Summary
The Notch signaling pathway constitutes an ancient and conserved mechanism for cell-cell communication in metazoan organisms, and has a central role both in development and in adult tissue homeostasis. Here, we summarize structural and biochemical advances that contribute new insights into three central facets of canonical Notch signal transduction: ligand recognition; autoinhibition and the switch from protease resistance to protease sensitivity; and the mechanism of nuclear-complex assembly and the induction of target-gene transcription. These advances set the stage for future mechanistic studies investigating ligand-dependent activation of Notch receptors, and serve as a foundation for the development of mechanism-based inhibitors of signaling in the treatment of cancer and other diseases.
Introduction
Notch receptors are modular, single-pass transmembrane proteins that receive signals from transmembrane ligands that are expressed on neighboring cells. The signals that are transduced by these receptors have a central role in cell-fate decisions both during embryonic development and in adult tissue homeostasis. The essential role of Notch signaling during development is evident from the embryonic lethality that is associated with deficiencies in Notch signaling in various model organisms, including worms, flies and mice. Notch signals are used iteratively at different decision points to guide functional outcomes that depend heavily on gene dose and signaling context. Significantly, both deficiencies and abnormal increases of Notch signaling are also associated with human developmental anomalies and cancer, again emphasizing the importance of precisely regulating the intensity and duration of Notch signals (see (Aster et al., 2008; Bray, 2006) for recent reviews).
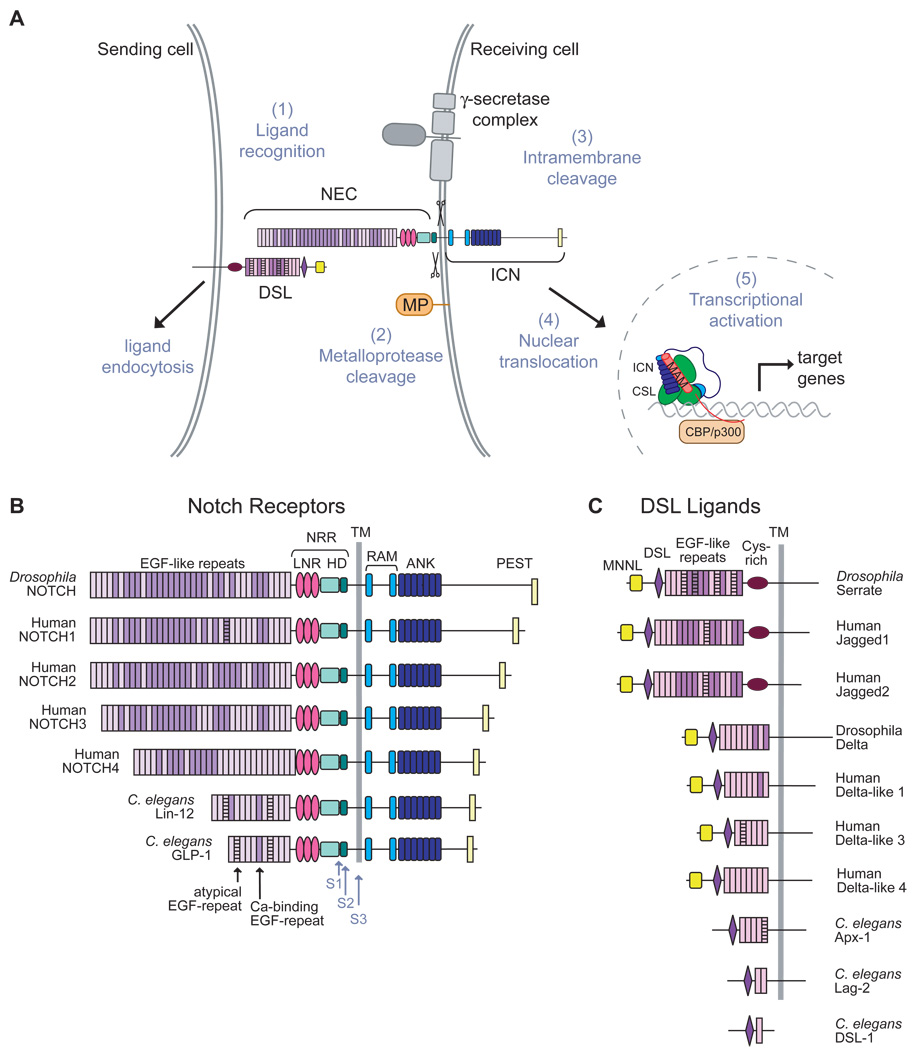
The core protein components of the Notch signaling circuit are present in metazoan organisms ranging from sea urchins to humans (Figure 1A). Canonical Notch signaling begins when a ligand of the Delta/Serrate/Lag-2 (DSL) family binds to Notch at the cell surface (Fehon et al., 1990). Ligand engagement initiates a process called regulated intramembrane proteolysis (RIP), in which the Notch receptor is first cleaved at a juxtamembrane extracellular site by a metalloprotease of the ADAM (a disintegrin and metalloprotease) family (Brou et al., 2000; Mumm et al., 2000). This ligand-dependent cleavage step renders the metalloprotease-processed, truncated receptor sensitive to subsequent intramembrane cleavage(s) by the γ-secretase multiprotein enzyme complex (De Strooper et al., 1999; Struhl and Greenwald, 1999; Ye et al., 1999). Processing by γ-secretase releases the intracellular part of the Notch receptor [ICN; also known as the Notch intracellular domain (NICD)] from the membrane, allowing it to translocate into the nucleus where it assembles into a transcriptional activation complex (Jarriault et al., 1995). The core components of this complex include the DNA-binding transcription factor CSL (C-promoter-binding factor (in mammals; also known as RBP-J)/Suppressor of hairless (in Drosophila melanogaster) /Lag1 (in Caenorhabditis elegans) (Fortini and Artavanis-Tsakonas, 1994; Jarriault et al., 1995; Tamura et al., 1995), the ICN, and a co-activator protein of the Mastermind (MAM)/Lag-3 family (Petcherski and Kimble, 2000a; Petcherski and Kimble, 2000b; Wu et al., 2000). Activation of transcription at CSL binding sites also appears to depend on the recruitment of additional co-activators such as p300 (Fryer et al., 2002; Wallberg et al., 2002), which may constitute the main link between the core Notch-containing complex and the general transcription machinery.
Figure 1.
Notch signaling pathway and domain organization of Notch receptors and DSL ligands. (A) Model for the major events in the Notch signaling pathway. Signals initiated by the engagement of ligand (1) lead to metalloprotease cleavage (MP) at site S2 (2). This proteolytic step allows the cleavage of Notch by the γ-secretase complex at site S3 within the transmembrane domain (3), and release of intracellular notch (ICN) from the membrane (4). ICN translocates to the nucleus where it enters into a transcriptional activation complex with CSL and Mastermind (MAM;5). (B, C) The domain organization of Notch receptors (B) and DSL-family ligands (C) from fly, human and worm.
A number of different proteins are known to modulate Notch signal transduction. The induction of Notch cleavage by ligands relies on E3 ligases such as Neuralized (Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001; Yeh et al., 2001) and Mindbomb (Itoh et al., 2003), which facilitate epsin-dependent endocytosis of ligands in the ligand-expressing cells (Lai et al., 2005; Le Borgne et al., 2005a; Le Borgne et al., 2005b; Overstreet et al., 2004; Wang and Struhl, 2004; Wang and Struhl, 2005). Other modulators of signaling exert their effects by regulating ligand responsiveness (e.g. Fringe and Rumi glycosyltransferases (Acar et al., 2008; Bruckner et al., 2000; Cohen et al., 1997; Johnston et al., 1997; Moloney et al., 2000; Panin et al., 1997)), by controlling ligand and ICN turnover (e.g. Sel-10 and related F-box proteins; (Gupta-Rossi et al., 2001; Hubbard et al., 1997; Mao et al., 2004; Oberg et al., 2001; Tsunematsu et al., 2004; Wu et al., 2001)), and by other less-well-characterized mechanisms (e.g. Deltex-1; (Busseau et al., 1994; Diederich et al., 1994; Matsuno et al., 1995; Matsuno et al., 1998; Mukherjee et al., 2005)). The importance of the C-terminal PEST domain in regulating ICN turnover has also taken on far greater significance in the light of newly identified mutations in human T-cell acute leukemias that delete the PEST region of ICN1 (Weng et al., 2004).
Because there are several recent reviews that describe the molecular events of normal and pathogenic signaling (e.g. (Aster et al., 2008; Bray, 2006)), here we will apply a structural and biochemical perspective to summarize progress towards understanding three central facets of canonical Notch signal transduction: ligand recognition; autoinhibition and the switch from protease resistance to protease sensitivity; and the mechanism of nuclear complex assembly and the induction of target gene transcription. A summary of structures that contain components of the Notch signaling pathway and their complexes is presented in Table 1.
Table 1.
Structures of Notch-pathway-related proteins and their complexes
| Description | PDB | Species | Method | Reference |
|---|---|---|---|---|
| Extracellular Notch | ||||
| LNR-A domain (Notch1) | 1pb5 | human | NMR | (Vardar et al., 2003) |
| Ligand-binding, EGF-like repeats11–13 (Notch1) |
1toz | human | NMR | (Hambleton et al., 2004) |
| LNR-HD (Notch2) | 2oo4 | human | X-ray | (Gordon et al., 2007) |
| Ligand-binding, EGF-like repeats11–13 (Notch1) |
2vj3 | human | X-ray | (Cordle et al., 2008a) |
| Notch Ligands | ||||
| Jagged-1, DSL domain and EGF-like repeats 1–3 | 2vj2 | Human | X-ray | (Cordle et al., 2008a) |
| Intracellular Notch | ||||
| ANK (Notch) | 1ot8 | fly | X-ray | (Zweifel et al., 2003) |
| CSL (Lag1)-DNA | 1ttu | worm | X-ray | (Kovall and Hendrickson, 2004) |
| ANK repeats 3–7 (Notch1) | 1ymp | mouse | X-ray | (Lubman et al., 2005) |
| ANK (Notch1) | 1yyh | human | X-ray | (Ehebauer et al., 2005) |
| ANK(Notch1) | 2f8x | human | X-ray | (Nam et al., 2006) |
| ANK(Notch1)-MAM-CSL(RBP-Jk)- DNA |
2f8y | human | X-ray | (Nam et al., 2006) |
| RAMANK(Lin12)-MAM(Sel8)- CSL(Lag1)-DNA | 2fo1 | worm | X-ray | (Wilson and Kovall, 2006) |
| ICN (Notch) + CSL (Su(H)) | fly | EM | (Kelly et al., 2007) | |
| Hydroxylated ANK (Notch1) | 2qc9 | mouse | X-ray | (Coleman et al., 2007) |
| RAM(Lin12)-CSL(Lag1)-DNA | 3brd | worm | X-ray | (Friedmann et al., 2008) |
| RAM(Lin12)-CSL(Lag1)-DNA | 3brf | worm | X-ray | (Friedmann et al., 2008) |
| CSL(RBP-Jk)-DNA | 3brg | mouse | X-ray | (Friedmann et al., 2008) |
| Extracellular Notch-associated molecules | ||||
| TACE | 1bkc | human | X-ray | (Maskos et al., 1998) |
| Maniac Fringe | 2j0a | mouse | X-ray | (Jinek et al., 2006) |
| Maniac Fringe + UDP/Mn | 2j0b | mouse | X-ray | (Jinek et al., 2006) |
| γ-secretase | EM | (Ogura et al., 2006) | ||
| γ-secretase | EM | (Lazarov et al., 2006) | ||
| Intracellular Notch-associated molecules | ||||
| Supressor of Deltex [Su(dx)] WW domains 3–4 | 1tk7 | fly | NMR | (Fedoroff et al., 2004) |
| Deltex ring-H2 finger | 1v87 | NMR | (Miyamoto et al., 2004) | |
| Deltex WWE domain | 2a90 | fly | X-ray | (Zweifel et al., 2005) |
| Su(dx) WW domain 4/phosphorylated Notch peptide |
2jmf | fly | NMR | (Jennings et al., 2007) |
| S-phase kinase-associated protein 1(Skp1)/F-box and WD repeat domain-containing 7(Fbw7) |
2ovp | human | X-ray | (Hao et al., 2007) |
| Skp1/Fbw7/CyclinE C-term degron | 2ovq | human | X-ray | (Hao et al., 2007) |
| Skp1/Fbw7/CyclinE N-term degron | 2ovr | human | X-ray | (Hao et al., 2007) |
| Factor Inhibiting HIF-1 (FIH)/Notch1 peptide | N/A | mouse | X-ray | (Coleman et al., 2007) |
| FIH/Notch1 peptide | N/A | mouse | X-ray | (Coleman et al., 2007) |
Ligand recognition
Domain organization
The N-terminal part of the Notch ectodomain consists of a series of epidermal growth factor (EGF)-like repeats that are responsible for ligand binding (Figure 1B). Each EGF-like repeat is about 40 residues long and contains six cysteine residues that form three disulfide bonds with a characteristic pairing. The number of EGF-like repeats varies among receptors from different species. The two Notch receptors in the worm, LIN-12 and GLP-1, have 14 and 11 N-terminal EGF-like repeats, respectively, whereas the Notch receptor in the fly and the four mammalian Notch receptors are much larger, having 29–36 EGF-like repeats (Fleming, 1998).
The extracellular domains of Notch ligands of the DSL family also have a modular architecture (Figure 1C). The N-terminal region of the ligands, with the exception of the worm proteins, which are the most divergent among the group, contains a conserved ∼100 amino acid MNNL (Module at the N-terminus of Notch Ligands) domain (Figure 1C). All ligands then contain a distinct cysteine-rich module called a DSL domain near the N-terminus, followed by a series of iterated EGF-like repeats that precede the transmembrane segment. Serrate and Jagged ligands also contain a cysteine-rich domain that bears some sequence similarity to von Willebrand Factor C domains between the EGF-like repeats and the transmembrane domain, whereas the Delta class of ligands does not (Figure 1C).
Notch-ligand interactions
Early studies that investigated the binding of the fly Notch receptor to its ligands used cell-aggregation assays to detect an interaction between cells that expressed Notch and those that expressed the ligand Delta (Fehon et al., 1990). This approach was then used to show that the minimal region of Notch that is necessary and sufficient for association with Delta comprised the EGF-like repeat pair 11–12 (Rebay et al., 1991). In a different assay, the Irvine group constructed a series of fusions between ectodomain fragments of the fly Notch receptor and alkaline phosphatase to assess the binding of these molecules to S2 cells that expressed either the Delta or Serrate ligands (Xu et al., 2005). These studies confirmed the essential importance of EGF-like repeats 11–12 for binding to both ligands, and pointed out that additional repeats between 6–36 also contribute to the strength of binding in this cell-based assay. These studies also investigated the influence of Notch receptor modification by the N-GlcNac transferase Fringe in modulating binding, showing that Fringe modification confers competence for the binding of Delta, but interferes with binding to Serrate. As far as the ligands are concerned, the conserved N-terminal MNNL and DSL domains both appear to participate in optimal binding to Notch (Parks et al., 2006; Shimizu et al., 1999).
We are aware of three published reports of binding assays that were performed in vitro with isolated receptor and ligand ectodomain fragments to quantify binding affinities. The Hirai group used an ELISA assay to detect the binding of soluble mouse Jagged1-Fc fusions to His-tagged mouse Notch2 ectodomain fragments that were immobilized to a surface (Shimizu et al., 1999). They determined a binding affinity of approximately 0.7 nM for the in vitro interaction between Jagged1 and an N-terminal fragment of Notch2 spanning EGF-like repeats 1–15. The Baker group also used an ELISA assay to assess interactions between secreted ectodomains of fly Notch EGF-like repeats 11–20 and Delta, estimating a Kd of 1.87 nM for this interaction from their solidphase binding assays. Interestingly, they also detected an association between a Notch polypeptide containing EGF-like repeats 11–20 and a separate Notch fragment comprising EGF-like repeats 21–30 (Pei and Baker, 2008). This latter region overlaps with the part of Notch that contains the Abruptex class of alleles, a complex group of dominant Notch mutations that appear genetically to result in enhanced Notch signaling. Finally, binding interactions between minimally interacting regions of receptor and ligand were examined by surface plasmon resonance, and a Kd of 130 µM was reported for the binding of bacterially expressed, biotinylated Notch1 EGF-like repeats 11–14 to a Delta-like 1 (Dll1) fragment consisting of the N-terminal domain, the DSL domain and the first three EGF-like repeats. In this study, site-directed mutagenesis of a key calcium-coordinating residue in EGF-like repeat 12 was then used to show that the structural integrity of the calcium binding site in this repeat is required to observe specific binding (Cordle et al., 2008b).
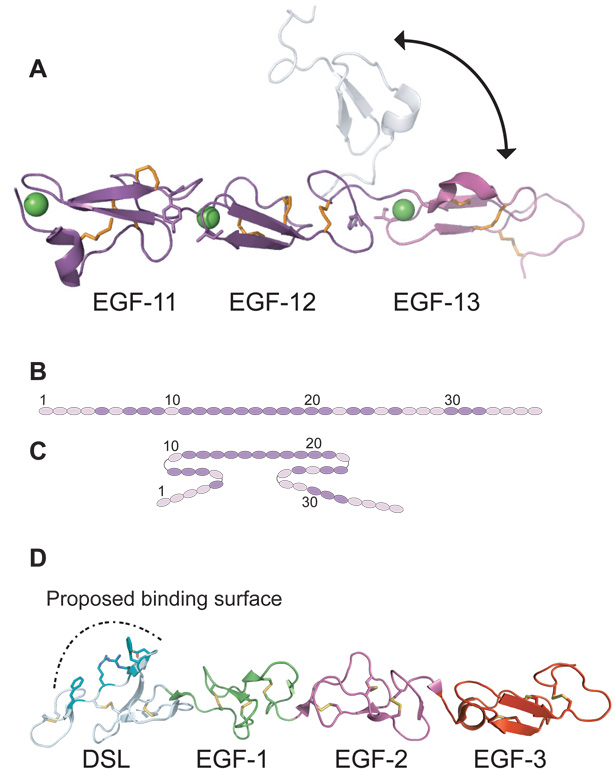
Despite the accruing evidence that multiple regions of both Notch receptors and ligands are involved in the binding interaction that leads to a signal, there is limited structural information available to guide mechanistic understanding of ligand recognition. Handford and colleagues have reported NMR and X-ray structures of a polypeptide comprising EGF-like repeats 11–13 from human NOTCH1 (Cordle et al., 2008a; Hambleton et al., 2004), and an X-ray structure of a region of JAGGED1 consisting of the DSL domain and EGF-like repeats 1–3 (Cordle et al., 2008a). In the structure of the Notch1 polypeptide, each EGF-like repeat in the structure adopts a characteristic EGF-like-repeat fold, which consists of a core two-stranded, antiparallel β-sheet that is oriented parallel to the axis between its N and C termini and three disulfide bonds. In the X-ray structure, the coordination of calcium fixes the orientation between adjacent repeats, creating a gently curving, rod-like structure (Cordle et al., 2008a). By contrast, in the NMR structure, the position of EGF-like repeat 13 is less well defined with respect to repeat 12, suggesting that the interdomain relationship between EGF-like repeats 12 and 13 is more dynamic, and that the position of repeat 13 is not rigidly fixed with respect to repeat 12 (Figure 2A). Based on the structure of these three calcium-binding EGF modules, the authors proposed that the ectodomain of Notch is in an extended conformation, with consecutive calcium-binding EGF-like repeats that form rigid rod-like structures, perhaps interrupted by non-calcium-binding repeats that impart some flexibility (Figure 2B). To account for the results from their studies, Irvine and colleagues proposed an alternative model, in which the Notch molecule adopts a compact global conformation for optimal interaction with DSL ligands (Xu et al., 2005) (Figure 2C). Their model postulates that the Notch ectodomain contains several potential hinge points, allowing the domain to fold onto itself to form a triple-stranded structure upon binding to ligand to afford extra avidity. The hinge points would presumably be derived from interdomain flexibility in the linkers that connect non-calcium-binding EGF-like repeats. The ability of EGF-like repeats 11–20 of the Drosophila Notch receptor to bind in trans to repeats 21–30 in solid-phase binding assays is consistent with this idea. The compact conformation model (Figure 2C) also rationalizes, at least in part, why there are so many EGF-like repeats in extracellular Notch, and creates a model for how variation in the glycosylation state of the receptor might modulate the affinity for different ligands by changing the flexibility of the hinges.
Figure 2.
Structures of NOTCH1 and JAGGED1 ectodomain fragments and models for the Notch ectodomain. (A) NMR structure of EGF-like repeats 11–13 from human NOTCH1 (PDB ID code 1TOZ). Two of 20 calculated structures in the ensemble are shown. The double-headed arrow indicates the range of positions that are occupied by repeat 13 among the 20 calculated structures. Disulfide bonds are colored orange, and hydrophobic residues engaged in interdomain contacts are shown as sticks. Bound calcium ions, placed by homology to other EGF-like-repeat structures, are shown as green spheres. (B, C) Two proposed models for the overall organization of the Notch1 ectodomain. Panel B shows a rod-like, extended model, whereas panel C illustrates one possible compact model. EGF-like repeats (ovals) are shaded dark purple when they contain a consensus calcium-binding site (per the definition used by UNIPROT), and light purple when they do not. (D). Cartoon representation of the X-ray structure for the JAGGED1 polypeptide (PDB code 2VJ2) comprising the DSL domain and EGF-like repeats 1–3. Disulfide bonds are yellow. Residues at the proposed Notch binding surface are rendered as colored sticks.
The X-ray structure of the JAGGED1 polypeptide (residues 187–335, corresponding to the DSL domain and the first three EGF-like repeats) represents the first glimpse of a Notch ligand at high resolution (Cordle et al., 2008a). This ligand-derived polypeptide adopts an extended structure reminiscent of the elongated conformation seen in EGF-like repeats 11–13 of Notch1 itself (Figure 2D). The structure of the DSL domain resembles an EGF-like fold with an N-terminal extension, but the disulfide bonding pattern is different, because one of the canonical disulfides from the EGF-like core is missing and a new disulfide bond is located within the N-terminal appendage. Accompanying pulldown experiments and functional studies in flies support the idea that a key contribution to Notch binding is derived from conserved residues that are exposed along one face of the DSL domain (Figure 2D), pinpointing for the first time a potential receptor binding interface on a Notch ligand.
Unanswered questions
Despite recent progress, the ligand recognition problem remains a persistent gap in the current understanding of Notch signaling, and many unanswered questions remain. First, how do the receptors actually recognize ligands, and what is the stoichiometry of ligand-receptor complexes once they are formed? How much do the binding affinities vary for different ligand-receptor pairs, and how much do regions that flank the key interacting domains contribute to binding affinity and specificity? Lastly, how do post-translational glycosylation events modulate ligand recognition and signaling? The low affinity (130 µM) of EGF-like repeats 11–14 for Dll1 fragments suggests that additional EGF-like repeats may be required on the Notch receptor side, the ligand side, or on both sides to increase avidity, as Irvine’s work suggests. In addition, clustering of ligand or Notch receptor may be necessary to mediate adhesive interactions in a recognition synapse between communicating cells. This idea is supported by atomic-force microscopy studies that measured strong adhesion forces between cells that expressed Notch and those that expressed ligand (Ahimou et al., 2004). Once ligand-receptor complexes are formed, the next step in receptor activation relies on proteolytic release of the intracellular portion of Notch from the membrane. Thus, we next focus on how the protease resistance of Notch is maintained in the absence of ligand, and how it may be released upon ligand engagement.
Activation switch
The negative regulatory region (NRR) of the Notch receptor, which is sandwiched between the ligand binding and transmembrane domains, harbors the structural machinery necessary to maintain metalloprotease resistance in the absence of ligand. The NRR contains three cysteine-rich Lin12/Notch repeats (LNRs) and a “heterodimerization domain” (HD) that contains both the S1 and S2 cleavage sites. Several lines of evidence accumulated over many years led to the conclusion that the NRR is the “activation switch” of the receptor. Receptors that lack the EGF-like repeats are functionally inert (Kopan et al., 1996; Lieber et al., 1993; Rebay et al., 1993; Struhl and Adachi, 1998). By contrast, deletion of the LNR modules or point mutations of key residues within them lead to gain-of-function phenotypes (Greenwald and Seydoux, 1990) and ligand-independent metalloprotease cleavage (Sanchez-Irizarry et al., 2004). The regulatory importance of the NRR is also highlighted by the fact that most Notch1 point mutations and insertions that are found in patients who have T-cell acute lymphocytic leukemia (T-ALL) are located within the NRR (Weng et al., 2004).
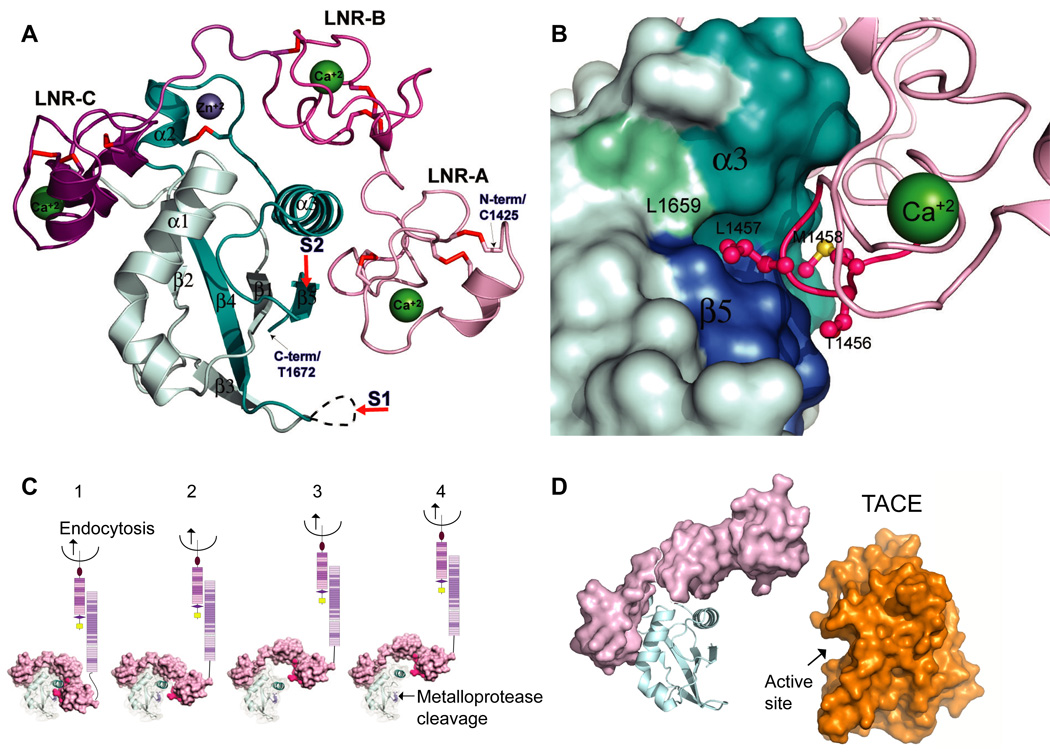
In the crystal structure of the NRR from human NOTCH2, the protein adopts an autoinhibited conformation in which the LNR and HD domains interact extensively to bury the metalloprotease site (Gordon et al., 2007). The LNR domain covers the HD domain, much like a mushroom cap protecting its stem (Figure 3A). Similar to the prototype LNR from human NOTCH1 that was solved by NMR (Vardar et al., 2003), each LNR module has an irregular fold with little secondary structure and is held together by three disulfide bonds and a calcium ion, coordinated by several acidic residues. The two subunits of the HD domain are intimately intertwined in an α-β sandwich, and constitute a single protein domain that resembles the SEA domains of mucins (Maeda et al., 2004). The poorly conserved loop that encompasses the S1 cleavage site, which was excised to facilitate crystallization, is distant from the metalloprotease-cleavage site S2.
Figure 3.
X-ray structure of the human NOTCH2 NRR in the autoinhibited conformation and models for signal activation. (A) Ribbon representation of the NRR. The LNR modules are colored different shades of pink and purple and the HD domain is colored in two shades of cyan; the light and dark cyan represent residues that are N- and C-terminal, respectively, to the furin cleavage loop. The three bound Ca++ ions are green, the bound Zn++ ion is blue, and the ten disulfide bonds are red. The positions of S1 and S2 cleavage are indicated with red arrows. (B) The LNR-AB linker sterically blocks access to the metalloprotease cleavage site. The hydrophobic pocket in the HD domain that houses the S2 site is rendered in a surface representation, and residues from the LNR-AB linker are in ball-and-stick representation. (C) Model for activation by mechanical force. Endocytosis of bound ligand generates a mechanical force that tugs on the LNR domain (panel 1) disengaging the hydrophobic plug from the hydrophobic pocket containing the S2 site (panel 2), and peeling the LNR repeats away from HD domain (panel 3). Partial or complete relaxation of the HD domain then allows access of metalloprotease to the S2 site, and cleavage of the scissile bond to trigger Notch activation (panel 4). (D) Peeling of the LNR repeats away from the HD domain may not confer sufficient exposure of the S2 site to allow cleavage by metalloprotease. The left panel depicts a hypothetical model for the negative regulatory region upon peeling of the first two LNR repeats away from the HD domain; the right panel shows the structure of the catalytic domain of the metalloprotease TACE (PDB ID code 1BKC). The deep active site cleft is indicated with an arrow. Panels A and B are adapted from reference (Gordon et al., 2007) and reproduced with permission (http://www.nature.com/nsmb/).
The interface between the LNR and HD domains buries 3000 Å2 of surface area and the molecular details of these interactions provide the structural basis for the metalloprotease resistance of Notch in the absence of ligand. Three highly conserved residues that are derived from the linker that connects LNRs A and B “plug” the small hydrophobic pocket that contains the metalloprotease site, thereby sterically occluding it (Figure 3B). The preceding helix from the HD domain, which is anchored over the S2 site by hydrophobic interactions with elements from the LNR domain, also precludes metalloprotease access. The structure also suggests that the LNR domain imparts global stabilization to the NRR via its extensive interactions with the HD domain and, most significantly, provides direct evidence that the LNR repeats must be displaced to unmask the S2 site of the HD and allow metalloprotease cleavage.
A key question that remains is how are the LNRs displaced to expose the S2 site? One model, first suggested by Muskavitch to rationalize trans-endocytosis of the Notch ectodomain into ligand-expressing cells, is that bound ligand induces mechanical strain on the Notch receptor (Parks et al., 2000). In light of the NRR structure, this process can be envisioned as one in which ligand stimulation exerts a mechanical force on the Notch receptor, pulling the LNR repeats away from the HD domain to expose the S2 site (Figure 3C). In a mechanotransduction “lift and cut” model with the NRR as the mechanosensor, endocytosis would then be the source of the mechanical force that is needed to peel the protective LNR modules away from the HD domain. This idea is consistent with the known requirement for endocytosis of ligands in Notch signal transduction, and with the finding that soluble ligands typically do not activate Notch receptors (Sun and Artavanis-Tsakonas, 1997). The alternative model for activation would be an allosteric model, in which ligand engagement trips an allosteric switch that disengages the LNR modules from the HD domain. Clearly, additional studies are needed to distinguish between these two models for activation.
In either an allosteric or mechanotransduction model for Notch activation, it is unlikely that the activating metalloproteases can gain access to the Notch S2 site after mere stripping of the LNR modules away from the HD domain, because the active sites of metalloproteases such as tumor-necrosis factor-α (TNF-α)-converting enzyme (TACE; also known as ADAM17) lie in a deep cleft (Figure 3D). Thus, we would expect that certain key secondary structural elements of the HD domain will also unravel after displacement of the LNR repeats, generating an “open” conformation that renders the S2 site accessible to the protease. Such conformational changes in the HD domain might include localized movement or melting of helix3, anchored above the cleavage site (Figure 3B), to release the strand that contains the S2 site, or even complete unfolding of the HD domain with accompanying subunit dissociation (Nichols et al., 2007).
The discovery that mutations are frequently found in the HD domain of NOTCH1 in human T-ALL moved NOTCH1 to the forefront in understanding disease pathogenesis and also pointed to the NOTCH1 NRR as a mechanism-based therapeutic target. These mutations, which map primarily to the highly conserved hydrophobic interior of the HD, lead to ligand-independent increases in signaling, suggesting that domain destabilization facilitates ligand-independent S2 cleavage and subsequent receptor activation (Malecki et al., 2006; Weng et al., 2004). Recently, inhibitory and activating antibodies against the NRR from human NOTCH3 were reported (Li et al., 2008); the epitope of the inhibitory antibody includes residues from both the LNR-A and HD domains, consistent with the notion that it clamps the NRR in its metalloprotease-resistant conformation. Importantly, the existence of modulatory antibodies constitutes proof of principle, showing that it is possible to identify mechanism-based therapeutics that turn Notch signaling on or off via the NRR. Additional studies to define the structural characteristics of the “on” state of the NRR in normal and disease-associated signaling should also help to guide the development of mechanism-based modulators of signaling.
Nuclear translocation and transcriptional regulation
The ICN, which is released from the membrane upon γ-secretase cleavage, is a potent inducer of target gene transcription. In canonical Notch signaling, transcriptional activation depends on the formation of a core protein-DNA complex that includes the ICN, the CSL transcription factor and a protein of the Mastermind-like (MAML) family.
The ICN comprises several functional regions, including an N-terminal RAM (recombination binding protein-Jκ-associated molecule) domain (Tamura et al., 1995), an ankyrin repeat (ANK) domain, and less-conserved regions including a variable transactivation domain (Kurooka et al., 1998) and a C-terminal PEST sequence (Figure 1A). The ANK domain, which comprises seven ankyrin repeats, is the most conserved region of the ICN and is essential for Notch-receptor function. Each ankyrin repeat is about 33 residues long, and typically folds into a pair of antiparallel helices followed by a β-hairpin that connects to the next repeat; together, the repeats stack to form a curved, elongated domain (Mosavi et al., 2004). In X-ray structures of isolated ANK domains from the fly Notch receptor and human NOTCH1, repeats 2–7 are ordered, whereas the first ankyrin repeat remains disordered (Ehebauer et al., 2005; Nam et al., 2006; Zweifel et al., 2003).
The structure of worm CSL (Lag-1) bound to a 13-mer DNA duplex corresponding to a CSL-binding site in the proximal promoter of HES-1, a well-characterized human target of Notch activation, revealed an unpredicted domain organization for CSL (Kovall and Hendrickson, 2004). The structured core of CSL consists of three domains: an N-terminal Rel-homology region (RHR-N); a central β-trefoil domain (BTD); and a C-terminal Rel-homology region (RHR-C). A long β-strand extends from the BTD to the RHR-C, connecting all three domains such that RHR-C sits atop the RHR-N. The DNA is bound by the BTD and the RHR-N and makes no contacts with the distal RHR-C domain. A large surface is buried at the protein-DNA interface with extensive non-specific contacts between the highly conserved and basic CSL face to the phosphate backbone and base-specific contacts with 6 of the 8 base pairs of the core CSL-binding consensus.
Genetic, molecular and biochemical studies of the interaction between CSL and ICN established that binding of ICN to CSL is bipartite, through a higher-affinity, stable interaction with the RAM region (Kovall and Hendrickson, 2004; Lubman et al., 2007; Tamura et al., 1995), and a much weaker, but detectable, interaction with the ANK domain (Aster et al., 1997; Tamura et al., 1995). By contrast, binding of the third key component for transcriptional regulation, MAML, to ICN-CSL complexes on DNA occurs independently of the RAM domain, whereas inclusion of the ANK domain of ICN is necessary and sufficient for recruitment of MAML1 to the CSL-ICN complexes (Nam et al., 2003). The assembly of this ternary complex only requires a short N-terminal region of MAML1, which acts as a potent dominant-negative inhibitor of Notch signaling in functional assays because it lacks the part of the protein that normally recruits the transcriptional machinery (Weng et al., 2003). Neither CSL nor the intracellular portion of Notch1 (ICN1) alone binds detectably to MAML1, but together the two proteins cooperate to bind MAML1 with high affinity. The cooperative assembly of the MAML1-ANK-CSL-DNA complex suggests that a primary function of the ANK domain of ICN1 is to render CSL competent for MAML loading, and leads to two possible recruitment mechanisms: allosteric exposure of a cryptic binding site on one partner of the ICN-CSL complex; or binding to a composite surface that is created upon complexation.
Insights from structures of Notch transcription complexes
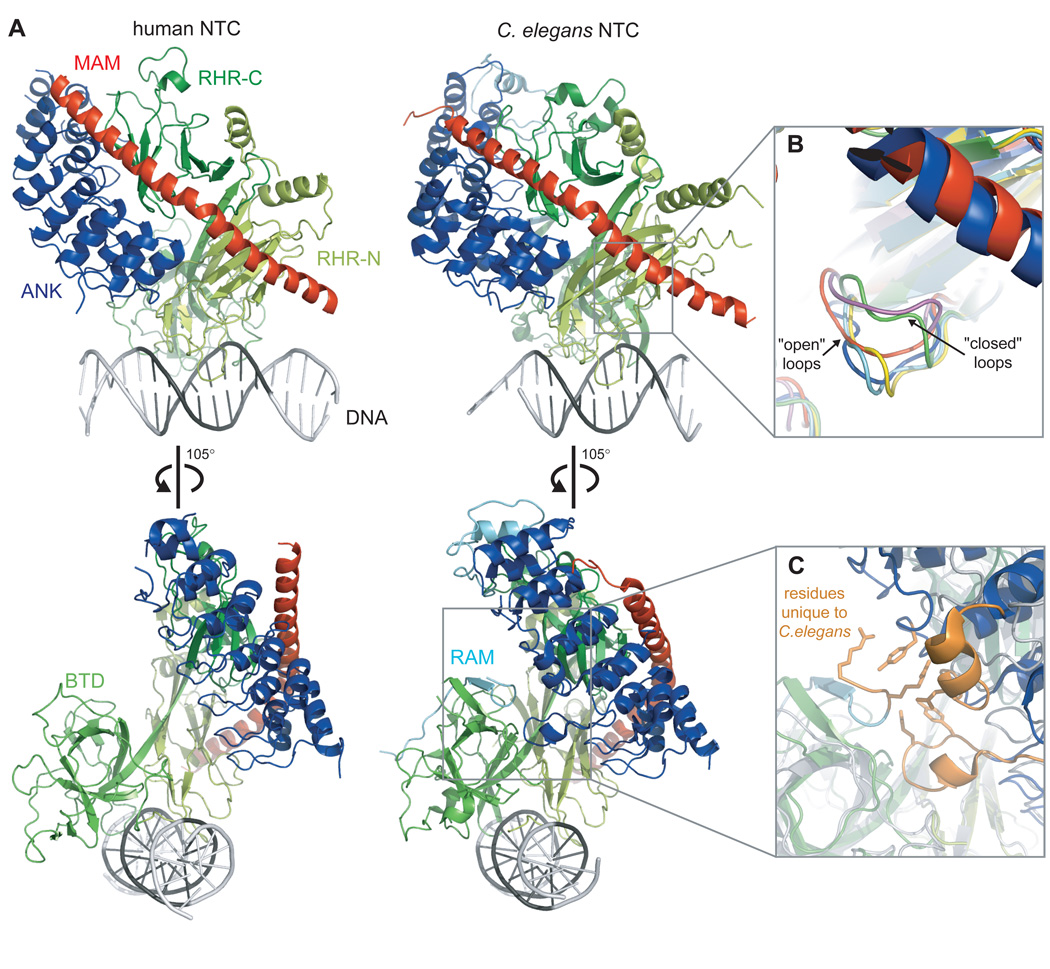
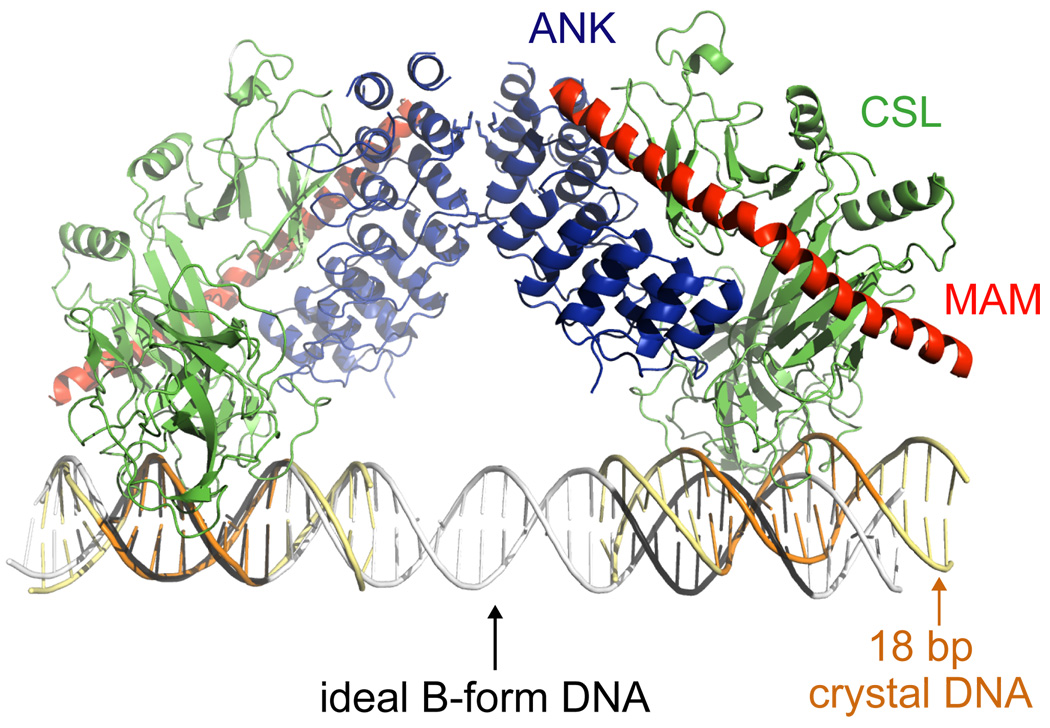
X-ray crystal structures of human and worm Notch transcription complexes (NTCs), which include portions of intracellular Notch, CSL and MAM bound to DNA, provide the structural basis for cooperativity in the recruitment of MAM to Notch transcription complexes (Nam et al., 2006; Wilson and Kovall, 2006). In both the human and worm complexes, the concave face of the Notch ANK domain packs against the two RHR domains of CSL, with the majority of the contact interface derived from RHR-C. The MAML polypeptide, which is disordered in solution in the case of human NOTCH1 (Nam et al., 2006), adopts a helical conformation in both complexes, and makes extensive contacts with a composite surface that is derived from the interface between the ANK and RHR-C domains. A kink in the MAML helix also allows it to wrap further around CSL to contact the RHR-N domain in both cases (Figure 4A).
Figure 4.
Ribbon diagrams representing the structure of human and worm Notch ternary complexes. (A, left) Human complex of the ANK domain of Notch1, CSL and the N-terminal region of Mastermind-like 1 (MAM) bound to an 18 base-pair DNA sequence from the hes1 promoter (PDB code 2F8X). (A, right) Worm complex of the RAMANK region of Lin12, Lag-1, and the N-terminal region of Sel-8 (PDB code 2FO1). The structures illustrate the cooperative binding of MAM to a composite surface that is created at the interface between the Notch ANK domain and CSL. Bottom panels show a 105° rotation around the vertical axis. (B) Superposition of CSL structures showing the difference in loop conformation between CSL-DNA complexes and complexes that include RAM, ANK domains or both. Mouse CSL-DNA (PDB code 3BRG, magenta) and worm CSL-DNA (PDB code 1TTU, green) structures have a “closed” loop. Worm RAM-CSL-DNA (PDB code 3BRF, yellow and PDB code 3BRD, cyan), human ANK-MAM-CSL-DNA (red), and worm RAMANK-MAM-CSL-DNA (blue) complexes have an “open” loop. (C) Superposition of worm (colors) and human (grey) ternary complex structures. Several unique insertions at the N-terminus of RAM and within the ANK domain are found in worm Lin12 but not in other Notch molecules (orange). These features may play a role in the more compact packing of the worm NTC structure when compared to the human NTC structure.
The first repeat of the isolated Notch ANK domain structures is disordered (Ehebauer et al., 2005; Nam et al., 2006; Zweifel et al., 2003), whereas this first repeat is ordered in both the human (Nam et al., 2006) and worm NTC structures (Wilson and Kovall, 2006). The worm ternary complex structure, which includes both RAM and ANK regions, also has electron density that corresponds to two short parts of the RAM region at its N- and C-termini, but the remainder of this region is not visible in the structure, which is consistent with structural disorder in the intervening region. A 20-residue sequence at the N-terminus of RAM, which includes a conserved WXP motif that is implicated in the binding of RAM to the β-trefoil domain of CSL in previous studies, binds to a groove in the BTD of CSL at a site that is distant from the CSL interface with the ANK domain and MAML. The C-terminal part of the RAM region adopts an ankyrin-like fold, which caps the N-terminus of ANK and may help to stabilize the first ankyrin repeat in the complex. Similar to MAM and the first ankyrin repeat of Notch, the N-terminal cap also appears to become ordered only upon complex formation.
When the CSL-DNA and ternary complexes from worm are directly compared, it is evident that CSL adopts a more closed conformation in the complex with RAMANK (constructs of Notch that encompass both the RAM and ANK domains) and MAM than it does alone (Kovall and Hendrickson, 2004; Wilson and Kovall, 2006). Areas of the RHR-N and BTD that are near the DNA interface superimpose well, but the RHR-C and parts of the BTD of CSL in the ternary complex are closer to each other, filling more of the cavity in the U-shaped CSL molecule. By contrast, the CSL molecule in the human NTC complex, which only includes the ANK domain of Notch, remains in a more open conformation that more closely resembles that of the worm CSL-DNA complex.
Role of the RAM region in transcription complex assembly
It had been speculated that this difference between the human and worm ternary complexes was due to the binding of the N-terminal RAM peptide in the worm structure, which was not present in the human structure (Barrick and Kopan, 2006). Alternatively, the conformational differences between the complexes could also be accounted for by species-specific differences in the RAM and ANK regions of the various proteins, or by intrinsic flexibility of the CSL protein (Figure 4). Three recently reported structures, including two worm CSL-RAM-DNA complexes in different crystallographic space groups and a mouse CSL-DNA complex, further clarify the influence of RAM on the conformation of CSL (Friedmann et al., 2008). A significant finding from these studies is that the RAM region does not induce global closure of CSL upon binding, indicating that the primary structural differences between the two ternary complexes are a consequence of interspecies variation or conformational flexibility.
One consistent conformational difference noted among different classes of CSL containing structures is in the position of a loop (connecting strands C and C’) of the RHR-N domain of CSL, which is located directly below the C-terminal portion of MAM (Friedmann et al., 2008). In all structures that contain either the ANK or RAM domain (or both domains, as in the case of the worm ternary complex), this loop is in an “open” conformation with the loop flipped away from MAM, whereas the loop is in a “closed” conformation in the two CSL-DNA structures that were solved in isolation from worm and mouse (Figure 4b). Because a side chain from the loop would clash with one of the MAML side chains when the loop is in the closed position, the authors propose that the binding of RAM to the β-trefoil domain of CSL may trigger opening of the loop in the RHR-N domain, but this possibility has not yet been tested directly. Examination of the structures suggests that a simple side-chain rotation around the β-carbon atom of the loop residue might prevent the clash, but perhaps a movement of the entire loop is the most energetically favored way to accommodate MAML.
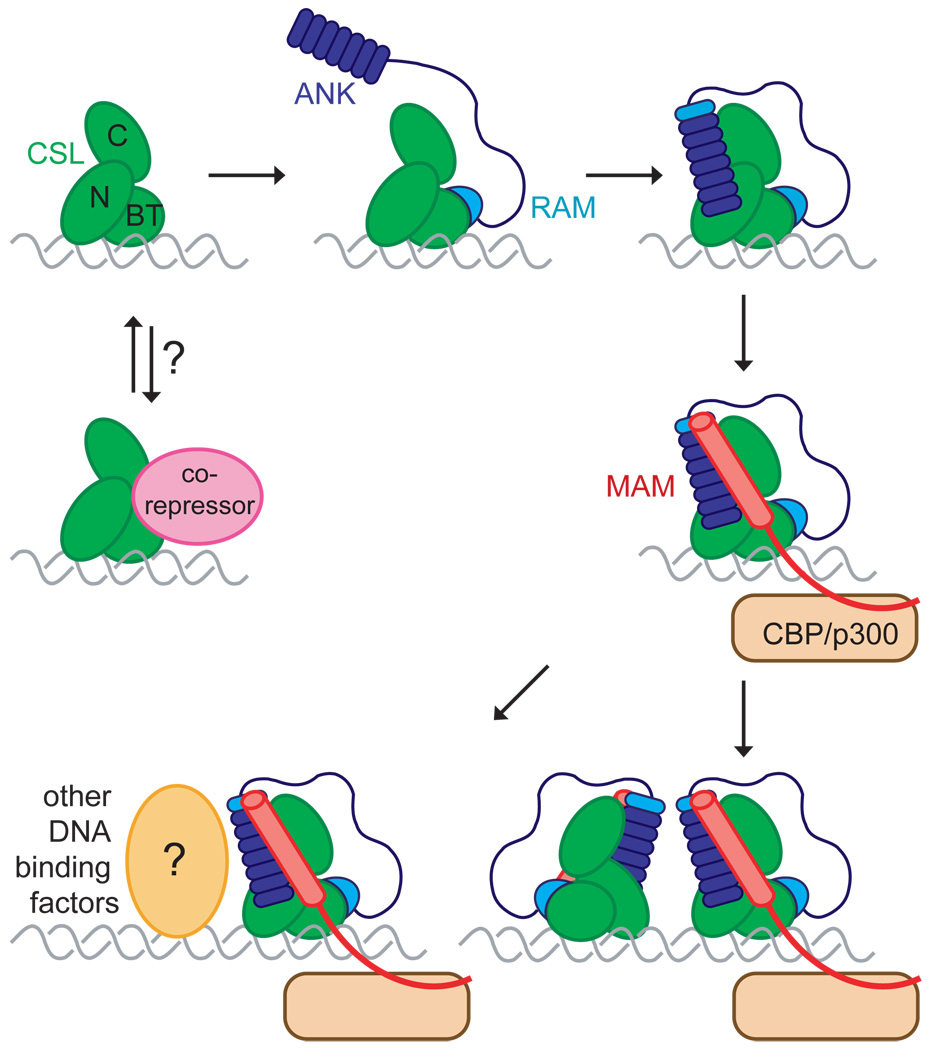
Recent biochemical studies, which complement these crystal structures, suggest that the primary role of the RAM region is to recruit intracellular Notch to CSL. Biophysical studies of the complete RAM region in isolation and in the context of RAMANK polypeptides have shown that RAM is natively unstructured, and density for 70 residues of this unstructured linker sequence between the 20-residue RAM sequence bound to the β-trefoil domain of CSL and the ANK domain remains disordered in the worm transcription complex (Wilson and Kovall, 2006). Modeling of the unstructured region between the bound RAM peptide and the N-terminal end of the ANK domain as a worm-like chain reveals that the most probable distance between the two CSL-binding domains is about 50 Å (Bertagna et al., 2008), which is in very good agreement with the distance between the binding sites in the ternary complex structures. The affinity of RAM peptides for various CSL proteins from different species, which has been determined in several different studies (Del Bianco et al., 2008; Friedmann et al., 2008; Lubman et al., 2007), ranges from 0.022–2 µM, whereas the affinity of the ANK domain for CSL is low (Nam et al., 2003), and has been estimated to be greater than 20 µM (Del Bianco et al., 2008). Above the concentrations that are needed for saturated RAM peptide binding to CSL, the worm-like chain approximation predicts an effective concentration for the ANK domain around 0.5 mM (Bertagna et al., 2008), consistent with fluorescence transfer experiments suggesting that recruitment via RAM leads to docking of the ANK domain onto its cognate site on CSL (Del Bianco et al., 2008). Together, the structural and biochemical studies best support a model for complex assembly in which the high-affinity RAM peptide interacts first with its binding site on CSL, increasing the effective local concentration of the ANK domain and allowing it to nestle onto the RHR part of CSL to create the composite interface for MAML recruitment.
Despite the structural and biochemical information that is now accumulating about the RAM and ANK domains of intracellular Notch, less is known about the overall structure of the entire ICN molecule that is released from the membrane upon γ-secretase cleavage. Recent single-particle electron microscopy analysis of ICN from the fly indicates that intact ICN can adopt either an extended or a compact conformation (Kelly et al., 2007). At low ionic strength, the compact form is favored, whereas increasing calcium concentrations induce a shift from a compact ICN form that cannot bind CSL to an extended form that binds CSL, and ultimately to a fibrous aggregated form that can no longer bind CSL (Kelly et al., 2007). Together, the biochemistry and electron microscopy suggest that modulation of the conformation of ICN may regulate its activity. However, given that the calcium concentrations used in these studies are unlikely to be physiologic, the triggers that may be responsible for such conformational rearrangements remain unknown.
Formation of higher order complexes on specialized promoter elements
A remaining challenge in understanding how ICN acts as an accessory transcription factor is to elucidate the transcriptional codes that constitute Notch response elements in different biological signaling contexts. One small step in that direction emerged from the consideration of the packing arrangement of symmetry-related complexes in the structure of the human Notch transcription complex on an 18-mer CSL consensus binding site from the hes1 promoter region (Nam et al., 2007; Nam et al., 2006). In a number of well-characterized Notch-responsive genes in flies and mammals, including hes-1, the proximal promoter contains dual Su(H)-paired sites or “sequence-paired" binding sites (SPSs), which consist of two CSL binding sites that are oriented head to head and typically separated by 16 or 17 nucleotides (Bailey and Posakony, 1995; Nellesen et al., 1999). Previous studies of genes containing SPS elements have shown that the integrity of the SPS is needed for proper Notch-dependent gene transcription in cell-based assays and transgenic flies (Cave et al., 2005; Ong et al., 2006). In the structure of the human NTC, crystal contacts between two copies of the ANK domain that are related by a two-fold symmetry axis orient the two 18-mer hes1 DNA duplexes head to head in a near-linear orientation that mimicks an inverted-repeat pair about 65 Å apart, corresponding approximately to the distance spanned by 19 base pairs of B-form DNA (Figure 5). Biochemical studies revealed that residues that are engaged in these crystal contacts help guide cooperative dimerization of Notch transcription complexes on the SPS from the hes-1 promoter region, and that disruption of the putative dimer interface within ANK prevents the induction of a luciferase reporter gene under the control of the same promoter region (Nam et al., 2007). The existence of clustered CSL sites in many other Notch responsive genes also raises the possibility that the assembly of higher-order Notch complexes may represent a more general mechanism for regulating target-gene transcription. Taken together with the structural and biochemical studies of complexes that are assembled on single sites, the potential for dimerization of Notch transcription complexes leads to a new working model for the assembly of various Notch nuclear complexes (Figure 6).
Figure 5.
Ribbon diagram of two symmetry-related copies of the human NTC (PDB code 2F8X), revealing the near-linear orientation of the two DNA elements that mimics the inverted repeat of the SPS element. Superposition of the symmetry-related pseudo-dimer (DNA in yellow, with CSL binding sites in orange) on ideal B-form DNA corresponding to 42 base-pairs of the hes1 promoter (grey, with CSL binding sites in black) reveals that the orientation and spacing between the two CSL sites in the crystal approximates, but does not match, the expected spacing and orientation in a natural SPS.
Figure 6.
Model for assembly of Notch ternary complexes. A high-affinity interaction between the N-terminal RAM peptide of Notch and the β-trefoil domain (BTD) of CSL is likely to be the first event in the assembly of Notch transcriptional activation complexes. This step allows the lower-affinity ANK domain to bind at its docking site, resulting in ordering of the ankyrin-like N-cap and first repeat of the ANK domain. The interface between the ANK domain and the RHR-N and RHR-C domains of CSL create a composite surface for the binding of MAM, which recruits CBP/p300. Higher-order homotypic assemblies of Notch complexes or heterotypic assemblies with other transcription factors may be required for transcription of specific Notch targets.
Conclusion/Perspectives
A number of significant structural and biochemical advances over the past several years have informed the current understanding of the molecular logic of Notch signaling, but the picture is by no means complete. There are representative structures of a ligand-binding fragment of human NOTCH1 and of a minimal Notch-binding fragment of human JAGGED1, yet the three-dimensional organizations of the complete Notch and ligand ectodomains remain unknown, as does the structural basis for ligand recognition. Similarly, the X-ray structure of the activation switch of human NOTCH2 has revealed the molecular basis for autoinhibiton, but a clear picture of how ligand stimulation relieves this autoinhibition has yet to emerge.
With regard to Notch transcription complexes, structural and biochemical studies of the core assemblies from human and worm provide substantial insight into how ICN and CSL combine to recruit MAM proteins. However, eukaryotic gene transcription usually requires the concerted binding of multiple DNA-binding transcription factors, and Notch-responsive genes are no exception. Understanding the structural basis for cooperativity between the Notch transcription complex and other DNA-binding transcription factors is likely to be one future research frontier. Another may be how the core transcription complexes engage other key transcriptional regulators, such as histone acetyl transferases and chromatin-remodeling complexes, to activate gene transcription. Deciphering the additional complexity that results from post-translational modifications, such as phosphorylation and hydroxylation, represents yet another future challenge. These problems, along with other unresolved issues in the biochemistry and signaling of the Notch pathway, should keep researchers busy for several years to come.
Abbreviations used
- ANK
The ankyrin repeat region of intracellular Notch.
- BTD
beta-trefoil domain.
- CSL
The nuclear transcription factor bound by activated Notch, also known as RBP-Jκ, recombination-signal-sequence-binding protein for Jκ genes, or CBF1 in mammals, Suppressor of Hairless in D. melanogaster, and LAG-1 in C. elegans.
- DSL
Canonical Notch ligand of the Delta, Serrate, or Lag-2 family.
- EGF
Epidermal Growth Factor, EGF-like domains are present in the extracellular domain of Notch and ligands
- GSI
Gamma-secretase inhibitor. Gamma secretase is the multisubunit membrane protease that cleaves Notch receptors at site S3 during receptor activation.
- HD
Heterodimerization domain. This domain immediately precedes the membrane, and contains the furin (S1) and metalloprotease (S2) cleavage sites.
- HES-1
Human Enhancer-of-split homolog 1, a Notch target gene containing sequence-paired sites in its proximal promoter region.
- ICN
Intracellular Notch, also called NICD, Notch intracellular domain.
- LNR
Lin12-Notch repeat. Each repeat is about 35–40 residues long, and contains three disulfide bonds.
- MAM
Mastermind, a specialized transcriptional co-activator that binds to ICN/CSL complexes. The three Mastermind-like proteins in mammals are designated MAML1–3.
- MNNL
Module at the N-terminus of Notch Ligands
- NEC
Notch extracellular subunit.
- NRR
Notch Negative regulatory region, which includes the three Lin12-Notch repeats (LNRs) and the heterodimerization domain.
- NTC
Core Notch transcription complex, which includes parts of Mastermind-like 1, Intracellular Notch, and CSL.
- NTM
Notch transmembrane subunit.
- RAM
RBP-Jκ associated molecule, the region of intracellular Notch immediately C-terminal to the transmembrane segment.
- RAMANK
Constructs of Notch encompassing both the RAM and ANK domains.
- RHR
Rel-homology region.
- SPS
Sequence paired site or Suppressor of Hairless paired site. Specialized Notch responsive elements with two CSL binding sites arranged head to head.
- TACE
TNF-alpha converting enzyme, a metalloprotease implicated in Notch cleavage at S2
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi Is a CAP10 Domain Glycosyltransferase that Modifies Notch and Is Required for Notch Signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahimou F, Mok LP, Bardot B, Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J Cell Biol. 2004;167:1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Robertson ES, Hasserjian RP, Turner JR, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Barrick D, Kopan R. The Notch transcription activation complex makes its move. Cell. 2006;124:883–885. doi: 10.1016/j.cell.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem Soc Trans. 2008;36:157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- Busseau I, Diederich RJ, Xu T, Artavanis-Tsakonas S. A member of the Notch group of interacting loci, deltex encodes a cytoplasmic basic protein. Genetics. 1994;136:585–596. doi: 10.1093/genetics/136.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave JW, Loh F, Surpris JW, Xia L, Caudy MA. A DNA transcription code for cell-specific gene activation by notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, Whiting E, Ryan D, Zinyk D, Boulianne G, et al. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, Cook KM, Cockman ME, Lancaster DE, Kessler BM, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Zi Yan, Tay J, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008a doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle J, Redfieldz C, Stacey M, van der Merwe PA, Willis AC, Champion BR, Hambleton S, Handford PA. Localization of the Delta-like-1-binding Site in Human Notch-1 and Its Modulation by Calcium Affinity. J Biol Chem. 2008b;283:11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich RJ, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 1994;120:473–481. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- Ehebauer MT, Chirgadze DY, Hayward P, Martinez Arias A, Blundell TL. High-resolution crystal structure of the human Notch 1 ankyrin domain. Biochem J. 2005;392:13–20. doi: 10.1042/BJ20050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff OY, Townson SA, Golovanov AP, Baron M, Avis JM. The structure and dynamics of tandem WW domains in a negative regulator of notch signaling, Suppressor of deltex. J Biol Chem. 2004;279:34991–35000. doi: 10.1074/jbc.M404987200. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Friedmann DR, Wilson JJ, Kovall RA. RAM induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008 doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Greenwald I, Seydoux G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature. 1990;346:197–199. doi: 10.1038/346197a0. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional Interaction between SEL-10, an F-box Protein, and the Nuclear Form of Activated Notch1 Receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, Handford PA, Downing AK. Structural and functional properties of the human notch-1 ligand binding region. Structure. 2004;12:2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Wu G, Kitajewski J, Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jennings MD, Blankley RT, Baron M, Golovanov AP, Avis JM. Specificity and autoregulation of Notch binding by tandem WW domains in suppressor of Deltex. J Biol Chem. 2007;282:29032–29042. doi: 10.1074/jbc.M703453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chen YW, Clausen H, Cohen SM, Conti E. Structural insights into the Notch-modifying glycosyltransferase Fringe. Nat Struct Mol Biol. 2006;13:945–946. doi: 10.1038/nsmb1144. [DOI] [PubMed] [Google Scholar]
- Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- Kelly DF, Lake RJ, Walz T, Artavanis-Tsakonas S. Conformational variability of the intracellular domain of Drosophila Notch and its interaction with Suppressor of Hairless. Proc Natl Acad Sci U S A. 2007;104:9591–956. doi: 10.1073/pnas.0702887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. Embo J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region [published erratum appears in Nucleic Acids Res 1999 Mar 1;27(5):following 1407] Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci U S A. 2006;103:6889–6894. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005a;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005b;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, Scoggin S, Fu T, Vien L, Histen G, et al. Modulation of Notch Signaling by Antibodies Specific for the Extracellular Negative Regulatory Region of NOTCH3. J Biol Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Lubman OY, Ilagan MX, Kopan R, Barrick D. Quantitative dissection of the Notch:CSL interaction: insights into the Notch-mediated transcriptional switch. J Mol Biol. 2007;365:577–589. doi: 10.1016/j.jmb.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubman OY, Kopan R, Waksman G, Korolev S. The crystal structure of a partial mouse Notch-1 ankyrin domain: repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005;14:1274–1281. doi: 10.1110/ps.041184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, Seki E, Matsuda T, Motoda Y, Kobayashi A, et al. Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16) J Biol Chem. 2004;279:13174–13182. doi: 10.1074/jbc.M309417200. [DOI] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, et al. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc Natl Acad Sci U S A. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Eastman D, Mitsiades T, Quinn AM, Carcanciu ML, Ordentlich P, Kadesch T, Artavanis-Tsakonas S. Human deltex is a conserved regulator of Notch signalling. Nat Genet. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Muto Y, Tochio N, Koshiba S, Inoue M, Yabuki T, Aoki M, Tomo Y, Seki E. Solution structure of the RING-H2 finger domain of mouse Deltex protein 2. Nippon Bunshi Seibutsu Gakkai Nenkai Puroguramu, Koen Yoshishu. 2004;27:815. [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The notch intracellular domain is ubiquitinated and negatively regulated by the mammalian sel-10 homolog. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Ogura T, Mio K, Hayashi I, Miyashita H, Fukuda R, Kopan R, Kodama T, Hamakubo T, Iwatsubo T, Tomita T, et al. Three-dimensional structure of the gamma-secretase complex. Biochem Biophys Res Commun. 2006;343:525–534. doi: 10.1016/j.bbrc.2006.02.158. [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Overstreet E, Fitch E, Fischer JA. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 2004;131:5355–5366. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Parks AL, Stout JR, Shepard SB, Klueg KM, Dos Santos AA, Parody TR, Vaskova M, Muskavitch MA. Structure-function analysis of delta trafficking, receptor binding and signaling in Drosophila. Genetics. 2006;174:1947–1961. doi: 10.1534/genetics.106.061630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- Pei Z, Baker NE. Competition between Delta and the Abruptex domain of Notch. BMC Dev Biol. 2008;8:4. doi: 10.1186/1471-213X-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000a;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J. Mastermind is a putative activator for Notch. Curr Biol. 2000b;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development. 1997;124:3439–3448. doi: 10.1242/dev.124.17.3439. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- Vardar D, North CL, Sanchez-Irizarry C, Aster JC, Blacklow SC. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry. 2003;42:7061–7067. doi: 10.1021/bi034156y. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF Act Cooperatively To Mediate Transcriptional Activation from Chromatin Templates by Notch Intracellular Domains In Vitro. Mol Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- Wang W, Struhl G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 2005;132:2883–2894. doi: 10.1242/dev.01860. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 Is an Inhibitor of Notch Signaling That Targets Notch for Ubiquitin-Mediated Protein Degradation. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- Xu A, Lei L, Irvine KD. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J Biol Chem. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. [DOI] [PubMed] [Google Scholar]
- Ye Y, Lukinova N, Fortini ME. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- Yeh E, Dermer M, Commisso C, Zhou L, McGlade CJ, Boulianne GL. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr Biol. 2001;11:1675–1679. doi: 10.1016/s0960-9822(01)00527-9. [DOI] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Barrick D. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure (Camb) 2005;13:1599–1611. doi: 10.1016/j.str.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Hughson FM, Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12:2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]