Summary
Exon skipping repair is a strategy being investigated in early stage clinical trials to treat Duchenne muscular dystrophy. This is most applicable to the majority of cases which arise when genetic defects cause frame shift mutations, and induced exon skipping of out-of-phase exons restores the reading frame. However, the consequences to the edited protein so produced have not been considered. In many cases alternative routes to restoring the reading frame are possible, and we show in a test case involving exon 44 that the resulting differently edited proteins greatly vary in stability, with one of them very similar to normal unskipped dystrophin, and the other much less stable as assessed by the thermodynamics of folding as well as resistance to proteolysis. This has implications for the design of optimal therapeutic exon skipping strategies, which presumably wish to result repairs with as much fidelity to normal dystrophin as possible.
Keywords: dystrophin, exon skipping, spectrin type-repeat, stability
Introduction
Dystrophin is the protein defective in the devastating genetic disease, Duchenne Muscular Dystrophy, DMD†[1]. Those afflicted are normal at birth, but are subject to progressive muscle deterioration leading to a seriously reduced quality of life in their second decade, inevitably culminating in death, typically in their twenties. This X-linked condition is unfortunately common, afflicting 1 in 3500 male births, largely because the dystrophin gene is huge and thus subject to a concomitantly high mutational load. It is in fact the largest human gene spanning ~2.5 Mbp or ~0.07% of the entire genome, and is composed of 79 exons and transcribed from at least 7 promoters yielding a variety of functional and tissue specific isoforms[2].
The major function of this protein appears to be the stabilization of the myocyte membrane by forming a mechanical linkage between it and costameric actin[3]. Because myocytes repeatedly change shape during contraction/relaxation cycles inherent in their function, the membrane is constantly in flux; dystrophin is thought to help control and bring order to this movement. In its absence, the membrane becomes leaky, and cell death ensues. The muscle tissue as a whole can regenerate these lost cells at first, but eventually (in the first decade of life, typically) the regenerative capacity diminishes, then is exhausted, and an increasingly rapid deterioration ensues, leading to death.
While the exact details of the manner in which dystrophin stabilizes the sarcolemma are currently uncertain, its long rod-like shape has long suggested that it functions as a molecular reinforcing rod. Dystrophin owes this shape to membership in the spectrin family of proteins[4], which consist of those proteins comprised predominantly of multiple tandem copies of the spectrin type repeat, STR[5]. This ~110 amino acid motif consist of a 3 α-helices bundled in a zigzag fashion into small ~5 nm rods; this arrangement places the N- and C-termini at opposite ends, so that tandem copies produce increasingly long rods. Dystrophin has 24 STRs, and thus a rod ~120 nm long.
At either end of these rods are binding domains of some sort. In dystrophin’s case, these are: an actin binding domain at the N-terminus that facilitates binding to F-actin in the muscle machinery[6]; and at the C-terminus, a variety of globular domains that facilitate interaction with a large multi-protein membrane complex embedded in the sarcolemma. This thus links the membrane to the contractile apparatus, providing the crucial stabilization.
The binding motifs have been sometimes viewed as the interesting bits, with the intervening rod simply a passive and perhaps dispensable structural element. However, the properties of the rod (in particular the presence of putatively flexible hinge regions) are crucial to its proper functioning, and the rod region also possesses secondary binding functions for both actin[7, 8] and membrane phospholipids[9, 10], which are proposed to be involved in its mechanical stabilization function[11] – so it may not be as dispensable as some models portray it, as least for full functionality. Another reason the rod region is significant is that this region, making up most of the sequence of the gene, suffers most of the mutations. In contrast to many genetic diseases in which a handful of mutations are known, there are thousands of dystrophin mutations known. Furthermore, 30–50% of patients present with de novo (in them or their immediate maternal lineage) mutations, and the list is growing.
Currently there is no cure and little effective treatment for DMD. However, the devastating outcome, relative frequency, single factorial nature, and sympathetic patient demographics have made this condition an attractive target for gene therapy. Aside from problems common to all gene therapies (delivery, integration, immune response) dystrophin suffers from two additional issues: the target tissue, muscle, makes up a very large fraction of the total body mass, and the gene is too large to be compatible with the leading delivery vectors. This last problem is of course again a function of its large size, and current clinical trials are considering an edited dystrophin rod[12, 13], based upon a fortuitously discovered natural deletion mutation from a patient with a very mild form of Becker’s muscular dystrophy (BMD, a related condition in which dystrophin is not absent, but merely dysfunctional to some extent)[14]. This deletion occurs in the rod region – as do most other edits being considered to ameliorate this size problem for gene therapy. The tacit assumption, again, is that the rod is largely dispensable. Some, but not all, minidystrophins seem to work well in mouse models[13, 15]. However, mice are much less severely affected than humans – whose large muscle loads and long lifespan place heavy demands on muscle resiliency and stability. It remains to be seen whether the edits chosen are optimal in human – or even if it the significant other hurdles of gene therapy can be overcome.
In the mean time, a second strategy is also being investigated: exon skipping therapy[16, 17]. This technique turns the large size of dystrophin to advantage: for most defects, only a small portion of the gene is damaged – typically by a small deletion, or less commonly by duplication or point mutation. Most of the defects that cause DMD are those that result in a frame shift, which inevitably causes premature polypeptide chain termination. These frame shifts typically arise with deletions that newly juxtapose exons with incompatible phases with respect to the genetic code. Since the major function of dystrophin involves linking binding domains at each end of the molecule, if it is truncated prematurely function is totally lost. In addition, nonsense mediated decay induced by premature stop codons often causes RNA destruction and total loss of even the truncated dystrophin.
On the other hand, deletions that juxtapose exons with compatible phases often result in BMD, sometimes - but importantly, not always - with very mild severity. The technology has been developed to specifically interfere with the splicing during the mRNA maturation process, and specifically skip and so eliminate almost any given exon. This suggests that it should be possible to convert out-of-phase DMD mutations to in-phase BMD with judicious in vivo editing. This strategy, too, works in animal models, and is currently being investigated in a seemingly successful Phase I human clinical trial[18].
However, it is unclear exactly how to proceed with the repair other than restoring the reading frame. For many defects there are several alternative strategies with different sets of “extra” exons to skip which restore the reading frame. This is further complicated by the very large range and number of DMD causing defects that might be approached with this strategy. How then do we decide upon an optimal strategy when alternatives exist? Or does it even matter, so long as the reading frame is restored?
We posit that not only the effect on the RNA structure – chiefly the restoration of the reading frame – but also the impact of this edit on the structure and stability of the dystrophin protein should be considered. Restoring the reading frame but producing a dysfunctional protein would be a pyrrhic victory at best. As such we have embarked upon a study to examine the impact of alternative exon repair events on the biophysical and biochemical properties of the rod domain. Our hypothesis is that some alternative exon skip therapies will produce edited proteins that more faithfully reproduce the properties of the native unskipped protein.
Our test case in this study focuses on one specific region, surrounding exon 44. Exon 44 begins in phase 0 (i.e. its boundary lies in between codons) but ends in phase 2 (i.e. it ends after the middle G in the GGG codon of amino acid Gly 2097 of Dp427m). Thus, if it is deleted – and this is a defect that has been commonly observed in patients (e.g. there are 97 entries currently listed for deletion of exon 44 in the Leiden database [19]) - it induces a frame shift when exon 43, ending on phase 0, is fused to exon 45, which begins in phase 2. Deletion of exon 44 is also an interesting region to test the above hypothesis because there are two distinct and disparate ways of repairing it by exon skipping‡: skipping in addition the preceding exon 43, producing a double deletion Δ43,44; or skipping in addition the following exon 45, producing Δ44,45. One of these, Δ44,45 removes a region nearly concordant with the 17th STR of the rod, whereas the other, Δ43,44, removes parts of the 16th and 17th STR leaving behind a hybrid type STR. Both of these restore the reading frame, but are they equivalent at the protein level?
Methods
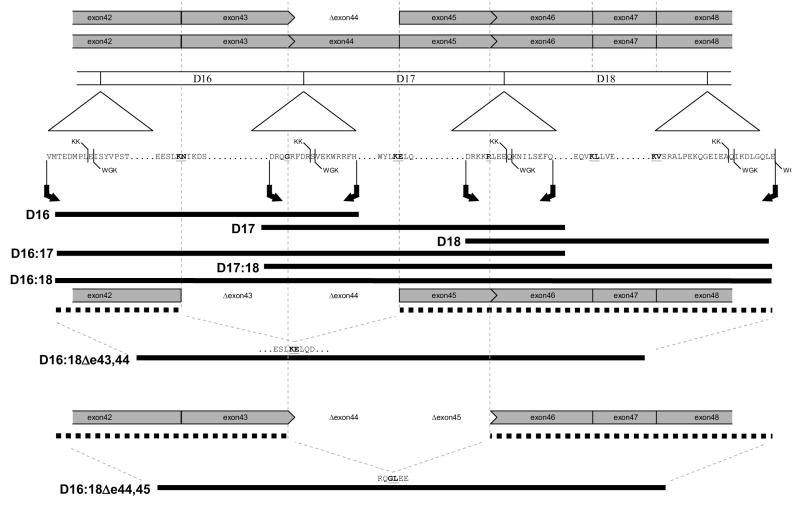
To answer this question we have produced versions of these alternative exon skipped rods in our recombinant dystrophin system (as well as various wild type, unskipped rods for comparison) based upon our previous work[20–22]. The most relevant proteins are the parent, unskipped D16:18 (denoting a protein spanning the 16th to the 18th STR of dystrophin) and the two alternative exon skipped variants D16:18Δe43,44 (denoting D16:18 missing the region coded for by exons 43 and 44) and D16:18Δe44,45. Since the exon skipped variants are smaller than the parent, by about one STR, we also created both 2-STR native unskipped fragments in this region, D16:17 and D17:18; and for further completeness, the single STR D16, D17 ands D18 motifs. The details of the target proteins constructed are given in Figure 1.
Figure 1. The nature of the various dystrophin fragments considered is shown, in relation to both the exon structure and STR motif structure of the region.
At the top, the shaded bars show the exon structure, with phase 0 exon boundaries (breaks between codons) shown as |, and phase 2 boundaries (breaks after the second base of a codon) as >. Deletion of exon 44 juxtaposes exons of incompatible phase and causes a frameshift, and thus DMD. The next, unshaded bar shows the STR motif structure. Due to limited sequence homology, the exact STR boundaries are uncertain, with two systems commonly in use: KK, and WGK. Fortunately, in this region the differences are each only one amino acid; however differences of up to 8 amino acids are found elsewhere. Due to this ambiguity, and also since short flanking sequences are sometimes needed to facilitate expression of motif, we have started each fragment 8 amino acids before (right pointing arrow) the KK boundary, and ended it (left pointing arrow) 8 amino acids after. The exon skipped motifs were made using D16:18 as a parent, with the appropriate sequences deleted as shown; exact sequence at the deletion junction is shown.
Our methods for producing these proteins have been reported elsewhere[20–22]. Briefly, genes were assembled by PCR, expressed in E. coli as fusion proteins with glutathione-S-transferase to facilitate affinity purification, released from this fusion partner by thrombin, and further purified by ion exchange and hydrophobic interaction chromatography. The only peculiarities specific to these proteins are the specific conditions used for the ion exchange chromatography of these specific target proteins, which is listed for each in Table 1. All genes were subject to DNA sequencing to verify that they were correctly constructed, the proteins were subject to MALDI mass spec to verify appropriate molecular mass, and the hydrodynamic radius was determined by dynamic light scattering using a Malvern Instruments Zetasizer Nano S to confirm that they were not aggregating in solution and had appropriate hydrodynamic properties. These quality control metrics are presented in Table I.
Table 1.
Purification parameters and quality control data for the target proteins
| target protein | purification | molecular mass | DLS sphere equivalent size | |||
|---|---|---|---|---|---|---|
| pI | purification pH and matrix type | theoretical | Maldi MS | Δ% | (nm) | |
| D16 | 5.45 | Q pH 5.5 | 15188 | 15246 | 0.38% | 4.82 |
| D17 | 9.27 | S pH 6.3 | 15424 | 15484 | 0.39% | 3.82 |
| D18 | 4.89 | Q pH 5.5 | 15000 | 15011 | 0.07% | 3.77 |
| D16:17 | 6.19 | Q pH 7.9 | 28006 | 28028 | 0.08% | 5.42 |
| D17:18 | 5.72 | Q pH 6.0 | 27923 | 27901 | −0.08% | 5.17 |
| D16:18 | 5.32 | Q pH 6.0 | 40504 | 40578 | 0.18% | 6.24 |
| D16:18Δe43,44 | 4.65 | Q pH 5.5 | 27422 | 27404 | −0.07% | 5.40 |
| D16:18Δe44,45 | 4.72 | Q pH 5.5 | 27354 | 27494 | 0.51% | 5.15 |
Purified proteins were subject to stability analysis by two techniques: thermal denaturation, and protease sensitivity analysis, both of which have also been previously described[20]. Briefly, the thermal denaturation was monitored by CD spectroscopy as the protein solution (~0.3 mg/ml in PBS, depending on the protein – each run was set to give an initial CD signal of ~50 millidegrees at 222 nm) was scanned up in temperature from ~13C to > 90 C using a Peltier type thermostated cell installed in a Jasco 715 spectrometer. The spectrometer was calibrated against a primary standard of camphor sulfonate, according to the manufacturer’s instructions.
To verify reversibility of the transition observed, the samples were cooled after the run back to the starting temperature, and the CD spectra re-acquired. In most cases, nearly full reversibility was observed. In other cases, the protein did not spontaneously refold, as judged by failure of the signal to return to the original φ222 value, and lack of regeneration of an α-helical CD spectrum. Visual examination of these samples at that time showed the presence of a white flocculence, suggesting macroscopic aggregation had occurred.
In these cases, we hypothesized that the system might be reversible through the transition region, but then slowly aggregated irreversibly during the prolonged incubation at higher temperatures needed to establish the high temperature unfolded baseline region. This was corroborated by examination of the highvoltage signal of the CD spectrometer. This signal is a proxy for the absorbance of the signal, and would be perturbed by the appearance of large aggregates which would scatter the beam. This signal showed an abrupt transition to a higher value (more absorbing) at some temperature above the transition temperature observed in the CD signal. This suggested that two separate events were occurring: a denaturation event followed by an aggregation event, and raised the possibility that we could obtain reversibility throughout most of the denaturation transition region. Thus, we conducted separate experiments in which identical samples were scanned to specific temperatures part way up the transition, but before the onset of aggregation, and then immediately cooled by immersion in a water bath at 37 °C for 2 minutes before being cooled to the starting temperature (generally 13 °C) and having the spectra re-acquired. Runs with increasing final temperatures were tested, at 1 °C intervals, to determine the maximum temperature to which we could heat the sample while retaining 80% reversibility upon cooling to 13 °C, as judged by return of φ222 to the original value. The point here is to validate the reversibility of the unfolding reaction through the crucial transition region, from which the thermodynamic parameters – especially Tm - are derived.
Thermal denaturation data was analyzed by application of a two state, constant CP model in conjunction with linear “folded” and “unfolded” pre and post-transition regions. The transition between these baselines is determined by the parameters Tm, ΔH and CP, which were obtained by fitting the experimental data using non-linear least squares method to the equation[23]
An alternative measure of stability is provided by protease sensitivity experiment as described[20]. Briefly, 10 ul aliquots containing 2.5 ug test proteins in PBS were exposed to a range of concentrations (a geometric dilution series) of a non-specific protease, Proteinase K, PK. A nonspecific protease is used so that the rates better reflect compactness of folding and stability of tertiary structure, rather than the idiosyncrasies of the primary sequence differences of each target[24–26]. The series was adjusted so that two steps were equivalent to a 10 fold dilution, and 7 concentrations between 68ng and 68 pg PK were typically used (protease: sample protein ratios of 1:37 to 1:37,000). Samples were incubated at 37 °C for 30 minutes, analyzed by SDS-PAGE, and the amount of undigested proteins assessed at each [PK] by densitometry using ScionImage, a windows port of NIH-Image. The fraction undegraded was fit to a first order exponential decay, and the concentration of PK needed to produce 50% destruction, PK50, was reported as a measure of resistance. In the case of D16:18Δe4344, this protein was very sensitive to PK, and we adjusted the PK dilution series, reducing the concentrations by a factor of 31.6 (=101.5, or three steps in our hemidecadal geometric dilution series) i.e. a range of 2.2 ng to 2.2 pg.
Results
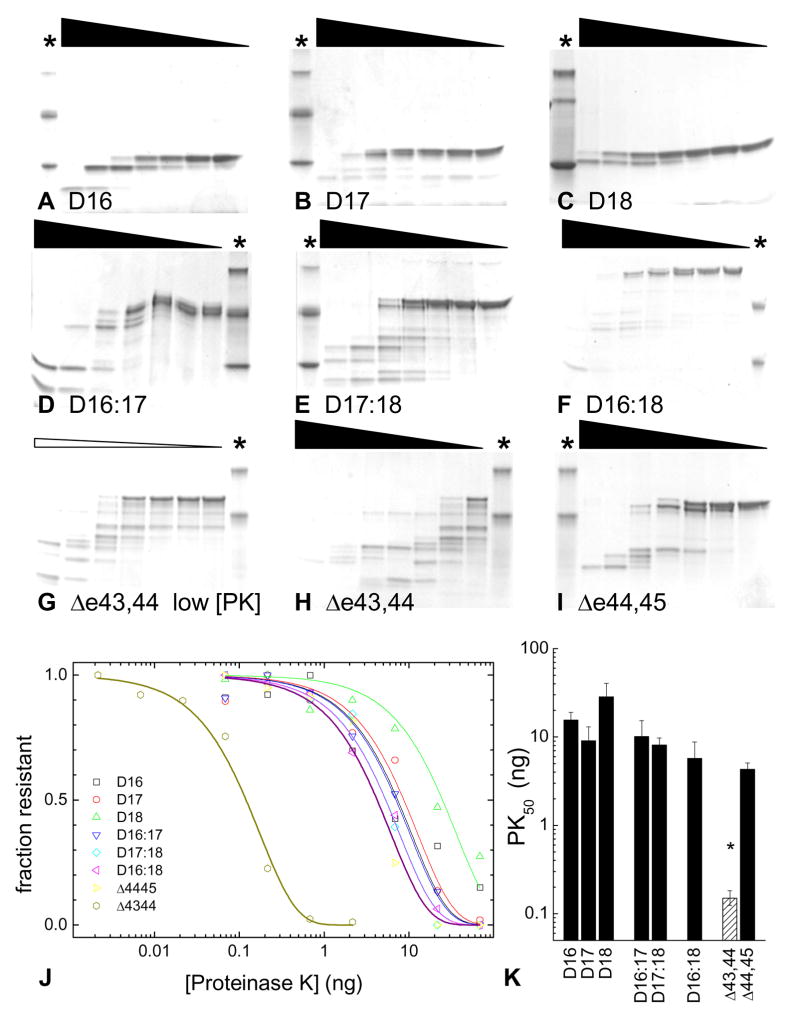
Proteins were purified to single band homogeneity by SDS PAGE; D16:18De43,44 was the sole exception. Preparations of this protein showed minor low molecular weight degradation bands despite our best efforts and the three-step chromatography procedure; an estimate of this can been seen in the right most lane of the proteolysis experiments (with insignificant PK) described below (i.e. Figure 3G). While this degradation was troubling, it did provide this first indication that this construct is much less stable than the other exon spliced construct, D16:18De44,45, or the other unskipped constructs.
Figure 3. Protease Susceptibility.
Proteins were assessed for their stability toward proteolysis by challenge with a non-specific protease, Proteinase K, PK. A constant amount of test protein was incubated with various dilutions of PK, as suggested by the wedges above each gel (although as discussed in the text, the dilution series was not linear but geometric, with two lanes equivalent to a ten-fold reduction in PK). One protein, D16:18Δe43,44 was so susceptible to PK, as seen in gel H, that separate experiments with a 31.6-fold diluted (= 101.5, or three lanes) initial PK were conducted. This is indicated by the thinner white wedge, gel G. Lanes containing molecular mass markers are indicated by a *; note that different sets of markers are used: F, C - (14, 40) A, B,D,E – (14, 27 40 kDa); G,H,I – (40,27 kDa). J Remaining intact protein was quantified by densitometry and fit to a first order decay model. The various unskipped proteins are shown using open symbols and thin lines, whereas the two exon skipped variants are shown with solid symbols and thick lines. Flanking sequences were used to accommodate ambiguities in alignment and facilitate protein expression (see figure 1), and we were not concerned with PK trimming these, but rather destruction of the STR motifs as a whole. As such, we considered all bands within ~2kDa of the undigested protein to be undegraded for purposes of this measurement, so that the measurement reports on PK sensitivity of the STR region, and not these flanking regions. It can be seen that the D16:18Δe43,44 is substantially less stable than all of the others, and that D16:18Δe44,45 falls very nearly within the region of the unskipped proteins. K Average PK50 values with error bars of one standard deviation of at least three independent experiments are shown. Statistical analysis of the entire data set by ANOVA indicates that there is a significant difference between the samples with P = 1.2*10−7; Tukey post hoc tests identify D16:18Δe43,44 as different from the rest by a wide margin, P< 3*10−5, which is indicated by an asterisk above that bar (which is also shaded lighter).
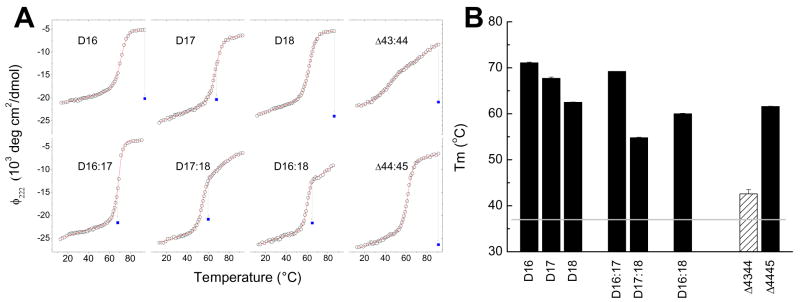
The first test quantitatively comparing the various proteins was thermal denaturation as monitored by CD spectroscopy, shown in Figure 2, which monitors the proper folding of the protein and provides some measure of the thermodynamic stability of the protein. It can be seen that the D16:18Δe43,44 variant is substantially less stable than any of the other proteins, whereas the alternative repair skipped proteins D16:18Δe44,45 has a Tm that lies within the range of the other unskipped rods, and in fact slightly above that of that parent D16:18. The transition of D16:18Δe43,44 is also much broader, indeed this protein is starting to unfold at physiological temperature of 37 °C. In contrast, all the other motifs – including the alternative repair exon skipped protein, D16:18Δe44,45 - exhibit much sharper transition at much higher temperatures, and appear to be essentially 100% folded at 37 °C.
Figure 2. Unfolding of the test proteins was monitored by CD spectroscopy.
A All proteins displayed a substantially α-helical signal (data not shown) so the ellipticity at 222 nm was used to monitor the unfolding as the temperature was scanned up at 1 C/min. The experimental data is shown by the open circles, and the transition was fit to a 2 state model as described. Reversibility of the transition was assessed by returning the sample to the start temperature (solid squares). In this case the temperature reflects the temperature from which it was cooled, not the temperature at which that ellipticity was measured, 13C. In some cases, reversibility was not seen after the complete scan; in this case separate experiments were conducted to determine the highest temperature from which at least 80% reversibility was seen, and those values are shown. B. Transition midpoints are shown for each protein; error bars show the standard deviation of three independent measurements. Note the low Tm of D16:18Δe43,44 in B, and the broad transition for this variant evident in A.
We can attempt to quantify this by examining the midpoint of unfolding and perhaps extracting thermodynamic stability parameters from these traces. This analysis relies on fitting the unfolding transitions to a thermodynamic model[23] and using nonlinear least squares fitting to extract the parameters, and implicitly assumes the transition is reversible. We have confirmed reversibility of the transitions of many of the proteins, including the two exon skipped variants, by returning the samples to the starting temperature and reacquiring the spectra. For most proteins reversibility was observed; however in two cases substantial irreversibility under standard conditions (scanning at 1 C/min to > 90 °C then cooling) was observed. In these cases, we were able to establish substantial reversibility (>80% signal returned) much of the way through the transition regions by the partial scan technique in which heating only proceeded partway through the transition before immediate rapid cooling. These highest temperatures of reversibility are indicated by the vertical lines in Figure 2, and the extent of return to more negative ellipticity upon cooling to 13 ° C indicated by the symbol at the lower terminus of this line. Since the transition region most strongly determines the thermodynamic parameters, reversibility through most of this region demonstrates that these parameters, reported in Table 2, are thus substantially reliable; the full, slow runs to high temperatures are needed only to establish reliable unfolded-baselines and this unfolded-baseline part of the scan does not greatly impact the thermodynamic parameters.
Table 2. Stability parameters of proteins studies.
Values reported are mean +/−standard deviation of at least 3 independent experiments.
| target protein | proteolysis | thermal unfolding | ||
|---|---|---|---|---|
| PK50 | Tm | ΔH | ||
| ng | log(ng) | °C | kJ/mol | |
| D16 | 15.5 | 1.19 +/− 0.15 | 71.1 +/− 0.1 | −299 +/− 8 |
| D17 | 9.0 | 0.96 +/− 0.32 | 67.6 +/− 0.3 | −250 +/− 11 |
| D18 | 28.4 | 1.45 +/− 0.26 | 62.5 +/− 0.1 | −271 +/− 3 |
| D16:17 | 7.0 | 0.85 +/− 0.22 | 69.2 +/− 0.1 | −505 +/− 12 |
| D17:18 | 8.1 | 0.91 +/− 0.14 | 54.8 +/− 0.1 | −341 +/− 14 |
| D16:18 | 5.7 | 0.76 +/− 0.32 | 59.1 +/− 0.1 | −432 +/− 23 |
| D16:18Δe43,44 | 0.15 | −0.82 +/− 0.14 | 42.6 +/− 0.9 | −179 +/− 26 |
| D16:18Δe44,45 | 4.7 | 0.68 +/− 0.15 | 61.6 +/− 0.1 | −326 +/− 6 |
However, this increasing irreversibility should be noted, and for those proteins, the ΔH values in particular might be considered less reliable. In any event, this is not a problem for either of the exon skipped variants which are fully reversible, and does not impact the major conclusion that the D16:18Δe43,44 is substantially less stable than either the alternative repair De44,45 or the wild type rods in this region, and in fact it is partially unfolded at physiological temperatures.
We also assessed the resistance of these proteins to proteolysis. Proteolysis of dystrophin is seen DMD and BMD[27], as well as in some certain other disease states involving dystrophin [28]. Even though protease challenge does not measure fundamental thermodynamic parameters as the thermal denaturation studies do, susceptibility to proteolysis is inversely correlated to compact folding, and so is also related stability[24, 29]. These experiments were conducted by challenging a constant bolus of test protein with a geometric dilution series of protease K, and assessing the extent of degradation by SDS PAGE, Figure 3. Again, protease challenge showed that D16:18Δe43,44 stood out as substantially less stable than the alternative exon skip repair D16:18Δe44,45. Furthermore, this best repair seems to be good enough to produce a rod with stability in the same range as unskipped rods in this region: although D16:18Δe44,45 exhibits a very slightly lower value PK50 =4.4 than wild type unskipped multi-STR rods, (D16:17, D17:18 and D16:18; 5.7 < PK50 < 10.1) this difference is not statistically significant (ANOVA P =0.33). Conversely, D16:18Δe43,44 is nearly two orders of magnitude less stable, at PK50 = 0.15, a difference that is strongly statistically significant. (ANOVA, P=1.2*10−7; post hoc Tukey tests identify D16:18Δe43,44 as the outlier, with P=2.7*10−5).
Discussion
We have thus demonstrated that: alternative repairs of the same underlying defect are not necessarily equivalent in the specific case of an exon44 deletion, a Δe44,45 repair results in a more faithful repair than a Δe43,44 repair.
We do note that some STRs in the related protein spectrin appear to have low stability when studied in isolation[30]. However these generally become more stable when studied in the context of larger rods, and both edits in this work were studied in the context of such larger tandem rods. As well, the specific result that the Δe43,44 repair produces a rod with significantly lower stability than Δe44,45 can be viewed in the context of the overall stability of all the STRs in this region, including the single STRs D16, D17 or D18. All of these, as well as the larger 2- and 3-STR unskipped tandem rods appear stable and well folded under physiological conditions (see Figures 2 and 3 and Table 2). Thus, this specific portion of the dystrophin rod appears to be a naturally stable region, and the Δe43,44 edit will change its character.
The Δe44,45 skip would produce a protein that roughly deletes the entirety of D17 whereas Δe43,44 leaves behind and juxtaposes the N terminal region of D16 and the C-terminal region of D17, creating a hybrid D(16,17) STR. While the feasibility of these types of hybrid STRs may have seemed to be questionable, we have recently shown that in some other cases they are quite stable and well folded[20]. In fact, that study was inspired by the observation that these types of hybrid STR producing exon skipping events predominate among naturally occurring alternative splicing in the dystrophin rod[31]. The test case in that study was Δe41,42, a naturally occurring event - so it is perhaps not surprising that it was viable. In fact, it is even tissue specific, suggesting that the resultant edited rod may occur naturally and have some biological relevance. Indeed, aside from therapy for DMD, the role of alternative splicing in dystrophin biology may be greater than appreciated, and may extend to other conditions such a X-linked dilated cardiomyopathy, a condition that is allelic with but distinct from DMD[32, 33].
Hybrid-STR type repairs were also the result of the first clinical test of exon skipping in humans[34], (skipping of exons 19 and 20, resulting in a D(4,5) hybrid) and are among the several outcomes of exon 51 skipping in the successful and ongoing Phase I clinical trial in humans[18] (i.e. the repair performed in patient 3 therein, Δe49-51 produces a hybrid STR, D(19,20)).
However, neither Δe43,44 nor Δe44,45 are among the limited cohort of naturally observed exon skipping events – nor are obviously the majority of possible exon skipping repairs envisioned for the many DMD mutations to which exon skipping repair offers hope. As such, our work here provides guidance in the design of optimal and effective strategies. While our work provides only information about the relative stability of two alternative skips designed to repair a Δe44 defect, this method could be applied to any given defect to produce and test a similar set of alternative skips in vitro before human or animal studies are employed, with their greater financial and ethical burdens.
The types of studies reported here are complementary to observations of naturally occurring dystrophin variants, mostly those involved in DMD or BMD. Such natural “experiments” are currently the main source of information about the consequences of various exon deletions, and large databases have been compiled such as the Leiden database, www.dmd.nl [19] and the Human Gene Mutation Database, www.hgmd.org [35]. However this clinical data reports a more complex and nuanced linkage between the underlying genetic defect and function than our direct studies, and in some cases might be misleading when considered in isolation. There are several reasons for this, including: 1) these large datasets are compiled from many sites worldwide over many decades, and are of varying accuracy; 2) clinical datasets are biased toward defects present in patients that are affected severely enough to warrant clinical treatment; 3) the severity of disease is the result not only of the defect in the patient, but also the genetic background of the patient.
The first two of these are defects of the database, and not of the underlying science. Simple errors in characterization of the underlying defects have been documented as analytical techniques have become more sophisticated[36], and represent part of the problem with relying exclusively on these sources. A more subtle problem concerns sample bias, which results in the underrepresentation of “good outcome” defects (i.e. subclinical, or very mild BMD) and an overestimation of the likelihood than any given defect might result in a “bad outcome” (i.e. DMD). For instance, every single one of the repairs being studied in the current Phase I/II clinical trial of exon skipping are linked to cases of DMD, BMD and IMD in the data bases, as are the two repairs studied in this work. (Actually, one the of the repairs in the clinical trial, De51,52, patient 4, is linked exclusively to DMD and IMD, and not at all to BMD – and so would seem to be especially problematic by this very narrow “examine the clinical databases” criterion.) However all those repairs seem to have produced some dystrophin production, and the trial is generally considered quite successful and is moving forward
Aside from simple error – case 1 above – a leading explanation of these “surprising outcomes” (DMD for in-frame defects, or BMD for out-of-frame; or more broadly heterogeneous outcomes for a single underlying defect) is variation in the mRNA maturation process due the differing genetic backgrounds of the patients yielding differing levels and alleles of various trans-acting splicing factors. It has recently been demonstrated that ~92% of human genes are alternatively spliced, and that this occurs in both a tissue- and individual-specific fashion[37]. It seems likely that the different genetic background of individuals as it pertains to mRNA maturation may lead them to react in different ways to various defects in the dmd gene – both underlying pathogenic defects, as well as therapeutically produced repairs. This process of alternative splicing of the dmd gene has long been known for its role in the production of low numbers of dystrophin positive revertant fibers in some DMD patients, and individual variation in alternative splicing of the dmd gene has been shown to occur[38], although the full extent of this phenomenon is still unclear. This process may be linked to the heterogeneity of clinical severity of specific defects, for instance if an individual has unusually active levels of some alternative splicing pathway that may naturally ameliorate some individual defect.
It should be noted that the vast majority of defects in these databases are characterized at either the DNA or RNA level, and have not been considered in the context of alternative splicing. However, the disease manifests itself though production (or lack thereof) of the dystrophin protein, so the effect of the defect on protein structure, stability and function is crucial. Unfortunately, full characterization of the specific defect at the protein level is very difficult and almost never done; typically only rough estimates of size differences are possible, and only for very large defects. It may eventually be necessary to take into account both a patient’s underlying defect as well as their status with respect to other factors involved in dmd mRNA as maturation to ensure production of functional dystrophin protein. However, that is a complex process that is currently not feasible. Our approach decouples some of this complexity, and allows us to clearly isolate effects of a specific edit on the dystrophin protein itself, without regard to these other process.
The Δe44 defect is particularly problematic, since if we cannot effectively repair it by a backwards skip to Δe43,44, or by the next in-frame forward skip, Δe44,45, the next available in-frame repair would necessitate deletion of 11 exons and produce Δe44,54. Although multiple exon skipping has been demonstrated, the more exons that need to be skipped, the more challenging the therapy becomes. A recent examination of the efficacy of a very similar skip Δe45–55 concluded that such multiexon skipping is currently not feasible[39]. Thus, those afflicted with this Δe44 defect are left with choosing between Δe44,43 or Δe44,45, both of these have been associated with DMD in the databases – yet one must be chosen if these patients are to be helped. We note that the Δe43,44 repair has been studied in ex vivo repair in cultured primary myoblasts from a DMD patient[16]. The specific defect being repaired in that case was not Δe44, but a point mutation in exon 43, and so Δe44,45 was not appropriate and was not attempted - so we can make no comparison. However, while the double exon skip repair was successful, and some Δe43,44 dystrophin was produced, the amount was low compared to normal levels – typical of AON mediated in vivo exon skipping, which is currently inefficient. However, this specific Δe43,44 variant also took longer to detect immunologically than the other skip tested in that study, involving a different exon, (Δe46–50→Δe45,51), Although other factors may be at work, such as the kinetics of the in vivo skipping, our study suggests that intrinsic instability this edit may contribute to the lower protein levels observed. In the only test of exon skipping in humans where comparisons can be made, involving the 4 patients treated with exon 51 skipping reagents, levels of dystrophin production also varied considerably. While many other factors including the previous progression of the disease and the extent of deterioration of the targeted muscle, and the specific genetic background of the patient, may have contributed, it seems likely that the different repairs were successful to differing extents. Unfortunately, alternative repairs in a single patient were not attempted, so again no direct comparison can be made.
This complexity suggests that empirical studies such we have conducted herein are valuable in helping us to understand the consequences of any given repair on the dystrophin protein eventually produced. While it seems highly likely that any sort of frame restoration will provide significant benefit to DMD patients who currently face a bleak outlook, it is also far from clear that all outcomes will be effective, and even less so that they will all be equivalent. This will become particularly relevant as patients hopefully experience expanded lifespan and improvements in quality of life which will place greater demands on muscle long-term stability and function – so that not only a minimal restoration, but rather an optimal restoration of the dystrophin gene is needed. We hope that the concept of testing and considering the impact on dystrophin protein structure and function of alternative exon skipping strategies can help design more effective therapeutic interventions for the devastating condition that is DMD.
Acknowledgments
This work was financially supported by the NIH/NIAMS through 5R01AR053970.
Footnotes
Abbreviations used: DMD: Duchenne muscular dystrophy; BMD Becker muscular dystrophy; STR: spectrin type repeat; Dx: a protein composed of the xth STR motif of dystrophin; Dx:y: a protein spanning the xth to the yth STR of dystrophin. D16:18Δe43,44 or equivalently in figures for brevity, Δe43,44: D16:18 with the region coding for exons 43 and 44 deleted; D16:18Δe44,45 or equivalently Δe44,45: D16:18 with the region coding for exons 44 and 45 deleted
To be sure, a much larger skip forward, deleting from exon 44 to 54 would also restore the reading frame. However the difficulty and efficiency decreases with number, and most exon skipping strategies envisioned in the near term involve only one or a few exons.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O’Brien KF, Kunkel LM. Dystrophin and muscular dystrophy: past, present, and future. Mol Genet Metab. 2001;74:75–88. doi: 10.1006/mgme.2001.3220. [DOI] [PubMed] [Google Scholar]
- 2.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–40. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 3.Judge LM, Haraguchiln M, Chamberlain JS. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J Cell Sci. 2006;119:1537–46. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
- 4.Koenig M, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265:4560–6. [PubMed] [Google Scholar]
- 5.Broderick MJ, Winder SJ. Spectrin, alpha-actinin, and dystrophin. Adv Protein Chem. 2005;70:203–46. doi: 10.1016/S0065-3233(05)70007-3. [DOI] [PubMed] [Google Scholar]
- 6.Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150:1209–14. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybakova IN, Humston JL, Sonnemann KJ, Ervasti JM. Dystrophin and utrophin bind actin through distinct modes of contact. J Biol Chem. 2006;281:9996–10001. doi: 10.1074/jbc.M513121200. [DOI] [PubMed] [Google Scholar]
- 8.Amann KJ, Renley BA, Ervasti JM. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J Biol Chem. 1998;273:2841923. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- 9.An X, Guo X, Gratzer W, Mohandas N. Phospholipid binding by proteins of the spectrin family: a comparative study. Biochem Biophys Res Commun. 2005;327:794–800. doi: 10.1016/j.bbrc.2004.12.063. [DOI] [PubMed] [Google Scholar]
- 10.Le Rumeur E, Fichou Y, Pottier S, Gaboriau F, Rondeau-Mouro C, Vincent M, Gallay J, Bondon A. Interaction of dystrophin rod domain with membrane phospholipids. Evidence of a close proximity between tryptophan residues and lipids. J Biol Chem. 2003;278:5993–6001. doi: 10.1074/jbc.M207321200. [DOI] [PubMed] [Google Scholar]
- 11.Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–17. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Watchko J, O’Day T, Wang B, Zhou L, Tang Y, Li J, Xiao X. Adeno-associated virus vector-mediated minidystrophin gene therapy improves dystrophic muscle contractile function in mdx mice. Hum Gene Ther. 2002;13:1451–60. doi: 10.1089/10430340260185085. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Li J, Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc Natl Acad Sci U S A. 2000;97:13714–9. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, Davies KE. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–2. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 15.Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF, Harper HA, Robinson AS, Engelhardt JF, Brooks SV, Chamberlain JS. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8:253–61. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 16.Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, van Ommen GJ, den Dunnen JT, van Deutekom JC. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet. 2004;74:83–92. doi: 10.1086/381039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, Danos O. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–9. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 18.van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, de Kimpe SJ, Ekhart PF, Venneker EH, Platenburg GJ, Verschuuren JJ, van Ommen GJ. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–86. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 19.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–44. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 20.Menhart N. Hybrid spectrin type repeats produced by exon-skipping in dystrophin. Biochim Biophys Acta. 2006;1764:993–9. doi: 10.1016/j.bbapap.2006.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saadat L, Pittman L, Menhart N. Structural cooperativity in spectrin type repeats motifs of dystrophin. Biochim Biophys Acta. 2006;1764:943–54. doi: 10.1016/j.bbapap.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Mirza A, Menhart N. Stability of dystrophin STR fragments in relation to junction helicity. Biochim Biophys Acta. 2008;1784:1301–9. doi: 10.1016/j.bbapap.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–77. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 24.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 25.Spolaore B, Bermejo R, Zambonin M, Fontana A. Protein interactions leading to conformational changes monitored by limited proteolysis: apo form and fragments of horse cytochrome c. Biochemistry. 2001;40:9460–8. doi: 10.1021/bi010582c. [DOI] [PubMed] [Google Scholar]
- 26.Polverino de Laureto P, Frare E, Gottardo R, Van Dael H, Fontana A. Partly folded states of members of the lysozyme/lactalbumin superfamily: a comparative study by circular dichroism spectroscopy and limited proteolysis. Protein Sci. 2002;11:2932–46. doi: 10.1110/ps.0205802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assereto S, Stringara S, Sotgia F, Bonuccelli G, Broccolini A, Pedemonte M, Traverso M, Biancheri R, Zara F, Bruno C, Lisanti MP, Minetti C. Pharmacological rescue of the dystrophin-glycoprotein complex in Duchenne and Becker skeletal muscle explants by proteasome inhibitor treatment. Am J Physiol Cell Physiol. 2006;290:C577–82. doi: 10.1152/ajpcell.00434.2005. [DOI] [PubMed] [Google Scholar]
- 28.Badorff C, Knowlton KU. Dystrophin disruption in enterovirus-induced myocarditis and dilated cardiomyopathy: from bench to bedside. Med Microbiol Immunol. 2004;193:121–6. doi: 10.1007/s00430-003-0189-7. [DOI] [PubMed] [Google Scholar]
- 29.Tsai CJ, Polverino de Laureto P, Fontana A, Nussinov R. Comparison of protein fragments identified by limited proteolysis and by computational cutting of proteins. Protein Sci. 2002;11:1753–70. doi: 10.1110/ps.4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An X, Guo X, Zhang X, Baines AJ, Debnath G, Moyo D, Salomao M, Bhasin N, Johnson C, Discher D, Gratzer WB, Mohandas N. Conformational stabilities of the structural repeats of erythroid spectrin and their functional implications. J Biol Chem. 2006;281:10527–32. doi: 10.1074/jbc.M513725200. [DOI] [PubMed] [Google Scholar]
- 31.Sironi M, Cagliani R, Pozzoli U, Bardoni A, Comi GP, Giorda R, Bresolin N. The dystrophin gene is alternatively spliced throughout its coding sequence. FEBS Lett. 2002;517:163–6. doi: 10.1016/s0014-5793(02)02613-3. [DOI] [PubMed] [Google Scholar]
- 32.Gualandi F, Rimessi P, Cardazzo B, Toffolatti L, Dunckley MG, Calzolari E, Patarnello T, Muntoni F, Ferlini A. Genomic definition of a pure intronic dystrophin deletion responsible for an XLDC splicing mutation: in vitro mimicking and antisense modulation of the splicing abnormality. Gene. 2003;311:25–33. doi: 10.1016/s0378-1119(03)00527-4. [DOI] [PubMed] [Google Scholar]
- 33.Ferlini A, Sewry C, Melis MA, Mateddu A, Muntoni F. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul Disord. 1999;9:339–46. doi: 10.1016/s0960-8966(99)00015-2. [DOI] [PubMed] [Google Scholar]
- 34.Takeshima Y, Yagi M, Wada H, Ishibashi K, Nishiyama A, Kakumoto M, Sakaeda T, Saura R, Okumura K, Matsuo M. Intravenous infusion of an antisense oligonucleotide results in exon skipping in muscle dystrophin mRNA of Duchenne muscular dystrophy. Pediatr Res. 2006;59:690–4. doi: 10.1203/01.pdr.0000215047.51278.7c. [DOI] [PubMed] [Google Scholar]
- 35.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 36.Zeng F, Ren ZR, Huang SZ, Kalf M, Mommersteeg M, Smit M, White S, Jin CL, Xu M, Zhou DW, Yan JB, Chen MJ, van Beuningen R, den Dunnen J, Zeng YT, Wu Y. Array-MLPA: comprehensive detection of deletions and duplications and its application to DMD patients. Hum Mutat. 2008;29:190–7. doi: 10.1002/humu.20613. [DOI] [PubMed] [Google Scholar]
- 37.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sironi M, Cagliani R, Comi GP, Pozzoli U, Bardoni A, Giorda R, Bresolin N. Trans-acting factors may cause dystrophin splicing misregulation in BMD skeletal muscles. FEBS Lett. 2003;537:30–4. doi: 10.1016/s0014-5793(03)00066-8. [DOI] [PubMed] [Google Scholar]
- 39.van Vliet L, de Winter CL, van Deutekom JC, van Ommen GJ, Aartsma-Rus A. Assessment of the feasibility of exon 45–55 multiexon skipping for duchenne muscular dystrophy. BMC Med Genet. 2008;9:105. doi: 10.1186/1471-2350-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]