Abstract
Both p53 and its repressor Mdm2 are subject to ubiquitination and proteasomal degradation. We show that knockdown of the deubiquitinating enzyme USP5 (isopeptidase T) results in an increase in the level and transcriptional activity of p53. Suppression of USP5 stabilizes p53, whereas it has little or no effect on the stability of Mdm2. This provides a mechanism for transcriptional activation of p53. USP5 knockdown interferes with the degradation of ubiquitinated p53 rather than attenuating p53 ubiquitination. In vitro studies have shown that a preferred substrate for USP5 is unanchored polyubiquitin. Consistent with this, we observed for the first time in a mammalian system that USP5 makes a major contribution to Lys-48-linked polyubiquitin disassembly and that suppression of USP5 results in the accumulation of unanchored polyubiquitin chains. Ectopic expression of a C-terminal mutant of ubiquitin (G75A/G76A), which also causes the accumulation of free polyubiquitin, recapitulates the effects of USP5 knockdown on the p53 pathway. We propose a model in which p53 is selectively stabilized because the unanchored polyubiquitin that accumulates after USP5 knockdown is able to compete with ubiquitinated p53 but not with Mdm2 for proteasomal recognition. This raises the possibility that there are significant differences in proteasomal recognition of p53 and Mdm2. These differences could be exploited therapeutically. Our study reveals a novel mechanism for regulation of p53 and identifies USP5 as a potential target for p53 activating therapeutic agents for the treatment of cancer.
Ubiquitination of proteins plays a key role in the regulation of many important pathways in the cell (1). It can act as a signal which targets proteins for degradation by the 26 S proteasome and can also control protein activity and localization (2). Alterations in the ubiquitin-proteasome system have been implicated in a range of diseases including cancer, and there is considerable interest in components of this pathway as targets for therapeutic intervention. Bortezomib, a direct inhibitor of the protease activity of the proteasome, is used in cancer therapy. It is a standard treatment for multiple myeloma. However, it is not effective as a single agent for the treatment of a number of other types of cancers, and trials are under way to test its efficacy in combination with other therapeutic agents (3, 4).
The 26 S proteasome is a large protein complex composed of one or two 19 S regulatory cap complexes and a 20 S core. The 19 S cap participates in ubiquitin recognition and mediates the unfolding of proteins targeted for degradation. The 20 S core carries out protein degradation (5, 6). The sequential action of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) mediates the conjugation of ubiquitin to target proteins (7). Proteins can be conjugated to one molecule of ubiquitin (monoubiquitinated) to multiple single ubiquitin molecules at different sites (multiply monoubiquitinated) and to chains of ubiquitin (polyubiquitinated). It is generally thought that a chain of at least four ubiquitin molecules is required for efficient recognition by the proteasome (8). However, there are examples where monoubiquitination is sufficient to target proteins for degradation by the proteasome (9-11). Ubiquitin is conjugated to proteins through the formation of an isopeptide bond between its C terminus (Gly-76) and most frequently the ∊-side chain amino group of a lysine residue in the target protein. Polyubiquitin chains are similarly generated by isopeptide bond formation between the C terminus of one ubiquitin and the ∊-side chain amino group of a lysine residue in the next ubiquitin in the chain. In free/unanchored polyubiquitin (polyubiquitin not conjugated to a target protein), one end of the chain, which is referred to as the proximal end, has a ubiquitin with a free C terminus. Any of seven lysines in ubiquitin can be used to form an isopeptide bond with another ubiquitin (12, 13). Lys-48-linked chains are predominantly involved in targeting proteins for proteasomal degradation.
Ubiquitin-ubiquitin and ubiquitin-protein bonds can be cleaved by the action of deubiquitinating enzymes (DUBs).4 There are five subclasses of DUBs, the largest of which is the ubiquitin-specific protease (USP) family (14). Some DUBs remove ubiquitin from substrates before proteasomal recognition, resulting in inhibition of substrate degradation. Another role of DUBs is to regulate the pools of unanchored ubiquitin and polyubiquitin. Unanchored isopeptide bond-linked polyubiquitin is generated as a result of deubiquitination of proteins and by de novo synthesis (15, 16). One source of free polyubiquitin is the deubiquitination of proteins at the proteasome. After recognition of the ubiquitinated protein by the proteasome, the ubiquitin is released. This is necessary for entry of proteins into the proteasome (17). Unanchored polyubiquitin is disassembled to regenerate free ubiquitin. The in vitro substrate specificity of USP5 (isopeptidase T) is consistent with an involvement of this enzyme in disassembly of free polyubiquitin. USP5 sequentially removes ubiquitin from the proximal end of unanchored polyubiquitin chains (15, 18-20). Homologues of USP5 are required for the dissociation of free polyubiquitin in Saccharomyces cerevisiae and Arabidopsis thaliana (Ubp14) (21, 22) and Dictyostelium discoideum (UbpA) (23). The consequences of USP5 suppression have not been previously investigated in a mammalian system.
The ubiquitin-proteasome system plays a major role in regulation of the p53 pathway. In most cases tumor progression requires loss of p53 function because of the protective role of p53 against tumor development. This can occur through inactivating mutations in p53. However, 50% of tumors retain wild-type p53, whose function is at least partially attenuated by other mechanisms. The activation of p53 by non-genotoxic agents is a therapeutic approach for the treatment of those cancers which express wild-type p53 (24, 25). Recent studies with animal models of cancer have clearly highlighted the therapeutic potential of restoring p53 function (26, 27). p53 is regulated by ubiquitination at several levels. p53 is ubiquitinated and undergoes proteasomal degradation. In addition, repressors of p53 including Mdm2 are also ubiquitinated and are degraded by the proteasome. Changes in the stability of p53 and Mdm2 participate in p53 activation after cellular stress (28, 29). Mdm2 can repress the transcriptional activity of p53 by binding to its transactivation domain. Mdm2 is also an E3 ubiquitin ligase for p53. Mdm2 undergoes “autoubiquitination” in vitro or when over-expressed; however, studies using mouse embryo fibroblasts expressing a knockin mutant of Mdm2 without intrinsic E3 ligase activity indicate that another E3 is involved in ubiquitination of Mdm2 in these cells (30). The effect of direct inhibitors of the proteolytic activity of the proteasome on the activity of wild-type p53 is complex. In some studies inhibitors have been reported to block p53 degradation and increase p53 trans-activation activity (31, 32). In other studies they have been shown to stabilize wild-type p53 without increasing its transcriptional activity (28, 33, 34). A likely reason for this is that by stabilizing Mdm2, direct inhibitors of the protease activity of the proteasome cause accumulation of sufficient Mdm2 to repress p53 by direct binding (28). In addition, a transcription-independent direct effect of p53 at the mitochondria has been implicated in cell death resulting from proteasome inhibition (35). Sensitivity to proteasome inhibition has been reported to be partially dependent on the p53 status in some cancer cells and to be independent of p53 in others (36).
p53 has been shown to be regulated by the DUBs herpesvirus associated ubiquitin specific protease (HAUSP) /USP7 (29, 37) and USP2a (38), which selectively deubiquitinate components of the p53 pathway. In this study we carried out a screen using an shRNA library targeting members of the DUB family (39) with the aim of identifying additional DUBs whose suppression results in p53 activation. USP5 was identified as a regulator of p53 in this screen. Knockdown of USP5 stabilizes p53 while having little or no effect on the stability of Mdm2. This results in an increase in the levels and transcriptional activity of p53. USP5 can make a major contribution to the disassembly of free polyubiquitin in mammalian cells. Our data indicate that the effects of USP5 knockdown on the p53 pathway are mediated by the accumulation of unanchored polyubiquitin. The differential effect of USP5 knockdown on the stability of Mdm2 and p53 can be explained by differences in sensitivity to competition with free polyubiquitin for recognition by the proteasome.
EXPERIMENTAL PROCEDURES
Cell Culture
ARN8 cells were derived from wild-type p53-expressing A375 human melanoma cells by stable transfection with a p53-responsive reporter construct (RGCΔFos-LacZ) (40). ARN8 cells were cultured in Dulbecco's modified Eagle's medium, HCT116 p53+/+ and p53−/− cells were cultured in McCoy's 5A medium, and H1299 cells were cultured in RPMI, each supplemented with 10% fetal calf serum and 50 μg/ml gentamycin. Cells were grown at 37 °C, 5% CO2 in a humidified atmosphere. HCT116 p53−/− cells were generated from the parental cell line by homologous recombination (41).
Plasmids and Synthetic siRNA Duplexes
For the His6-ubiquitin construct, a single copy of human ubiquitin was cloned into the HindIII and XhoI sites of pcDNA3. The G75A/G76A ubiquitin mutant was generated from wild-type pcDNA3 His6-ubiquitin using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The shRNA library targeting 50 deubiquitinating enzymes was constructed as described previously using the pSUPER vector (39) and was generously provided by Professor René Bernards (The Netherlands Cancer Institute). Non-targeting control duplex #2 and synthetic siRNA duplexes A (D-006095-02) and B (D-006095-03) complementary to USP5 were purchased from Dharmacon. There are two known alternatively spliced forms of USP5 which differ by an insertion of 23 amino acids. The substrate specificity of the isoforms appears to be identical in vitro (42). The siRNA USP5 (A) and (B) used in this study target both splice forms. The sequences of p53 siRNA (A) and (B) were GACTCCAGTGGTAATCTAC (43) and GCATGAACCGGAGGCCCAT (44), respectively.
Transfections
For the screen with the shRNA library, ARN8 cells were seeded onto 24-well tissue culture plates (1.5 × 104 cells/well) and transfected in duplicate with 0.8 μg of empty pSUPER vector or each pool of DUB shRNA using FuGENE 6 transfection reagent (Roche Applied Science) following the manufacturer's instructions. β-Galactosidase activity was assayed 72 h after transfection. For transfection with synthetic siRNA ARN8 or H1299 cells, well were seeded onto 6-well plates (6 × 104 cells/well) or 24-well plates (1.5 × 104 cells/well). HCT116 cells were seeded onto 6-well plates (0.5–1 × 105 cells/well). Transfection was carried out using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The final concentration of each siRNA was 30 nm. Cells were harvested 72 h post-transfection. For transfection with wild-type or mutant ubiquitin, cells were trypsinized, and 1 × 106 ARN8 cells in suspension were nucleofected with 10 μg of plasmid using nucleofection kit V (Amaxa) and program X-001 according to the manufacturer's instructions. Cells were seeded onto 6-well plates (1.5 to 3 × 105 cells/well) or 24-well plates (3 × 104 cells/well) and harvested 24 h after transfection.
Quantitative Detection of β-Galactosidase
Cells in 24-well plates were washed twice with phosphate-buffered saline and lysed with 100 μl of passive lysis buffer (Promega). 150 μl of substrate solution containing 80 μg/ml chlorophenol red β-d-galactopyranoside (Roche Applied Science), 0.5 mm MgCl2, 23 mm β-mercaptoethanol in 0.1 m sodium phosphate buffer, pH 7.5, was added to 40 μl of extract in a 96-well plate. The assay was quantified by absorbance measurements at a wavelength of 590 nm in a plate reader.
Gel Electrophoresis and Western Blotting
Cells were washed twice with phosphate-buffered saline at 4 °C. Cell extracts were prepared by direct lysis into SDS-urea electrophoresis sample buffer: 100 mm Tris, pH 6.8, 4% SDS, 8 m urea, 20% glycerol, 20 mm EDTA, 0.014% bromphenol blue. DNA was sheared by passage through a 25-gauge needle, and protein concentrations were measured using the BCA protein assay (Pierce). DTT was added to a final concentration of 100 mm, samples were heated for 5 min at 95 °C, and proteins were resolved by SDS-PAGE. Gels were transferred onto nitrocellulose for 16 h at 25 mA, and membranes were processed as described previously (45). To expose epitopes in ubiquitin, membranes to be probed for ubiquitin were boiled in deionized water for 30 min before blocking. Peroxidase-conjugated secondary anti-mouse antibodies (Jackson ImmunoResearch Laboratories) were used at a dilution of 1/10,000 and anti-rabbit antibodies (Pierce) at a dilution of 1/2,000. Bound antibodies were detected by enhanced chemiluminescence (Amersham Biosciences) or using Supersignal West Dura extended duration substrate (Pierce). The primary antibodies used were 4B2 for Mdm2, DO-1 for p53, Ab-1 for β-actin (Calbiochem), and Ubi-1 (Novus) and P4D1 (Upstate) for ubiquitin. Rabbit anti-USP5 and anti-Ser-15-phosphorylated p53 polyclonals were obtained from Proteintech Group Inc. and Cell Signaling Technology, respectively. A mouse monoclonal anti-His6 antibody was obtained from Clontech.
RNA Preparation and Real Time PCR
RNA was extracted using RNeasy columns (Qiagen), and real time PCR was carried out as described previously (45). Probes and primers for p53, Mdm2 P1 and P2, Bax, p21 (Waf1/Cip1), and β-actin were as described previously (38, 45). The USP5-specific primers and probes were as follows: 5′-CGGGACCAGGCCTTGAA-3′, 5′-TCGTCAATGTGACTGAAGATCCA-3′; probe, 6-FAM-CGCTGCGGGCCACGAACAATA-TAMRA.
Immunofluorescence
Cells were seeded on glass slides. After transfection with synthetic siRNA duplexes, cells were fixed with ice-cold methanol-acetone and incubated with primary antibodies followed by Alexa Fluor 488 dye-conjugated anti-mouse and Alexa Fluor 594 dye-conjugated anti-rabbit secondary antibodies (Invitrogen) as described previously (45). Monoclonal antibody 4B2 was used to detect Mdm2 and rabbit polyclonal CM1 for p53.
Immunoprecipitations
To preserve ubiquitination after lysis immunoprecipitation was carried out essentially as described previously (46). After transfection with siRNA duplexes, cells were lysed in SDS immunoprecipitation lysis buffer: phosphate-buffered saline containing 1% Nonidet P-40, 1% SDS, 5 mm EDTA, 10 mm iodoacetamide, 1 mm DTT, and protease inhibitor mixture (Roche Applied Science). The lysates were incubated at 60 °C for 10 min. DNA was sheared by passage through a 25-guage needle, and protein concentrations were measured using the BCA protein assay (Pierce). The pooled lysates from one 6-well plate were used for each immunoprecipitation (total volume 300 μl). The samples normalized for protein were diluted 10-fold with immunoprecipitation wash buffer: phosphate-buffered saline containing 1% Nonidet P-40, 5 mm EDTA, 1 mm DTT, and protease inhibitor mixture (Roche Applied Science). The diluted lysates were pre-cleared with a 200-μl packed volume of Sepharose beads by incubation for 45 min at 4 °C. The lysates were then incubated with 10 μl of CM1 or 10 μl of preimmune rabbit serum for 2 h at 4 °C. 20 μl packed volume of protein G-Sepharose beads were added, and the samples incubated for a further hour. The beads were washed five times with immunoprecipitation wash buffer and eluted in SDS-urea sample buffer. The immunoprecipitates were analyzed by Western blotting for p53 and ubiquitin using antibodies DO-1 and P4D1, respectively.
In Vitro Deubiquitination Assay
After transfection with siRNA, cells were lysed in Nonidet P-40 buffer: 0.1% Nonidet P-40, 50 mm Tris, pH 7.5, 150 mm NaCl, 5% glycerol, 1 mm DTT protease inhibitor mixture (Roche Applied Science). The lysates were centrifuged (14,000 × g, 10 min, 4 °C). The indicated amount of the extracts was added to 15 μl of assay buffer: 50 mm Tris, pH 7.5, 150 mm NaCl, 2 mm DTT. Reactions were initiated by the addition of 0.75 μg of recombinant Lys-48-linked polyubiquitin (Biomol). Samples were incubated at 37 °C for the indicated time, and the reactions were terminated by the addition of 15 μl of SDS-urea electrophoresis sample buffer. Polyubiquitin levels were determined by Western blotting for ubiquitin.
RESULTS
USP5 Knockdown Activates p53
To identify DUBs whose suppression activates p53, we carried out a screen using a library consisting of pools of 4 shRNA targeting 50 members of the DUB family (39). The screen was carried out in a derivative of the wild-type p53 expressing A375 melanoma cell line (ARN8), generated by stable transfection with a p53-responsive reporter construct (RGCΔFos-LacZ) that drives expression of β-galactosidase (40). In this screen transfection with the pool of shRNA complementary to USP5 was observed to result in an increase in β-galactosidase reporter activity (Fig. 1A). To validate USP5 as a target we looked at the effects of two synthetic siRNA complementary to different sequences in USP5. Transfection with either of these siRNA resulted in an increase in β-galactosidase activity (Fig. 1B). This confirms that USP5 knockdown genuinely causes an increase in reporter activity. We also looked at the effect of USP5 knockdown on the level of p53 and Mdm2. Suppression of USP5 resulted in an increase in p53 and Mdm2 protein expression (Fig. 1C).
FIGURE 1. USP5 knockdown regulates the p53 pathway.
A, ARN8 melanoma cells stably transfected with a p53-responsive reporter driving the expression of β-galactosidase were transfected with pools of pSUPER shRNA targeting 50 deubiquitinating enzymes. The shRNA pool targeting USP5 increases p53-responsive reporter activity. β-Galactosidase activity was normalized to total protein levels and expressed as a percentage of control (empty pSUPER vector). The values are the means of duplicate determinations ± a range of values. A representative experiment is shown. B–E, ARN8 cells were mock-transfected (−) or transfected with a non-targeting siRNA (Control) or siRNA complementary to two different sequences in USP5 (USP5 A and B). Knockdown of USP5 increases p53-reporter activity (B), p53 and Mdm2 protein levels (C), and p53 target gene expression (D) without effecting levels of the p53-independent P1 Mdm2 transcript. E, suppression of USP5 does not alter p53 mRNA levels. β-Galactosidase activity was normalized to total protein levels and expressed as a percentage of control (non-targeting siRNA). The values are the means ± S.D. of three experiments. mRNA expression was quantified by real-time PCR. mRNA levels were normalized to actin and expressed as a percentage of control (non-targeting siRNA). The values are the means ± S.D. of four experiments.
To investigate whether USP5 knockdown regulates the expression of endogenous p53 target genes, ARN8 cells were transfected with siRNA complementary to USP5, and mRNA levels of p53-responsive genes were assayed by real-time PCR. Mdm2 is itself a transcriptional target of p53. The Mdm2 gene has two promoters. One of these is p53-independent (P1), and the other is p53-responsive (P2). These give rise to different mRNA due to variations in the 5′-untranslated region. Consistent with specific transcriptional activation of p53, suppression of USP5 increased the level of Mdm2 P2 mRNA but had no affect on the level of Mdm2 P1 mRNA (Fig. 1D). In addition, USP5 knockdown increased mRNA expression of the p53-responsive genes p21 and Bax. Suppression of USP5 did not affect p53 mRNA levels (Fig. 1E), confirming that USP5 knockdown does not have a general effect on transcription and showing that changes in p53 protein expression are not due to changes in p53 mRNA levels.
To determine whether these effects of USP5 suppression are p53-dependent, ARN8 cells were co-transfected with siRNA targeting p53 and USP5. Transfection with either of two siRNA complementary to different sequences in p53 resulted in a reduction in p53 protein expression without affecting USP5 knockdown by co-transfected USP5 siRNA (Fig. 2A). p53 knockdown inhibited the increase in β-galactosidase reporter activity (Fig. 2B) and mRNA levels of the endogenous p53 target genes Mdm2 and p21 resulting from suppression of USP5 (Figs. 2, C and D). The increase in Mdm2 protein expression was also attenuated by suppression of p53 (Fig. 2A). To further investigate the p53 dependence of the affects of USP5 knockdown, HCT116 p53+/+ and p53−/− cells were transfected with USP5 siRNA. In HCT116 p53+/+ cells USP5 knockdown resulted in an increase in p53 and Mdm2 protein levels (Fig. 2E) and an increase in Mdm2 P2 mRNA expression (Fig. 2F). No increase in Mdm2 protein or mRNA levels was observed after USP5 knockdown in HCT116 p53−/− cells. The effects of USP5 knockdown in p53-null H1299 cells were also determined. Again in the absence of p53, siRNA-mediated knockdown of USP5 did not increase Mdm2 protein levels and did not affect the mRNA levels of p53 target genes (supplemental Figs. 1, A and B). These data indicate that siRNA-mediated suppression of USP5 results in an increase in p53 transactivation activity.
FIGURE 2. The effects of USP5 knockdown are p53-dependent.
A–D, ARN8 melanoma cells were transfected with non-targeting siRNA (Control) or siRNA targeting USP5 in combination with siRNA complementary to two different sequences in p53. Where appropriate, total siRNA levels were maintained constant by the addition of non-targeting siRNA. The increases in Mdm2 protein expression (A), p53-responsive transcriptional reporter activity (B), Mdm2 P2 mRNA (C), and p21 mRNA levels (D) are attenuated by p53 knockdown. β-Galactosidase and mRNA levels were normalized to total protein and actin mRNA, respectively, and expressed as a percentage of control (non-targeting siRNA). The values are the means ± S.D. of three experiments. E and F, HCT116 p53+/+ or HCT116 p53−/− cells were mock-transfected (−) or transfected with the indicated siRNA. E, knockdown of USP5 increases p53 and Mdm2 protein levels in HCT116 p53+/+ cells but has no effect on Mdm2 levels in HCT116 p53−/− cells. F, knockdown of USP5 increases Mdm2 P2 mRNA expression in HCT116 p53+/+ cells but not in HCT116 p53−/− cells. mRNA levels were normalized to TBP and expressed as a percentage of control (non-targeting siRNA). The values are the means ± S.D. of three experiments.
We examined the effect of USP5 knockdown on the localization of p53 and Mdm2 by immunofluorescence. In mock or control siRNA-transfected samples the occasional cell displayed a high level of nuclear p53 and Mdm2 (Fig. 3A). Consistent with the Western blotting data and with increased p53 transcriptional activity, USP5 knockdown caused an increase in staining for p53 and Mdm2. p53 accumulated in the nucleus, and Mdm2 co-localized with p53.
FIGURE 3. USP5 knockdown causes the accumulation of nuclear p53 without promoting Ser-15 phosphorylation of p53.
A, ARN8 cells were mock-transfected (−) or transfected with the indicated siRNA and analyzed by indirect immunofluorescence using CM1 to detect endogenous p53 and 4B2 to detect endogenous Mdm2. Suppression of USP5 results in the accumulation of p53 and Mdm2 in the nucleus. B, ARN8 cells were incubated with mitomycin C (MMC) or transfected with the indicated siRNA. UT, untreated. Total p53 expression and levels of Ser-15 phosphorylation were determined by Western blotting. USP5 knockdown does not increase the stoichiometry of Ser-15 phosphorylation of p53.
To assess whether USP5 knockdown is genotoxic, we looked at its effects on Ser-15 phosphorylation of p53. Phosphorylation of p53 at this site is used as a marker for genotoxicity (47). ARN8 cells were treated with the DNA cross-linker mitomycin C as a positive control. Incubation with mitomycin C resulted in an increase in the ratio of Ser-15 phosphorylated p53 to total p53, indicating that it increased the stoichiometry of p53 phosphorylation at this site (Fig. 3B). In contrast, USP5 knockdown did not promote Ser-15 phosphorylation of p53. USP5 knockdown increased the total p53 level to a greater extent than it increased the level of Ser-15-phosphorylated p53. This indicates that USP5 knockdown activates p53 without activating genotoxic pathways.
USP5 Knockdown Results in a Differential Effect on p53 and Mdm2 Protein Stability
To further investigate the mechanism of p53 activation, the effect of USP5 knockdown on the stability of p53 and Mdm2 was examined. We determined the rates of p53 and Mdm2 protein degradation in cells transfected with control or USP5 siRNA by the addition of cycloheximide, which inhibits protein synthesis. In ARN8 cells USP5 knockdown increased the stability of p53 (Fig. 4A). In contrast, the stability of Mdm2 was not affected by suppression of USP5. Elevated Mdm2 protein expression can be accounted for by the p53-dependent increase in Mdm2 mRNA levels (Figs. 1 and 2). USP5 knockdown also had a striking effect on the stability of p53 in HCT116 p53+/+ cells while having little or no effect on the stability of Mdm2 (supplemental Figs. 2, A and B). Treatment of ARN8 cells with MG132, which directly inhibits the proteolytic activity of the proteasome, stabilized both p53 and Mdm2 (Fig. 4B). The magnitude of the effect of MG132 treatment indicates that the proteasome is the major route for degradation of both p53 and Mdm2 in ARN8 cells. USP5 knockdown, thus, inhibits proteasomal degradation of p53 without affecting the proteasomal degradation of Mdm2. There is a precedent for this differential effect on protein stability. In yeast, knock-out of the homologue of USP5 also inhibits the proteasomal degradation of some proteins but not others through an as yet undefined mechanism (48).
FIGURE 4. USP5 knockdown inhibits the proteasomal degradation of p53 without effects on the proteasomal degradation of Mdm2.
ARN8 cells were transfected with the indicated siRNA and incubated with cycloheximide (CHX, 20 μg/ml) to inhibit protein synthesis before harvesting. A, the upper panel shows Western blots (different exposures are shown so that protein levels in the absence of cycloheximide are matched). The lower panel shows quantification of the Western blots. USP5 knockdown stabilizes p53 without affecting the stability of Mdm2. B, ARN8 cells were incubated with carrier or the proteasome inhibitor MG132 for 4 h before the addition of cycloheximide (20 μg/ml). Protein expression was analyzed by Western blotting and quantified. MG132 blocks the degradation of both p53 and Mdm2.
Mdm2 is a key repressor of p53 that can act both through direct binding to p53 and by promoting its proteasomal degradation. Because Mdm2 is also a p53 target gene, a negative feedback loop can arise. Such a feedback loop is in operation in ARN8 cells. In this cell line disruption of the Mdm2-p53 interaction by nutlin-3 (49) or siRNA-mediated knockdown of Mdm2 causes transcriptional activation of p53 (data not shown). This indicates that Mdm2 plays a major part in repressing the activity of p53. Furthermore, suppression of p53 decreases basal Mdm2 expression, and p53 activation increases Mdm2 protein levels (Fig. 2). By stabilizing p53 and not Mdm2, knockdown of USP5 can alter the set point of the feedback loop resulting in increased transcriptional activity of p53.
We next looked at the effect of USP5 knockdown on the pattern of p53 ubiquitination. In this and subsequent experiments to assess protein ubiquitination or levels of free ubiquitin, cells were lysed under denaturing conditions to prevent post-lysis deubiquitination. In ARN8 and HCT116 p53+/+ cells knockdown of USP5 resulted in an increase in the level of high molecular weight species of p53, consistent with the accumulation of ubiquitinated p53 (Fig. 5A and supplemental Fig. 3A). To further investigate the nature of these conjugates, p53 was immunoprecipitated from lysates of ARN8 cells transfected with control or USP5 siRNA, and the immunoprecipitates were analyzed by Western blotting for p53 and ubiquitin. Immunoprecipitated conjugates detected with the anti-p53 antibody were also recognized by the anti-ubiquitin antibody (Fig. 5B). This confirms that the conjugates contain ubiquitinated p53. The relative intensities of the species detected with the anti-p53 and anti-ubiquitin antibodies were different. The smear of ubiquitinated p53 migrating above the distinct ladder of ubiquitinated p53 was more intense in the Western blots for ubiquitin. This reflects the greater number of ubiquitin molecules per molecule of p53 in the higher molecular weight species. These results indicate that suppression of USP5 inhibits the degradation of ubiquitinated p53 rather than interfering with p53 ubiquitination.
FIGURE 5. Suppression of USP5 causes the accumulation of ubiquitinated p53.
ARN8 cells were mock-transfected (−) or transfected with the indicated siRNA. A, cells were lysed under denaturing conditions, and p53 levels were determined by Western blotting. A short exposure (lower panel) and an extended exposure (middle panel) of the Western blot for p53 are shown. USP5 knockdown causes the accumulation of high molecular weight conjugates of p53. B, lysates of ARN8 cells prepared under denaturing conditions were immunoprecipitated (IP) with preimmune rabbit serum or anti-p53 antibody CM1. Immunoprecipitates were analyzed by Western blotting for p53 (left panel) or ubiquitin (right panel). This confirms that the p53 conjugates detected after USP5 knockdown contain ubiquitinated p53.
USP5 Is Required for Disassembly of Free Polyubiquitin in ARN8 Melanoma Cells
Studies in vitro show that a preferred substrate for USP5 is unanchored/free polyubiquitin. That is polyubiquitin linked by isopeptide bonds which is not conjugated to a substrate protein (15, 18, 19). Consistent with this, knock-out of USP5 homologues in S. cerevisiae, Dictyostelium, and Arabidopsis (21-23) causes accumulation of free polyubiquitin. Previous studies have shown that unanchored polyubiquitin can inhibit proteasome-mediated degradation in vitro (8, 50, 51). The accumulation of ubiquitinated p53 after USP5 knockdown could be a result of competition with unanchored polyubiquitin for recognition by the proteasome. To investigate whether changes in the level of free polyubiquitin could be involved in the stabilization of p53, the effect of USP5 knockdown on the general pattern of ubiquitination in ARN8 and HCT116 p53+/+ cells was examined. USP5 knockdown resulted in the accumulation of a ladder of low molecular weight ubiquitin species which co-migrated with purified Lys-48-linked unanchored polyubiquitin (Fig. 6A and supplemental Fig. 3B). This indicates that USP5 knockdown results in an increase in the level of unanchored polyubiquitin. In contrast to the other multimers of free ubiquitin, the major ubiquitinated species which co-migrated with purified trimeric free polyubiquitin was slightly reduced in intensity by USP5 knockdown. This is likely to be ubiquitinated histone H2A, which obscures the endogenous trimeric ubiquitin. We observed that ubiquitinated histone H2A co-migrated with this band (data not shown). Histone H2A has been shown to be a major ubiquitinated protein in cells which can be detected in anti-ubiquitin Western blots (52). Treatment with direct inhibitors of the proteolytic activity of the proteasome dramatically reduces histone H2A ubiquitination due to depletion of free ubiquitin (53-55).
FIGURE 6. USP5 makes a major contribution to unanchored polyubiquitin disassembly.
A, ARN8 cells were mock-transfected (−) or transfected with the indicated siRNA. Cells were lysed under denaturing conditions, and the general pattern of ubiquitination was analyzed by Western blotting for ubiquitin. The left and right panels show low and high exposures respectively of the same blot. The right panel also shows Western blots for USP5 and actin. USP5 knockdown resulted in the accumulation of a ladder of low molecular weight ubiquitin species which co-migrated with purified Lys-48-linked free polyubiquitin. The position of migration of multimers of unanchored ubiquitin is indicated (UB). B and C, ARN8 cells were transfected with the indicated siRNA. Cells were lysed under non-denaturing condition. B, protein expression was analyzed by Western blotting. C, 0.15–5 μg of the ARN8 cell extracts were incubated with recombinant Lys-48-linked free polyubiquitin for the indicated times. Polyubiquitin levels were determined by Western blotting. USP5 knockdown reduced the free polyubiquitin disassembly activity of ARN8 extracts.
After suppression of USP5 we also observed a relatively modest increase in the level of high molecular weight ubiquitinated species. These co-migrated with ubiquitinated species in mock and control siRNA-transfected cells and are most likely to be ubiquitin-protein conjugates. The increase in high molecular weight ubiquitinated conjugates is consistent with a general defect in the proteasomal degradation of at least a subpopulation of ubiquitinated proteins after knockdown of USP5.
We also examined the effect of USP5 knockdown on the polyubiquitin disassembly activity in soluble extracts of ARN8 cells. After transfection of ARN8 cells with control or USP5 siRNA, extracts were prepared by lysis in Nonidet P-40 buffer. Knockdown of USP5 was confirmed by Western blotting (Fig. 6B). Purified Lys-48-linked unanchored polyubiquitin chains were incubated with the cell extracts for 5 min at 37 °C. Under these conditions undiluted control extract caused complete disassembly of the polyubiquitin (Fig. 6C). In contrast, little disassembly occurred with extract from cells in which USP5 expression was suppressed. A dilution of between 1 in 3 and 1 in 10 was required to reduce the polyubiquitin disassembly activity of control lysates to that observed with the lysates from cells in which USP5 had been knocked down. This confirms that USP5 makes a major contribution to the free polyubiquitin disassembly activity of ARN8 cells.
Expression of Ubiquitin G75A/G76A Recapitulates the Effects of USP5 Knockdown
To investigate the effect of free polyubiquitin on the p53 pathway, we employed a mutant of ubiquitin in which the C-terminal diglycine motif was substituted by alanine residues (G75A/G76A). The ectopic expression of this ubiquitin mutant would be predicted to result in the accumulation of free polyubiquitin for the following reasons. The diglycine motif of ubiquitin is required for conjugation to the side chain amino group of lysine residues in target proteins and other ubiquitin molecules (21, 56). However, a wild-type ubiquitin molecule can be conjugated via its C terminus to lysine residues in mutant ubiquitin lacking the diglycine motif. The mutant can, thus, become the proximal ubiquitin in a chain of free polyubiquitin which cannot be conjugated to other proteins via its C terminus. In addition, polyubiquitin with a proximal ubiquitin which lacks an intact diglycine motif is a very poor substrate for USP5 (18). Expression of mutant ubiquitin lacking the conserved C-terminal diglycine motif has been shown to mimic the effect of knock-out of the USP5 homologue Ubp14 in S. cerevisiae (21, 57). ARN8 cells were transfected with empty vector or a vector expressing His6-tagged wild-type or mutant ubiquitin. Ectopic ubiquitin expression was detected by Western blotting using an anti-His6 antibody. This confirmed that expression of ubiquitin mutant G75A/G76A resulted in the accumulation of multimeric unanchored ubiquitin (Fig. 7A). Wild-type His6-ubiquitin but not ubiquitin G75A/G76A was incorporated into high molecular weight conjugates. This indicates that, as predicted, the ubiquitin mutant is not conjugated to proteins in vivo. Like USP5 knockdown, expression of ubiquitin G75A/G76A caused an increase in protein levels of p53 and Mdm2 (Fig. 7B), p53-responsive reporter activity (Fig. 7C), and mRNA levels of the p53 target genes Mdm2 and p21 (Fig. 7D). No effect was seen after ectopic expression of wild-type ubiquitin on the levels or transcriptional activity of p53. These data indicate that accumulation of free polyubiquitin can increase p53 transactivation activity. To determine whether ectopic expression of ubiquitin G75A/G76A differentially affected the stability of p53 and Mdm2, ARN8 cells were transfected with empty vector or a plasmid expressing the ubiquitin mutant and treated with cycloheximide for the indicated time before harvesting. Strikingly ectopic expression of ubiquitin G75A/G76A also resulted in stabilization of p53 without stabilization of Mdm2 (Fig. 8A). In addition, like USP5, knockdown expression of ubiquitin G75A/G76A caused the accumulation of ubiquitinated p53 (Fig. 8B).
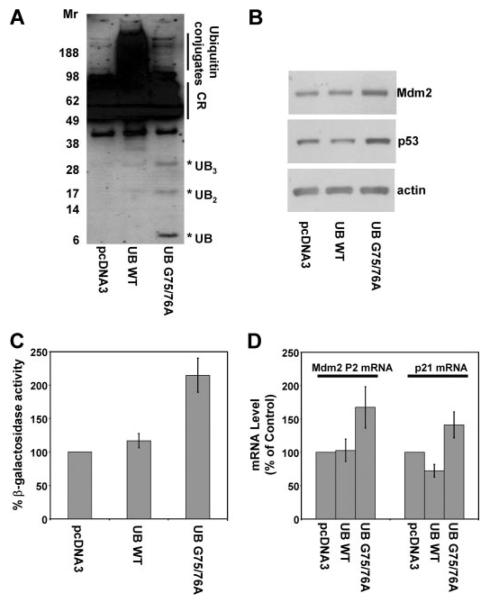
FIGURE 7. Expression of ubiquitin mutant G75A/G76A recapitulates the effects of USP5 knockdown.
ARN8 cells were transfected with empty vector (pcDNA3) or a vector expressing His6-tagged wild-type ubiquitin (UB WT) or ubiquitin mutant G75A/G76A (UB G75A/G76A). A, anti-His6 Western blot for His6-tagged ubiquitin. The position of migration of high molecular weight ubiquitin conjugates, cross-reacting proteins detected in the absence of His6-tagged ubiquitin (CR) and free ubiquitin and ubiquitin multimers (*UB) is indicated. Ectopic expression of UB G75A/G76A results in accumulation of unanchored polyubiquitin. Expression of ubiquitin mutant G75A/G76A but not wild-type ubiquitin results in an increase in p53 and Mdm2 protein levels (B), p53-responsive reporter activity (C), and p53 target gene expression (D). β-Galactosidase and mRNA levels were normalized to total protein and actin, respectively, and expressed as a percentage of control (non-targeting siRNA). The values are means ± S.D. of three experiments.
FIGURE 8. G75A/G76A ubiquitin expression stabilizes p53 but not Mdm2.
A, ARN8 cells were transfected with empty vector (pcDNA3) or a vector expressing His6-tagged ubiquitin mutant G75A/G76A (UB G75A/G76A). Cycloheximide (CHX) was added for the indicated time before harvesting. The upper panel shows Western blots (different exposures are shown so that protein levels in the absence of cycloheximide are matched). The lower panel shows quantification of the Western blots. Ubiquitin G75A/G76A stabilizes p53 without preventing the degradation of Mdm2. B, ARN8 cells were transfected with the indicated plasmid, and protein expression was analyzed by Western blotting. A short exposure (lower panel) and an extended exposure (middle panel) of the Western blot for p53 are shown. Like USP5, knockdown expression of ubiquitin G75A/G76A causes the accumulation of high molecular weight conjugates of p53.
Taken together the data strongly support a model in which the effects of USP5 knockdown on p53 are mediated by free polyubiquitin. Inhibition of proteasomal degradation of p53 without a defect in p53 ubiquitination is consistent with a mechanism involving the competition of free polyubiquitin with ubiquitinated p53 for proteasomal recognition. The differential effect of USP5 knockdown/free polyubiquitin on the stability of p53 and Mdm2 can be explained by these proteins having different sensitivities to inhibition of proteasomal recognition by unanchored polyubiquitin.
DISCUSSION
In this study we show that the deubiquitinating enzyme USP5 (isopeptidase T) can regulate the activity of p53. Knockdown of USP5 causes the accumulation of nuclear p53 and an increase in p53 transcriptional activity. Activation of p53 can be accounted for by the ability of USP5 suppression to inhibit the proteasomal degradation of p53 without affecting the degradation of Mdm2. We propose that the differential effect of USP5 knockdown on protein stability is due to differences in the sensitivity of p53 and Mdm2 to inhibition of proteasomal recognition by free/unanchored polyubiquitin which accumulates after USP5 knockdown.
The control of the stability of both p53 and Mdm2 by ubiquitination and proteasomal degradation is a complicating factor in therapeutic approaches aimed at increasing the transcriptional activity of wild-type p53 by targeting components of the ubiquitin-proteasome system. Direct inhibition of the proteolytic activity of the proteasome (28, 33, 34) and E2 ubiquitin-conjugating enzyme knockdown (45) can block p53 degradation without increasing its transcriptional activity. One possible reason for this is that in addition to stabilizing p53, these interventions also stabilize Mdm2 and cause the accumulation of sufficient Mdm2 to repress p53 by direct binding (28, 45). The differential effect of USP5 knockdown on the stability of p53 and Mdm2 provides a mechanism for p53 activation. Stabilization of p53 without stabilization of Mdm2 can alter the set point of the p53:Mdm2 feedback loop resulting in increased transcriptional activity of p53.
USP5 knockdown reduced the unanchored Lys-48-linked polyubiquitin disassembly activity in ARN8 cell lysates and increased the level of free polyubiquitin in intact cells. This demonstrates for the first time that USP5 makes a major contribution to free polyubiquitin disassembly in a mammalian system. Deletion of USP5 homologues in lower organisms similarly causes accumulation of free polyubiquitin (21-23). This is consistent with the substrate specificity of USP5. In vitro studies have shown that isopeptide bond-linked unanchored polyubiquitin chains are a preferred substrate for USP5 (18, 20). USP5 catalyzes the sequential release of ubiquitin from the proximal end of the unanchored polyubiquitin chain (the end with a ubiquitin subunit with a free C terminus). Alterations to the C terminus of the proximal ubiquitin such as conjugation to proteins and mutation or deletion of the conserved diglycine motif (Gly-75/76) inhibit cleavage by USP5. The crystal structure of the zinc finger ubiquitin binding domain (ZnF UBP) of USP5 in complex with ubiquitin has revealed the structural basis for this (58). The free C-terminal diglycine motif is inserted into a deep binding pocket in the zinc finger ubiquitin binding domain of USP5, and extensive contacts are made between USP5 and the diglycine motif of ubiquitin.
To assess the effect of free polyubiquitin on the p53 pathway, we employed a G75A/G76A mutant of ubiquitin. The properties of this mutant are such that its expression results in the accumulation of unanchored polyubiquitin. Ectopic expression of ubiquitin G75A/G76A recapitulated the effect of USP5 knockdown. The ubiquitin mutant stabilized p53 without stabilization of Mdm2 and caused an increase in p53 transcriptional activity. In addition, both USP5 knockdown and ectopic expression of ubiquitin G75A/G76A resulted in the accumulation of ubiquitinated p53. These observations strongly suggest that the effects of USP5 knockdown on the p53 pathway are mediated by unanchored polyubiquitin. This could act at several levels in the ubiquitin-proteasome system. There are multiple ubiquitin-binding proteins involved in a range of processes (59, 60). The accumulation of ubiquitinated species of p53 indicates that suppression of USP5 inhibits the degradation of ubiquitinated p53 rather than interfering with p53 ubiquitination. Our data are consistent with a model in which the free polyubiquitin that accumulates after USP5 knockdown competes for recognition of ubiquitinated p53 by the proteasome. This represents a previously unknown mechanism for the regulation of p53. USP5 knockdown also caused a modest increase in the total level of high molecular weight ubiquitinated species, indicating a general defect in the degradation of at least a sub-population of ubiquitinated proteins. The knock-out of USP5 homologues in lower organisms similarly causes defects in degradation of proteins targeted to the proteasome, and it was also suggested that a likely mechanism was competition of free polyubiquitin with ubiquitinated proteins for recognition by the proteasome (21-23). In support of this, unanchored polyubiquitin chains have been shown to bind with relatively high affinity to the proteasome (8, 61, 62) and to adaptor proteins involved in targeting ubiquitinated proteins to the proteasome (60, 63). In addition, unanchored polyubiquitin inhibits ubiquitin-dependent proteolysis in cell free systems (8, 50, 51). Conversely, USP5 can promote proteasomal degradation in vitro, putatively by preventing the build up of free polyubiquitin (15).
Deubiquitination of Mdm2 by the DUBs HAUSP or USP2a results in Mdm2 stabilization (37, 38). In addition, mutations of the ring finger of Mdm2, which prevent autoubiquitination, inhibit degradation of ectopically expressed Mdm2 (64). These data indicate that ubiquitination of Mdm2 is required for its degradation. Inhibition of the proteolytic activity of the proteasome showed that the major route for degradation of both Mdm2 and p53 is by the proteasome in ARN8 cells. These observations raise the question of how proteasomal recognition of p53 could be inhibited by USP5 knockdown/free polyubiquitin without inhibiting the proteasomal recognition of Mdm2. Ubiquitin-dependent recognition of proteins by the proteasome involves ubiquitin binding proteins which associate with the proteasome. Rpn10 (S5a/PSMD4) (61) and Rpt5 (S6′/Tbp-1) (65) are ubiquitin-binding proteins that are stoichiometric subunits of the proteasome. The proteasomal subunit Rpn13 (ADRM1/ARM1) has also recently been identified as a ubiquitin-binding protein (62, 66). Another class of proteins that are involved in proteasomal recognition are those containing a ubiquitin-like domain (UBL), which binds to the proteasome, and a ubiquitin binding domain (UBA), which binds to ubiquitinated substrates (67, 68). These adaptor proteins include hHR23A and -B and hPlic-1 and -2, which are mammalian homologues of S. cerevisiae Rad23 and Dsk2. A ubiquitin-binding protein lacking a ubiquitin-like domain has also been implicated in mediating proteasomal recognition (69). In the case of p53 and Mdm2, a common ubiquitin-binding protein could be involved in recognition of both proteins. There could be variations in the sensitivity to competition with free polyubiquitin for binding to the ubiquitin receptor because of differences in the affinity of receptor binding by ubiquitinated p53 and Mdm2. Alternatively, it is possible that there are distinct ubiquitin-binding proteins involved in the recognition of p53 and Mdm2. Again, the affinity of binding of ubiquitinated p53 and Mdm2 may be a contributory factor to the differential sensitivity to free polyubiquitin, or the ubiquitin receptor proteins may differ in their ability to bind the unanchored polyubiquitin that accumulates after USP5 knockdown. Studies in yeast have shown that there is substrate selectivity in recognition of ubiquitinated proteins, distinct ubiquitin-binding proteins being involved in the proteasomal recognition of particular ubiquitinated proteins (62, 68, 70).
Variations in the nature of the ubiquitin tag attached to p53 and Mdm2 such as differences in ubiquitin chain length and the nature of the cross-links could underlie the differences in sensitivity to free polyubiquitin. There is evidence that polyubiquitination of Mdm2 involves cross-links between lysine residues in addition to Lys-48 (71, 72). The nature of the tag could influence the affinity of the interaction with the ubiquitin receptor. It could also be responsible for the involvement of different receptors in the proteasomal recognition of p53 and Mdm2. There are variations in the ubiquitin interaction properties of known ubiquitin-binding proteins both in terms of the degree of polyubiquitination required for binding and in the linkage specificity (62, 63, 73). An additional possibility is that stable interaction with a ubiquitin receptor could require direct interaction of the receptor with p53 or Mdm2 in addition to binding to the ubiquitin conjugated to these proteins. This could also account for variations in the affinity of binding of p53 and Mdm2 to their ubiquitin receptor(s) or for the involvement of alternative receptors in their proteasomal recognition.
No clear picture has emerged with respect to the mechanism of recognition of p53 and Mdm2 by the proteasome. There is evidence that Mdm2 itself plays a post-ubiquitination role in recognition of p53 by the proteasome (74-76). In one investigation siRNA-mediated knockdown of hHR23 A and B caused accumulation of p53, consistent with a role in transport of p53 to the proteasome (77). In another study knockdown of hHR23 A and B caused a decrease in p53 levels (78). The discrepancy between these observations may be a result of variations in the relative expression of hHR23 proteins compared with other cellular components. hHR23 proteins have multiple activities, and their effects are critically dependent on their concentration (68). Unconjugated p53 binds to the Rpt6 (S8/Sug1) subunit of the 19 S proteasome (33). Mdm2 has been shown to bind independently of ubiquitination to the α7 (C8) subunit of the 20 S proteasome (79), to the Rpn1 (S2) subunit of the 19 S proteasome (80), and to the proteasome-binding protein gankyrin (81). Further investigation is required to determine the precise role of these interactions in the proteasomal recognition of p53 and Mdm2.
USP5 represents a potential target for p53-activating drugs for the treatment of cancer. Even though direct inhibition of the protease activity of the proteasome is regarded as being a non-specific means to target the ubiquitin-proteasome system, tumor cells are generally more sensitive to this than untransformed cells. USP5 suppression provides an alternative mechanism for the inhibition of degradation of ubiquitinated proteins by causing the accumulation of free polyubiquitin and blocking their recognition by the proteasome. Our data and observations in yeast (48) indicate that suppression of USP5 has a more selective effect on proteasomal degradation than direct inhibition of the proteolytic activity of the proteasome. Given this and the ability of USP5 knockdown to increase the trans-activation activity of p53, it will be of interest to further investigate the therapeutic efficacy of USP5 inhibition.
This study raises the possibility of differences in the recognition of p53 and Mdm2 by the proteasome. These differences could be important in the physiological regulation of p53 activity, and an improved understanding of the mechanism of p53 and Mdm2 recognition by the proteasome may identify additional therapeutic targets. For example, a ubiquitin receptor selectively involved in proteasomal recognition of ubiquitinated p53 would represent a potential target for p53 activating therapies.
Supplementary Material
Acknowledgments
We thank Professor René Bernards (The Netherlands Cancer Institute) for donating the shRNA DUB library and Dr. Dimitris P. Xirodimas for critical reading of the manuscript.
Footnotes
This work was funded by Cancer Research UK.
Publisher's Disclaimer: The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
The abbreviations used are: DUB, deubiquitinating enzymes; USP, ubiquitin-specific protease; shRNA, short hairpin RNA; siRNA, small interfering RNA; DTT, dithiothreitol.
REFERENCES
- 1.Ciechanover A. Exp. Biol. Med. (Maywood) 2006;231:1197–1211. doi: 10.1177/153537020623100705. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson KD, Ventii KH, Friedrich KL, Mullally JE. EMBO Rep. 2005;6:815–820. doi: 10.1038/sj.embor.7400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dicato M, Boccadoro M, Cavenagh J, Harousseau JL, Ludwig H, San Miguel J, Sonneveld P. Oncology. 2006;70:474–482. doi: 10.1159/000099284. [DOI] [PubMed] [Google Scholar]
- 4.Orlowski RZ, Kuhn DJ. Clin. Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 5.Wolf DH, Hilt W. Biochim. Biophys. Acta. 2004;1695:19–31. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Nandi D, Tahiliani P, Kumar A, Chandu D. J. Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 8.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam YA, Xu W, DeMartino GN, Cohen RE. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 10.Guterman A, Glickman MH. J. Biol. Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 11.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 12.Pickart CM, Fushman D. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda F, Dikic I. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Hadari T, Warms JV, Rose IA, Hershko A. J. Biol. Chem. 1992;267:719–727. [PubMed] [Google Scholar]
- 16.Shabek N, Iwai K, Ciechanover A. Biochem. Biophys. Res. Commun. 2007;363:425–431. doi: 10.1016/j.bbrc.2007.08.185. [DOI] [PubMed] [Google Scholar]
- 17.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, III, Koonin EV, Deshaies RJ. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson KD, Tashayev VL, O'Connor LB, Larsen CN, Kasperek E, Pickart CM. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 19.Stein RL, Chen Z, Melandri F. Biochemistry. 1995;34:12616–12623. doi: 10.1021/bi00039a017. [DOI] [PubMed] [Google Scholar]
- 20.Falquet L, Paquet N, Frutiger S, Hughes GJ, Hoang-Van K, Jaton JC. FEBS Lett. 1995;359:73–77. doi: 10.1016/0014-5793(94)01451-6. [DOI] [PubMed] [Google Scholar]
- 21.Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. Plant J. 2001;27:393–405. doi: 10.1046/j.1365-313x.2001.01106.x. [DOI] [PubMed] [Google Scholar]
- 23.Lindsey DF, Amerik A, Deery WJ, Bishop JD, Hochstrasser M, Gomer RH. J. Biol. Chem. 1998;273:29178–29187. doi: 10.1074/jbc.273.44.29178. [DOI] [PubMed] [Google Scholar]
- 24.Woods YL, Lane DP. Hematol. J. 2003;4:233–247. doi: 10.1038/sj.thj.6200260. [DOI] [PubMed] [Google Scholar]
- 25.Dey A, Verma CS, Lane DP. Br. J. Cancer. 2008;98:4–8. doi: 10.1038/sj.bjc.6604098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 27.Beraza N, Trautwein C. Hepatology. 2007;45:1578–1579. doi: 10.1002/hep.21789. [DOI] [PubMed] [Google Scholar]
- 28.Stommel JM, Wahl GM. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Mol. Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Williams SA, McConkey DJ. Cancer Res. 2003;63:7338–7344. [PubMed] [Google Scholar]
- 32.Concannon CG, Koehler BF, Reimertz C, Murphy BM, Bonner C, Thurow N, Ward MW, Villunger A, Strasser A, Kogel D, Prehn JH. Oncogene. 2007;26:1681–1692. doi: 10.1038/sj.onc.1209974. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Q, Wani G, Yao J, Patnaik S, Wang QE, El-Mahdy MA, Praetorius-Ibba M, Wani AA. Oncogene. 2007;26:4199–4208. doi: 10.1038/sj.onc.1210191. [DOI] [PubMed] [Google Scholar]
- 34.Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair VD, McNaught KS, Gonzalez-Maeso J, Sealfon SC, Olanow CW. J. Biol. Chem. 2006;281:39550–39560. doi: 10.1074/jbc.M603950200. [DOI] [PubMed] [Google Scholar]
- 36.Boccadoro M, Morgan G, Cavenagh J. Cancer Cell Int. 2005;5:18. doi: 10.1186/1475-2867-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks CL, Gu W. Cell Cycle. 2004;3:895–899. [PubMed] [Google Scholar]
- 38.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 40.Blaydes JP, Hupp TR. Oncogene. 1998;17:1045–1052. doi: 10.1038/sj.onc.1202014. [DOI] [PubMed] [Google Scholar]
- 41.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 42.Gabriel JM, Lacombe T, Carobbio S, Paquet N, Bisig R, Cox JA, Jaton JC. Biochemistry. 2002;41:13755–13766. doi: 10.1021/bi026096m. [DOI] [PubMed] [Google Scholar]
- 43.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 44.Martinez LA, Naguibneva I, Lehrmann H, Vervisch A, Tchenio T, Lozano G, Harel-Bellan A. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14849–14854. doi: 10.1073/pnas.222406899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP. J. Biol. Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 46.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 47.Lavin MF, Gueven N. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 48.Eisele F, Braun B, Pfirrmann T, Wolf DH. Biochem. Biophys. Res. Commun. 2006;350:329–333. doi: 10.1016/j.bbrc.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 49.Cheok CF, Dey A, Lane DP. Mol. Cancer Res. 2007;5:1133–1145. doi: 10.1158/1541-7786.MCR-07-0161. [DOI] [PubMed] [Google Scholar]
- 50.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Proc. Natl. Acad. Sci. U. S. A. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. J. Biol. Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 52.Fujimuro M, Sawada H, Yokosawa H. Eur. J. Biochem. 1997;249:427–433. doi: 10.1111/j.1432-1033.1997.00427.x. [DOI] [PubMed] [Google Scholar]
- 53.Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- 54.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. J. Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Q, Farah M, Webster JM, Wojcikiewicz RJ. Mol. Cancer Ther. 2004;3:1263–1269. [PubMed] [Google Scholar]
- 56.Perroy J, Pontier S, Charest PG, Aubry M, Bouvier M. Nat. Methods. 2004;1:203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- 57.Hodgins RR, Ellison KS, Ellison MJ. J. Biol. Chem. 1992;267:8807–8812. [PubMed] [Google Scholar]
- 58.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 59.Kirkin V, Dikic I. Curr. Opin. Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Hicke L, Schubert HL, Hill CP. Nat. Rev. Mol. Cell. Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 61.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 62.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raasi S, Varadan R, Fushman D, Pickart CM. Nat. Struct. Mol. Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 64.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. J. Biol. Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 65.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 66.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elsasser S, Finley D. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 68.Verma R, Oania R, Graumann J, Deshaies RJ. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 69.Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. EMBO J. 2006;25:554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Mol. Cell. Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Badciong JC, Haas AL. J. Biol. Chem. 2002;277:49668–49675. doi: 10.1074/jbc.M208593200. [DOI] [PubMed] [Google Scholar]
- 72.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. J. Biol. Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 74.Zhu Q, Yao J, Wani G, Wani MA, Wani AA. J. Biol. Chem. 2001;276:29695–29701. doi: 10.1074/jbc.M102634200. [DOI] [PubMed] [Google Scholar]
- 75.Blattner C, Hay T, Meek DW, Lane DP. Mol. Cell. Biol. 2002;22:6170–6182. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lukashchuk N, Vousden KH. Mol. Cell. Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Mol. Cell. Biol. 2003;23:8960–8969. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brignone C, Bradley KE, Kisselev AF, Grossman SR. Oncogene. 2004;23:4121–4129. doi: 10.1038/sj.onc.1207540. [DOI] [PubMed] [Google Scholar]
- 79.Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, Allday MJ, Xiao ZX. Mol. Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 80.Jin Y, Zeng SX, Sun XX, Lee H, Blattner C, Xiao Z, Lu H. Mol. Cell. Biol. 2008;28:1218–1229. doi: 10.1128/MCB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, Dawson S, Shimada Y, Mayer RJ, Fujita J. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.