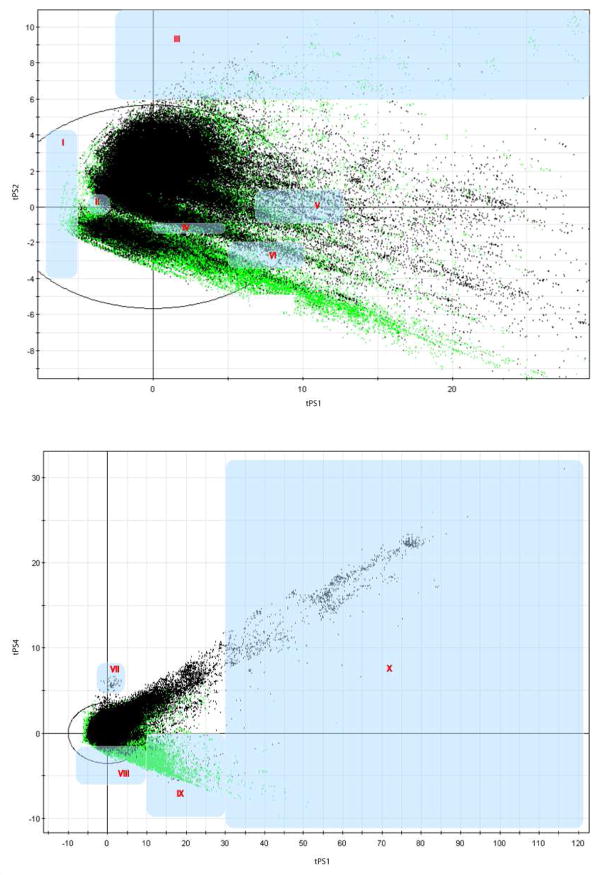
Figure 2.
An overview of the analyzed low density regions (NPs in green, medicinally chemistry compounds in black). Briefly, the first four dimensions of ChemGPS-NP can be interpreted as follows; size increases in the positive direction of PC1; compounds are increasingly aromatic in the positive direction of PC2; lipophilic compounds are situated in the positive direction of PC3 and polar in the negative direction; compounds are increasingly flexible in the positive direction of PC4 and more rigid in the negative direction. Regions I and II enclose smaller compounds than average. Region III holds compounds with increased aromaticity. In regions IV, V and VI reside compounds with increasing size in positive direction of PC1, and less aromatic features in negative direction of PC2. Region VII contains rather flexible, average sized compounds, while region VIII encloses fairly rigid, average sized compounds. Compounds in region IX are increasingly rigid and large. Region X contains compounds that are generally larger than average, and increasingly flexible in the positive direction of PC4.
The basic interpretation of the first four dimensions of ChemGPS-NP can be as follows: size increases in the positive direction of principal component one (PC1); compounds are increasingly aromatic in the positive direction of PC2; lipophilic compounds are situated in the positive direction of PC3; and predominantly polar compounds are located in the negative PC3 direction; compounds are increasingly flexible in the PC4 positive direction and more rigid in its negative direction. As can be interpreted from Figure 2, a majority of the NPs are found in the negative direction of PC4, while the medicinal chemistry compounds are encountered in the positive direction. This indicates that NPs are generally more structurally rigid than the medicinal chemistry compounds. Figure 2 also reveals that NPs tend to be situated in the negative direction of PC2, indicating lower degree of aromaticity than the medicinal chemistry compounds that are frequently drawn towards the positive direction of PC2. The distribution of size addressed in PC1 (see e.g. Figure 2), and lipophilicity and polarity addressed in PC3 (to some extent interpretable from Figure 1) appears to be very similar between the two sets. These results are in agreement with the recent results from Ertl and Schuffenhauer19.
NPs were found to cover CSSM regions that lack representation in medicinal chemistry compounds, indicating that these regions have yet to be investigated in drug discovery. These, by medicinal chemistry compounds, sparsely populated regions were subsequently analyzed. A subset of these regions, referred to as low density regions, are highlighted and numbered in Figure 2. Each of the regions was analyzed in terms of occupancy with regard to both NPs and medicinal chemistry compounds. Typical examples of compounds from the different regions are presented in Table 1. Some regions had low density for the simple reason that their location implies an impossible combination of properties, e.g. there are limits for individual properties, and a compound cannot simultaneously be small, highly lipophilic, and have several H-bond donors and acceptors. Regions I and II enclose smaller compounds than average. Region III holds compounds with increased aromaticity. Regions IV, V and VI contain compounds with a combination of increasing size in positive direction of PC1, and less aromatic features in negative direction of PC2. Region VII contains flexible, average sized compounds, while region VIII encloses fairly rigid, average sized compounds. Compounds in region IX are increasingly rigid and large. Region X contains compounds that are generally larger than average, and increasingly flexible in positive direction of PC4.
Table 1.
Scores specifications of the low density regions and with typical structure examples.
| Region | tPS1†† | tPS2‡‡ | tPS3§§ | tPS4*** | Example |
|---|---|---|---|---|---|
| I | <−5 | - | - | - |
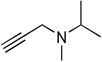
|
| II | −3.5 – −2.5 | −0.5 – 0.5 |
|
||
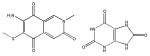
| III | - | >6 | - | - |
|
| IV | 0 – 4 | −1.5 – −1 | - | - |
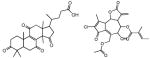
|
| V | 7.5 – 12.5 | −1 – 1 | - | - |
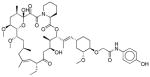
|
| VI | 5 – 10 | −2 – −3.5 | - | - |
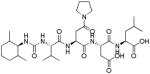
|
| VII | <5 | - | - | >4 |
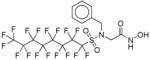
|
| VIII | <10 | - | - | <−1.5 |
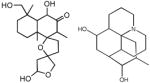
|
| IX | 10–30 | - | - | <0 |
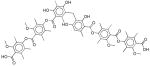
|
| X | >30 | - | - | - |
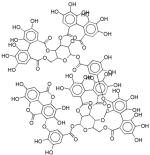
|
Predicted scores in component 1
Predicted scores in component 2
Predicted scores in component 3
Predicted scores in component 4
Lead-like NPs in low density regions
The low density regions were subsequently investigated with the purpose to identify possible (or tangible)34 so called lead-like34, 35 NPs from these regions. To distinguish lead-like compounds the following computational cut-off criteria were used, based on previous studies34, 35: molecular weight (MW) less than or equal to 460, the logarithm of the octanol/water partition coefficient (LogP) between −4 and 4.2, the logarithm of the intrinsic aqueous solubility (LogSw) larger than −5, number of rotatable bonds (RTB) less than or equal to 10, number of rings (RNG) less than or equal to 4, number of H-bond donors (HDO) fewer than or equal to 5, number of H-bond acceptors (HAC) fewer than or equal to 9. NPs occupying the low-density regions were investigated in terms of above-mentioned criteria and it was concluded that regions I, II, IV, and VIII (see Figure 2) contained lead-like compounds and were in fact mainly covered by NPs. In total, we found 40,348 unique DNP compounds to match the lead-like criteria; of these, 336 NP lead-like compounds are in region I, whereas region II holds 356, region IV contains 112, and region VIII 652 unique lead-like NPs, respectively.
NP close neighbours of registered drugs
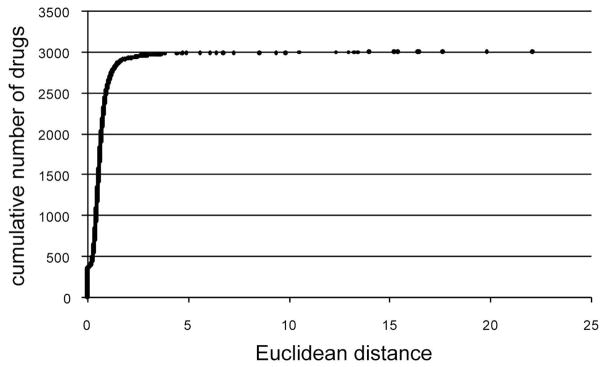
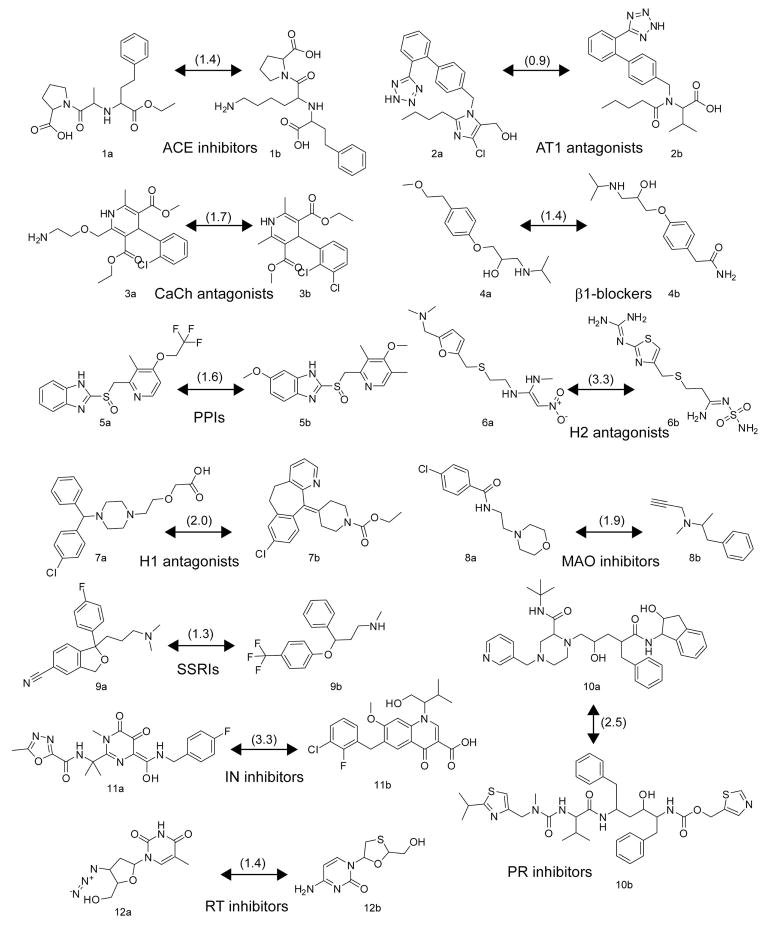
To study the chemical space covered by approved drugs, the GVKBIO Drug Database (GVKBIO_DD) was used36. GVKBIO_DD contains data on drugs approved by the FDA and other authorities extracted from pharmacological journals and other sources. The 3,211 compounds in GVKBIO_DD were mapped together with the DNP compounds using ChemGPS-NP. The resulting predicted scores in the eight dimensions were listed for all the compounds and Euclidean distances (EDs) over eight dimensions were calculated between the compounds in the two datasets. Thereby all NPs were assigned with 3,211 EDs, one ED to each drug. The NP/drug pairs were subsequently sorted in order of increasing EDs. In Figure 3 the 3,211 drugs are plotted against the ED to their closest NP neighbour. Interestingly 99.5 percent of all drugs have a NP neighbour closer than ED=10, and 85 percent of the drugs have a NP neighbour closer than the ED=1. This forms a strong argument that NPs has the potential to serve as an important source of inspiration for medicinal chemists. As a comparison, “within group” EDs were calculated between known drug pairs (1a–12b) exhibiting the same mode of action. Plots illustrating distinct clustering of these respective bioactivity groups using ChemGPS-NP are provided as supporting information. The within group EDs and the chemical structures of these drugs are given in Figure 4. The average within group ED was 1.8, the median was 1.6, and the standard deviation was 0.9. We found that 313 drug/NP pairs had ED equal to 0. To find exact matches between drugs and NPs was expected since many drugs are of natural origin. These were visually inspected and it could be verified that all of these pairs, disregarding stereochemistry, were identical compounds.
Figure 3.
Distribution of ED to the nearest NP neighbour for the drugs in GVKBIO_DD. The cumulative number of drugs is plotted against the ED to the closest NP neighbour.
Figure 4.
Chemical structures of drugs with shared mode of action used to calculate within group EDs; the angiotensin-converting enzyme (ACE) inhibitors enalapril (1a) and lisinopril (1b), the angiotensin receptor 1 (AT1) antagonists losartan (2a) and valsartan (2b), the calcium channel (CaCh) blockers amlodipine (3a) and felodipine (3b), the β1–adrenergic receptor (β1) blockers metoprolol (4a) and atenolol (4b), the proton pump inhibitors (PPIs) lansoprazole (5a) and omeprazole (5b), the histamine 2 (H2) receptor antagonists ranitidine (6a) and famotidine (6b), the H1 receptor antagonists cetirizine (7a) and loratidine (7b), the monoamine oxidase (MAO) inhibitors moclobemide (8a) and selegeline (8b), the selective serotonin re-uptake inhibitors (SSRIs) citalopram (9a) and fluoxetine (9b), the HIV-1 protease (PR) inhibitors indinavir (10a) and ritonavir (10b), the HIV-1 integrase (IN) inhibitors raltegravir (11a) and elvitegravir (11b), and the HIV-1 reverse transcriptase (RT) enzyme inhibitors zidovudine (12a), and lamivudine (12b). EDs are given in parentheses.
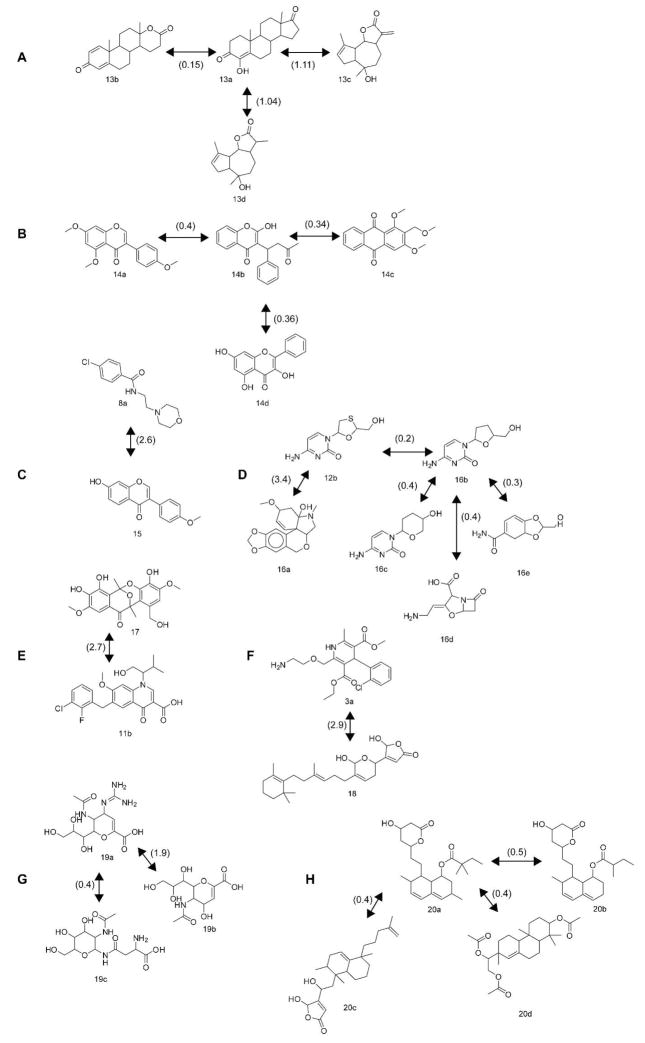
Non-identical NPs with very short EDs to any of the approved drugs are proposed for further analysis as potential lead compounds against the target in question. Among the NPs with relatively short EDs to any of the drugs we found a number of NPs that, in fact, had confirmed similar biological activity as the corresponding drug neighbour, which supports the use of near neighbours as a good starting point for drug discovery. The drugs in the examples presented in Figure 5 were selected to represent a wide array of different indications of general interest. For each of the selected drugs the EDs to all members of DNP were compared. The NPs with the shortest ED to the drug were surveyed in literature for publications regarding their bioactivity. This was repeated until an NP with interpretable activity corresponding to that of the drug was retrieved. In some cases the search was expanded slightly to incorporate additional examples. If no such compounds were found, structurally interesting not yet examined NPs were used as examples. Finally, at this stage, the proportion of NPs with similar or shorter EDs than the selected example in DNP was calculated. These numbers are given in the legend of Figure 5. In some of the cases the surveyed bioactivity was found in the NP with the shortest ED to the drug. In other cases a considerably larger portion of the NPs had to be checked before a compound with a corresponding activity was found. This does not necessary indicate that there are no closer actives – only that these compounds have not yet been assayed with regard to this activity. In these cases highly potent NPs might exist with much shorter EDs than the examples in Figure 5. The close ED members, with yet unknown biological activities, provide a wealth of suggestions and inspiration that could help overcome possible problems with synthetic feasibility, and e.g. indicate paths to more easily synthesized molecules. Examples are given below and chemical structures are given in Figure 5A–H. The drug/NP pair formestane (13a)/testolactone (13b) is one interesting drug/NP pair captured by this method. Testolactone (13b) from the DNP set, transformed from e.g. progesterone by the fungi Aspergillus tamarii, had the ED 0.15 to formestane (13a) from the GVKBIO_DD set. Testolactone (13b) is, just as its close and structurally very similar neighbour, an approved aromatase inhibitor used to treat e.g. breast cancer37. Also the two NPs 10-epi-8-deoxycumambrin B (13c) and 11βH,13-dihydro-10-epi-8-deoxycumambrin (13d) both isolated from Stevia yaconensis had short EDs, of 1.11 and 1.04 respectively, to the approved aromatase inhibitor formestane (13a). The compound 13c is moderately active while 13d has been found to have a pronounced activity38 as aromatase inhibitor. Structures of formestane and its NP neighbours are given in Figure 5A.
Figure 5.
Chemical structures of drug/NP pairs. The ED between the compounds is given in parentheses under the corresponding drug/NP pair. The proportion of NPs in DNP with similar or shorter EDs than the selected examples is given in parentheses, in percent, after each NP example, where 0% means that this was the single closest NP. (A) The aromatase inhibitor formestane (13a) and its NP neighbours 13b–d (0, 0.6, and 0.4% respectively). (B) The anticoagulant drug warfarin (14b) and its NP neighbours 14a (0%), 14c (0%), and 14d (0%). (C) The antidepressant drug moclobemide (8a) with NP neighbour 15 (6.4%). (D) The HIV-1 RT inhibiting drugs lamivudine (12b) and zalcitabine (16b) and their NP neighbours 16a (14%), 16c–e (0%). (E) The investigational new HIV-1 IN inhibiting drug (in phase III clinical trials) (11b) with NP neighbour 17 (8.4%). (F) The antihypertensive drug amlodipine (3a) and it NP neighbour 18 (7.3%). (G) The antiviral drug zanamivir (19a) with close NP neighbours 19b (0.05%), from which zanamivir was derived, and 19c (0%). (H) The antilipemic drug simvastatin (20a) and its close NP neighbours 20b (0.03%), from which simvastatin was originally derived, 20c (0.01%), and 20d (0.01%).
Another example of an interesting drug/NP pair captured by this method is 4′,5,7-trimethoxyisoflavone (14a), isolated from Ouratea hexasperma which has the ED 0.4 to the well known anticoagulant drug warfarin (14b). 14a has been shown to exhibit anticoagulant activities39, just like its drug neighbour. Also, both 1,3-dimethoxy-2-(methoxymethyl)-anthraquinone (14c), isolated from Coussarea macrophylla and galangin from e.g. Helichrysum nitens (14d) are close neighbours to warfarin (14b) (ED=0.34 and 0.36 respectively). Any studies performed regarding anticoagulant properties of these two compounds could not be found in literature. Structures of warfarin and its NP neighbours are given in Figure 5B.
The antidepressive drug moclobemide (8a), which acts by inhibiting the enzyme monoamine oxidase (MAO) has an active close NP neighbour in formononetin (15), isolated from Sophora flavescens. Formononetin (15) has been shown to inhibit MAO40. The ED between the two compounds is 2.6 and their structures are given in Figure 5C.
The HIV-1 RT inhibiting drug lamivudine (12b) has an active NP neighbour in littoraline A (16a), isolated from Hymenocallis littoralis. The ED between the compounds in this drug/NP pair is 3.4, and just like its neighbour, littoraline A inhibits HIV-1 RT41. Littoraline A (16a) is also a close neighbour (ED=3.3) of the HIV-1 RT inhibiting drug zalcitabine (16b). Zalcitabine (16b) also had three close NP neighbours that, to our knowledge, has not yet been tested for HIV-1 RT inhibiting activity; the structurally very similar NPs pentopyranine A (16c) isolated from Streptomyces griseochromogenes (ED=0.4); clavinimic acid (16d), isolated from Streptomyces clavuligerus (ED=0.4); and dioxolide A (16e) isolated from Streptomyces tendae (ED=0.3). The ED between zalcitabine (16b) and lamivudine (12b) is 0.2. Structures of these drugs and their close NP neighbours are given in Figure 5D.
Also the investigational new HIV-1 IN inhibiting drug elvitegravir (in phase III clinical trials) (11b) has a close NP neighbour with similar mode of action; integrastatin A (17), isolated from Ascochyta sp., inhibits HIV-1 IN42 and the ED between the two compounds is 2.7. Structures are given in Figure 5E.
The antihypertensive drug amlodipine (3a) acts by blocking calcium channels. The employed method captured an NP neighbour of this drug, the compound manoalide (18) isolated from the sponge Luffariella variablis, that also has been shown to block calcium channels43. The ED between the two compounds is 2.9 and their structures are given in Figure 5F. Numerous interesting drug/NP pairs with short EDs, where the activity of the NP remains to be investigated, were highlighted by this method. The neuraminidase inhibitor zanamivir (19a), used to treat e.g. avian flu, was derived from the NP 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (19b)44, 45, a NP widely distributed in animal tissues as well as in bacteria. The ED between these two compounds is 1.9. Zanamivir (19a) has a close NP neighbour, N-[2-(Acetylamino)-2-deoxy-β-D-glucopyranosyl]-L-asparagine (19c), within ED 0.4 (Figure 5G). These two structures do have very similar fragments, but their relative arrangement is very different. The antilipemic drug simvastatin (20a) were derived from the NP mevastatin (20b) (ED=0.5), an antifungal metabolite from Penicillium brevicopactum. Also simvastatin (20a) has several close and structurally similar NP neighbours, e.g. dysidiolide (20c) and 8(14)-pimarene-3,15,16-triol (20d), both within ED 0.4, that are not yet investigated for antilipemic activity. Structures are given in Figure 5H.
CONCLUSION
Author Les Brown famously said: Shoot for the moon. Even if you miss, you’ll land among the stars46. It might sound like close enough, but considering the vastness of chemical space, exploration and drug discovery needs to be more precise and focused than that. To make the navigation in chemical space easier, this can be advantageously divided into smaller sections or neighbourhoods. A first step is to reduce the vast theoretical chemical space by looking at the region encompassing only small molecules, i.e. CSSM. A second challenge for drug discoverers is to identify biologically relevant regions of chemical space, where we can, with a higher probability, find future leads for drug discovery. In this paper we have used ChemGPS-NP to steer through the vastness of chemical space and to further partition biologically relevant chemical space. Investigation of the coverage of chemical space by medicinal chemistry compounds revealed several low density regions. Naturally, some of these regions have low density because they correspond to intangible combination of properties, as well as technical and methodological difficulties. Some areas are subsequently extensively explored due to historical reasons and work focused around certain targets. Subsequently the coverage of chemical space by NPs was studied. The difference in coverage of biologically relevant chemical space by NPs and medicinal chemistry compounds was found to be noteworthy. Interestingly, several of the low density regions, with regard to medicinal chemistry compounds, had been evolutionary explored by Nature and covered by tangible lead-like NPs that could be of interest in drug discovery. Last but not least a number of close neighbours to approved drugs were identified from the NP dataset through calculation of EDs based on ChemGPS-NP coordinates. The central premise of medicinal chemistry, often referred to as the similarity principle47, that compounds with similar molecular properties often have similar biological activities, points towards an increased hit rate when screening these NPs for the biological activity in question. Several of the NPs in the drug/NP pairs revealed by this method, were also confirmed to exhibit the same activity as its drug neighbour. The method we have used here to identify the drug/NP pairs is derived from ChemGPS-NP scores and thus property based, in contrast to the frequently used fingerprint based similarity search methods. Fingerprints (e.g. Daylight48 and UNITY49) are vectors where the elements encode some aspect of the molecular structure, generated solely from the molecular structure. While some of the drug/NP pairs revealed by this property based method are structurally very similar, others are not. Methods based on structural fingerprints would risk missing some of the compound pairs which are structurally dissimilar, but here show up as property neighbours with similar biological activities. One highly appealing feature of property based methods would be the ability to assist in finding new scaffolds for scaffold-hopping or solely as inspiration. Since the revealed neighbours not necessarily are structurally similar it could be possible to overcome toxicological problems, synthetic feasibility issues, and unfavourable ADME properties. Examples of interesting drug/NP pairs revealed here that are not obviously similar with regard to chemical structure are amlodipine (3a)/manoalide (18) and zalcitabine (16b)/littoraline A (16a). Such identification of potential leads from an NP starting point may prove a useful strategy for drug discovery, in the search for novel leads and compounds with unique properties.
MATERIALS AND METHODS
Data sets
Three different data sets were used in this study; The WOMBAT database29, version 2007.01, the Dictionary of Natural Products (DNP) released October 200433, and GVKBIO Drug Database version June 200836.
Descriptor calculation
The molecular descriptors of ChemGPS-NP, and four of the descriptors used to distinguish lead-like compounds (LogP, RTB, RNG, HDO, HAC) were calculated with Dragon Professional 5.350. LogSw was calculated using the on-line software ALOGPS 2.151, 52. All descriptors were calculated from SMILES. Before analyses duplicates, salts, hydration information, and counter-ions were removed and the remaining charges were neutralised. The differences in stereochemistry were ignored since ChemGPS-NP uses only 2D descriptors to map the chemical space.
Database software
Filemaker Pro 8.553 and ISIS/Base54 were used to organize data.
Chemical structures
Chemical structures were drawn using ChemDraw Ultra 11.055.
Data analysis
PCA and PCA score prediction25 were performed using the software SIMCA P+ 11.556, with the training set ChemGPS-NP26. Prior to PCA all data were centred and scaled to unit variance.
Calculation of Euclidean distances
Euclidean distances based on ChemGPS-NP scores between the compounds in GVKBIO_DD and DNP were calculated using an in-house script written in awk, a simple and elegant pattern scanning and processing language. The Euclidean distance was calculated between points P = (p1, p2,….pn) and Q= (q1, q2,….qn) in Euclidean n-space, as defined by:
Supplementary Material
Acknowledgments
The authors are grateful to Theres Meinhard for help with writing the awk script used for calculation of EDs. Part of this work was supported by NIH grant 1U54MH084690-01 (TIO), and Helge Ax:son Johnssons stiftelse (AB).
Footnotes
Supporting Information Available: Clustering of a number of compound sets based on biological activity using ChemGPS-NP. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations: NP, natural product; ChemGPS, chemical global positioning system; WOMBAT, World of Molecular BioAcTivity; CSSM, chemical space of small molecules; COX, cyclo-oxygenase; Ro5, Rule of Five; DNP, dictionary of natural products; PCA, principal component analysis; PC, principal component; MW, molecular weight; LogP, logarithm of the octanol/water partition coefficient; LogSw, logarithm of the intrinsic aqueous solubility; RTB, number of rotatable bonds; RNG, number of rings; HDO, number of H-bond donors; HAC, number of H-bond acceptors; GVKBIO_DD, GVKBIO drug database; EDD, Euclidean distance; ACE, angiotensin-converting enzyme; AT1, angiotensin receptor I; CaCh, calcium channel; PPI, proton pump inhibitor; MAO, monoamine oxidase; SSRI, selective serotonin reuptake inhibitor; HIV-1, human immunodeficiency virus type 1; RT, reverse transcriptase; PR, protease; IN, integrase.
References
- 1.Adams D. The hitch-hiker’s guide to the galaxy. Pan Books; London: 1979. [Google Scholar]
- 2.Bohacek RS, McMartin C, Guida WC. The art and practice of structure-based drug design: a molecular modeling perspective. Med Res Rev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Pollock SN, Coutsias EA, Wester MJ, Oprea TI. Scaffold topologies. 1. Exhaustive enumeration up to eight rings. J Chem Inf Model. 2008;48:1304–1310. doi: 10.1021/ci7003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wester MJ, Pollock SN, Coutsias EA, Allu TK, Muresan S, Oprea TI. Scaffold topologies. 2. Analysis of chemical databases. J Chem Inf Model. 2008;48:1311–1324. doi: 10.1021/ci700342h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cragg GM, Newman DJ, Snader KM. Natural products in drug discovery and development. J Nat Prod. 1997;60:52–60. doi: 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- 6.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 7.Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Rouhi AM. Rediscovering natural products. Chem Eng News. 2003;81:77–91. [Google Scholar]
- 9.Rouhi AM. Betting on natural products for cures. Chem Eng News. 2003;81:93–103. [Google Scholar]
- 10.Larsson J, Gottfries J, Bohlin L, Backlund A. Expanding the ChemGPS chemical space with natural products. J Nat Prod. 2005;68:985–991. doi: 10.1021/np049655u. [DOI] [PubMed] [Google Scholar]
- 11.Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 12.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 13.Quinn RJ, Carroll AR, Pham NB, Baron P, Palframan ME, Suraweera L, Pierens GK, Muresan S. Developing a Drug-like Natural Product Library. J Nat Prod. 2008;71:464–468. doi: 10.1021/np070526y. [DOI] [PubMed] [Google Scholar]
- 14.Lipinski CA. Chris Lipinski discusses life and chemistry after the Rule of Five. Drug Discov Today. 2003;8:12–16. doi: 10.1016/s1359-6446(02)02556-4. [DOI] [PubMed] [Google Scholar]
- 15.Ganesan A. The impact of natural products upon modern drug discovery. Curr Opin Chem Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Grabowski K, Schneider G. Properties and architecture of drugs and natural products. Curr Chem Biol. 2007;1:115–127. [Google Scholar]
- 17.Henkel T, Brunne RM, Müller H, Reichel F. Statistical Investigation into the Structural Complementarity of Natural Products and Synthetic Compounds. Angew Chem Int Ed Engl. 1999;38:643–647. doi: 10.1002/(SICI)1521-3773(19990301)38:5<643::AID-ANIE643>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Lee ML, Schneider G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: application in the design of natural product-based combinatorial libraries. J Comb Chem. 2001;3:284–289. doi: 10.1021/cc000097l. [DOI] [PubMed] [Google Scholar]
- 19.Ertl P, Schuffenhauer A. Cheminformatics analysis of natural products: lessons from nature inspiring the design of new drugs. Prog Drug Res. 2008;66(217):219–235. doi: 10.1007/978-3-7643-8595-8_4. [DOI] [PubMed] [Google Scholar]
- 20.WDI, Derwent World Drug Index, http://www.derwent.com/products/lr/wdi/
- 21.MDDR, MDL Drug Data Report, http://www.prous.com/product/electron/mddr.html.
- 22.Jacoby E, Schuffenhauer A, Popov M, Azzaoui K, Havill B, Schopfer U, Engeloch C, Stanek J, Acklin P, Rigollier P, Stoll F, Koch G, Meier P, Orain D, Giger R, Hinrichs J, Malagu K, Zimmermann J, Roth HJ. Key aspects of the Novartis compound collection enhancement project for the compilation of a comprehensive chemogenomics drug discovery screening collection. Curr Top Med Chem. 2005;5:397–411. doi: 10.2174/1568026053828376. [DOI] [PubMed] [Google Scholar]
- 23.Brickmann K. Compound collection enhancement (CCE) increasing quality and size of the AZ compound collection. Presented at the 4th International conference on compound libraries; Düsseldorf, Germany. October, 2008. [Google Scholar]
- 24.Shelat AA, Guy RK. The interdependence between screening methods and screening libraries. Curr Opin Chem Biol. 2007;11:244–251. doi: 10.1016/j.cbpa.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemometr Intell Lab Syst. 1987;2:37–52. [Google Scholar]
- 26.Larsson J, Gottfries J, Muresan S, Backlund A. ChemGPS-NP: tuned for navigation in biologically relevant chemical space. J Nat Prod. 2007;70:789–794. doi: 10.1021/np070002y. [DOI] [PubMed] [Google Scholar]
- 27.Oprea TI, Gottfries J. Chemography: the art of navigating in chemical space. J Comb Chem. 2001;3:157–166. doi: 10.1021/cc0000388. [DOI] [PubMed] [Google Scholar]
- 28.Rosén J, Lövgren A, Kogej T, Muresan S, Gottfries J, Backlund A. ChemGPS-NPweb - chemical space navigation online. J Comput Aided Mol Des. 2009 doi: 10.1007/s10822-008-9255-y. In Press. [DOI] [PubMed] [Google Scholar]
- 29.WOMBAT database v. 2007.01. Sunset Molecular Discovery LLC. http://www.sunsetmolecular.com/
- 30.Olah M, Mracec M, Ostopovici L, Rad RF, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Mracec M, Oprea TI. WOMBAT: World of Molecular Bioactivity. In: Oprea TI, editor. Cheminformatics in Drug Discovery. Wiley -VCH; New York: 2005. pp. 223–239. [Google Scholar]
- 31.Oprea TI, Tropsha A. Target, chemical and bioactivity databases - integration is key. Drug Discov Today: Technologies. 2006;3:357–365. [Google Scholar]
- 32.Weininger D. SMILES, a chemical language and infromations system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28:31–36. [Google Scholar]
- 33.Dictionary of Natural Products, Chapman & Hall/CRC Press LLC, London, 2004. http://www.ramex.com/title.asp?id=1795/
- 34.Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Oprea TI, Allu TK, Fara DC, Rad RF, Ostopovici L, Bologa CG. Lead-like, drug-like or “Pub-like”: how different are they? . J Comput Aided Mol Des. 2007;21:113–119. doi: 10.1007/s10822-007-9105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The GVKBIO Drug Database. http://www.gvkbio.com/informatics.html/.
- 37.Cocconi G. First generation aromatase inhibitors--aminoglutethimide and testololactone. Breast Cancer Res Treat. 1994;30:57–80. doi: 10.1007/BF00682741. [DOI] [PubMed] [Google Scholar]
- 38.Blanco JG, Gil RR, Bocco JL, Meragelman TL, Genti-Raimondi S, Flury A. Aromatase inhibition by an 11,13-dihydroderivative of a sesquiterpene lactone. J Pharmacol Exp Ther. 2001;297:1099–1105. [PubMed] [Google Scholar]
- 39.Holzer G, Esterbauer H, Kronke G, Exner M, Kopp CW, Leitinger N, Wagner O, Gmeiner BM, Kapiotis S. The dietary soy flavonoid genistein abrogates tissue factor induction in endothelial cells induced by the atherogenic oxidized phospholipid oxPAPC. Thromb Res. 2007;120:71–79. doi: 10.1016/j.thromres.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Hwang JS, Lee SA, Hong SS, Lee KS, Lee MK, Hwang BY, Ro JS. Monoamine oxidase inhibitory components from the roots of . Sophora flavescens Arch Pharm Res. 2005;28:190–194. doi: 10.1007/BF02977714. [DOI] [PubMed] [Google Scholar]
- 41.Lin LZ, Hu SF, Chai HB, Pengsuparp T, Pezzuto JM, Cordell GA, Ruangrungsi N. Lycorine alkaloids from Hymenocallis littoralis. Phytochemistry. 1995;40:1295–1298. doi: 10.1016/0031-9422(95)00372-e. [DOI] [PubMed] [Google Scholar]
- 42.Singh IP, Sandip BB, Bhutani KK. Anti-HIV natural products. Current Science. 2005;89:269–287. [Google Scholar]
- 43.Wheeler LA, Sachs G, De Vries G, Goodrum D, Woldemussie E, Muallem S. Manoalide, a natural sesterterpenoid that inhibits calcium channels. J Biol Chem. 1987;262:6531–6538. [PubMed] [Google Scholar]
- 44.von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 45.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Phan TV, Smythe ML, White HF, Oliver SW, Colman PM, Varghese JN, Ryan DM, Woods JM, Bethell RC, Hotham VJ, Cameron JM, Penn CR. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 46.Brown L. http://www.lesbrown.com.
- 47.Johnson M, Maggiora GM. Concepts and Applications of Molecular Similarity. Wiley; New York: 1990. [Google Scholar]
- 48.Daylight Chemical Information Systems Inc. http://www.daylight.com/.
- 49.UNITY; Tripos Inc. http://www.tripos.com/
- 50.Dragon Professional 5.3 software, Talete srl, Milano, Italy. http://www.talete.mi.it/dragon.htm/.
- 51.VCCLAB, Virtual Computational Chemistry Laboratory, http://www.vcclab.org/
- 52.Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. Virtual computational chemistry laboratory--design and description. J Comput Aided Mol Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 53.Filemaker Pro 8.5. Filemaker Inc. http://filemaker.com/
- 54.ISIS/Base 2.5 SP2. MDL Informations Systems, Inc. http://www.mdl.com/
- 55.ChemDraw Ultra 11.0. CambridgeSoft. http://cambridgesoft.com/
- 56.SIMCA-P+ 11.5 software, Umetrics AB, Umeå, Sweden. http://www.umetrics.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.