Abstract
Infection with West Nile virus (WNV) causes a febrile illness that can progress to meningitis or encephalitis, primarily in humans that are immunocompromised or elderly. For successful treatment of WNV infection, accurate and timely diagnosis is essential. Previous studies have suggested that the flavivirus non-structural protein NS1, a highly conserved and secreted glycoprotein, is a candidate protein for rapid diagnosis. Herein, we developed a capture enzyme-linked immunosorbent assay (ELISA) to detect WNV NS1 using two anti-NS1 monoclonal antibodies (mAbs) that map to distinct sites on the protein. The capture ELISA efficiently detected as little as 0.5 ng/ml of soluble NS1 and exhibited no cross-reactivity for yellow fever, Dengue, and St. Louis encephalitis virus NS1. The capture ELISA reliably detected NS1 in plasma at day 3 after WNV infection, prior to the development of clinical signs of disease. As the time course of infection continued, the levels of detectable NS1 diminished, presumably because of interference by newly generated anti-NS1 antibodies. Indeed, treatment of plasma with a solution that dissociated NS1 immune complexes extended the window of detection. Overall, the NS1-based capture ELISA is a sensitive readout of infection and could be an important tool for diagnosis or screening small molecule inhibitors of WNV infection.
Keywords: flavivirus, diagnostic, pathogenesis, plasma, antibody
INTRODUCTION
West Nile virus (WNV) is a single stranded, positive sense enveloped RNA virus that is maintained in nature through a mosquito–bird–mosquito transmission cycle. A member of the Flaviviridae family, WNV is closely related to other significant human pathogens including yellow fever (YFV), dengue (DENV), tick-borne encephalitis (TBEV), Japanese encephalitis (JEV), Murray Valley encephalitis (MVEV), and St. Louis encephalitis (SLEV) viruses. WNV has been endemic in parts of Africa, Europe, the Middle East, Asia, and in Australia, where the more benign Kunjin virus (KUNV) variant circulates [Hall et al., 2003]. However, since 1999, WNV infections occur annually in North America. Humans, which are dead-end hosts for transmission, develop a febrile illness that progresses to meningitis, encephalitis or acute flaccid paralysis in a subset of individuals [Hubalek and Halouzka, 1999; Petersen et al., 2003; Sejvar et al., 2003]. Although treatment is supportive and no vaccine exists for humans, recent studies suggest that passive transfer of antibodies against WNV could have therapeutic potential [Ben-Nathan et al., 2003; Engle and Diamond, 2003; Gould et al., 2005; Julander et al., 2005; Oliphant et al., 2005; Chung et al., 2006; Morrey et al., 2006; Throsby et al., 2006]. As there appears to be a narrow treatment window for therapeutic efficacy, rapid diagnosis of WNV infection will be essential [Agrawal and Petersen, 2003].
Classically, flavivirus infection has been diagnosed by indirect immunofluorescence staining of infected cells, a plaque reduction neutralization assay, or virus isolation from patient serum samples [Yamada et al., 2002; Martin et al., 2004; Oceguera et al., 2007]. However, these assays are labor-intensive, require a biosafety level (BSL)-3 facility, and do not provide diagnostic information rapidly. Antibody-based serological assays are useful but may be limited because of a several day lag between infection and seroconversion [Tardei et al., 2000; Petersen et al., 2003; Ratterree et al., 2004]. Moreover, because of cross-reactivity of anti-flavivirus antibodies, prior exposure to related viruses or vaccines could limit the utility of antibody-based diagnostic tests [Koraka et al., 2001]. Although detection of viral RNA in blood samples by reverse transcriptase-PCR (RT-PCR) or nucleic acid amplification techniques provides a specific diagnosis at early time points, they are relatively expensive and require trained personnel and equipment. In addition, the amplitude and duration of viremia during human WNV infection are relatively low and short [Busch et al., 2005 a, b] compared to other flaviviruses, such as DENV, [Vaughn et al., 2000], resulting in a smaller window of detection of WNV nucleic acid in serum or plasma samples.
An alternative diagnostic approach is to measure antigenemia, which can only occur during an active infection. Previous studies have suggested that the secreted glycoprotein NS1, may be a useful diagnostic marker [Young et al., 2000; Alcon et al., 2002; Macdonald et al., 2005]. NS1 is a conserved 48-kilodalton (kDa) non-structural glycoprotein. Within infected cells, NS1 is believed to function as a co-factor in viral RNA replication [Mackenzie et al., 1996; Muylaert et al., 1996; Lindenbach and Rice, 1997; Khromykh et al., 1999]. Unlike the other non-structural proteins, NS1 is secreted [Winkler et al., 1988, 1989; Mason, 1989; Macdonald et al., 2005] and high levels are detected in the serum of flavivirus-infected patients [Young et al., 2000; Alcon et al., 2002; Libraty et al., 2002], and correlate with the development of severe disease in DENV infection. In this study, we used two mAbs to NS1 to develop a highly sensitive and specific diagnostic capture ELISA for WNV infection.
MATERIALS AND METHODS
Cells and Viruses
BHK21-15 cells were cultured as previously described [Diamond et al., 2000b]. The majority of experiments were performed with the WNV strain (3000.0259, passage 2) that was isolated in New York in 2000 [Ebel et al., 2001]. Some experiments were also performed with a lineage II WNV (strain 956 [Wengler and Gross, 1978]), DENV-2 (strain 16681 [Russell and Nisalak, 1967]), YFV (17D vaccine strain), or SLEV (strain GHA3 [Kramer and Chandler, 2001], Tampa Bay, FL, 1962) and were obtained from colleagues (E. Harris (Berkeley, CA), T. Chambers (St. Louis, MO), and A. Barrett (Galveston, TX)). For mouse experiments, viruses were diluted in Hank’s Balanced Salt Solution with 1% heat-inactivated serum and injected as described [Diamond et al., 2003a].
WNV NS1 Protein
Expression and purification of recombinant WNV NS1 from insect cells was described previously [Chung et al., 2006]. Briefly, the complete NS1 gene with an initiator methionine and 72 last nucleotides of WNV E (endogenous signal sequence) was cloned into pFastBac1 (Invitrogen, Carlsbad, CA), and expressed in baculovirus-infected SF9 insect cells under serum free conditions. Soluble NS1 was purified to homogeneity after sequential nickel affinity, size exclusion, and Mono Q ion-exchange chromatography. Protein concentration was determined by measuring the optical density at 280 nm wavelength using an MBA 2000 spectrometer (Perkin Elmer, Waltham, MA) with a coefficient of extinction of 2.026.
For NS1 protein quantitation of cell culture supernatants, BHK cells were plated into 6-well plates at a cell density of 6 × 105 cells per well with 1.5 ml of Dulbecco’s Modified Eagle Medium (DMEM) media with 10% FBS. Subsequently, cells were infected with lineage I or II WNV strains at a given multiplicity of infection (MOI). Cell supernatants from infected or uninfected cells were collected at 6, 12, 24, and 48 hr, centrifuged, and stored at 4°C prior to analysis. The amount of NS1 was calculated after subtracting the amount of NS1 in the original infective viral stock.
Flow Cytometry
After infection (MOI of 0.2) with YFV, SLEV, DENV-2 or WNV, immunoreactivity against intracellular NS1 levels was analyzed by flow cytometry of permeabilized cells as described previously [Diamond et al., 2000a]. Two or three days after infection, infected BHK cells were washed three times in PBS, fixed in PBS with 4% paraformaldehyde for 10 min at room temperature, washed twice in PBS, and permeabilized in Hank’s balanced salt solution (Sigma Chemical Co., St. Louis, MO) containing 10 mM HEPES (pH 7.3), 0.1% saponin (Sigma Chemical Co.), and 0.02% NaN3 (HHSN). For indirect immunofluorescence experiments, cells were resuspended in HHSN and individual mAbs, incubated for 1 hr at 4°C, washed three times in HHSN (4°C), resuspended in a 1/500 dilution of Alexa-Fluor 647-labeled goat anti-mouse IgG (Invitrogen), and incubated for 1 hr on ice in the dark. Cells were subsequently washed three times in HHSN (4°C), fixed in 1% paraformaldehyde in PBS, and stored prior to analysis on a Becton Dickinson FACSCaliber flow cytometer.
Primary Cell Cultures and Virus Infection
Bone marrow-derived macrophages (BM-Mφ) and dendritic cells (BM-DC) were generated as described previously [Samuel et al., 2006]. Briefly, bone marrow cells from C57BL/6 wild-type mice were cultured in 12-well plates at a density of 1.5 × 105 cells per well. BM-Mφ were differentiated in DMEM containing 40 ng/ml macrophage colony-stimulating factor (M-CSF; PeproTech, Inc., Rocky Hill, NJ), and BM-DCs were cultured in RPMI medium supplemented with 20 ng/ml of granulocyte-M-CSF and 20 ng/ml of interleukin-4 (PeproTech, Inc.). Following 7–8 days of differentiation, cell purity was determined by flow cytometry after staining with an anti-F4/80 antibody for BM-Mφ (Serotec, Inc., Raleigh, NC) or an anti-CD11c antibody for BM-DC (BD Pharmingen, San Diego, CA). Primary cortical neurons were prepared from day 15 C57BL/6 mouse embryos essentially as described [Klein et al., 2005], and experiments were performed using neurons cultured for 3–4 days. The purity (~98–99%) of cortical neuron cultures was determined via staining with anti-MAP2 (Chemicon, Temecula, CA). To determine the level of viral infection in primary cells, BM-Mφ, BM-DC, and primary cortical neurons were infected at an MOI of 0.01 for 1 hr and then washed extensively. Supernatants were harvested at 72 hr postinfection, and infectious virus was measured by plaque assay on BHK21-15 cells.
Monoclonal and Polyclonal Antibodies Against NS1
MAbs against WNV NS1 were generated, purified, and biotinylated as described previously [Chung et al., 2006]. Polyclonal antibody to WNV NS1 was produced in BALB/c mice after five injections (3–4-week intervals) with insect cell-generated, purified WNV NS1 protein complexed with adjuvant (RIBI Immunochemical, Hamilton, MI). The titer of pooled polyclonal antisera was greater than 1/400,000.
Mouse Experiments and Sampling of Plasma
Wild-type C57BL/6 mice were purchased from a commercial source (Jackson Laboratories, Bar Harbor, ME). Mice were inoculated subcutaneously with a given plaque forming unit (PFU) dose of WNV by footpad injection after anesthetization with xylazine and ketamine. Mouse experiments were approved and performed according to the guidelines of the Washington University School of Medicine Animal Safety Committee.
To harvest plasma, whole blood (300–500 μl) was collected by phlebotomy of the axillary vein immediately prior to euthanasia into tubes containing 45 μl of heparin (103 units/ml; Elkins-Sinn Inc., Cherry Hill, NJ) and 5 μl of 100 mM EDTA. After centrifugation, plasma samples were aliquotted and stored at −80°C.
Quantitation of Antibodies and Viral RNA
The levels of WNV-specific IgM and IgG in plasma were determined using an ELISA as previously described [Diamond et al., 2003b]. In brief, purified WNV NS1 protein was adsorbed overnight at 4°C to microtiter plates (Maxi-Sorp; Nunc, Rochester, NY). Non-specific binding was blocked after incubation with Blocking buffer (25 mM Tris-base (pH 7.5), 150 mM NaCl, 1.5% BSA, 3% heat-inactivated horse serum, 0.025% NaN3, and 0.025% NP-40) for 1 hr at 37°C. Plates were incubated with serial dilutions of heat-inactivated plasma from infected mice for 1 hr at 4°C. After extensive washing, plates were incubated serially with biotin-conjugated goat anti-mouse IgM (5 μg/ml) or anti-mouse IgG (2 μg/ml; Sigma Chemical Co.) and horse-radish peroxidase-conjugated streptavidin (2 μg/ml; Sigma Chemical Co.) for 1.5 hr. Enzyme activity was detected using the substrate 3, 3′,5, 5′-tetramethyl-benzidine (DakoCytomation, Carpinteria, CA), and 1 M H2SO4 was added to stop the enzymatic reaction. Optical densities were determined with an ELISA plate reader at 450 nm (Molecular Devices, Sunnyvale, CA).
For quantitation of viral RNA, serum samples were obtained from whole blood by phlebotomy of the axillary vein immediately prior to sacrifice. Viral RNA was harvested from aliquots (50 μl) of serum using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA). Viral RNA was quantitated by real-time fluorogenic reverse transcriptase PCR (RT-PCR) using an ABI 7000 sequence detection system (Applied Biosystems, Foster City, CA) according to a published protocol [Lanciotti et al., 2000].
Generation of NS1-Antibody Complexes
To determine the effect of NS1-antibody immune-complex on the performance of the capture ELISA, we generated NS1-antibody complexes in vitro using purified NS1 from baculovirus-infected SF9 cells and mouse polyclonal antibody against WNV NS1. Purified NS1 (10 ng/ml) was incubated with serially diluted polyclonal antibody for 3 hr at room temperature.
NS1 Capture ELISA
Purified 10NS1 capture mAb was adsorbed overnight at 4°C to Maxi-Sorp microtiter plates (Nalge Nunc International, Rochester, NY) in coating buffer (100 mM NaHCO3, 100 mM Na2CO3, pH 9.3) at 50 μg/ml and 55 μl/well. Non-specific binding was blocked after incubation with Blocking buffer for 1 hr at 37°C and rinsing with Washing buffer (25 mM Tris-base (pH 7.5), 150 mM NaCl, 0.5% BSA, 0.025% Sodium NaN3, and 0.025% NP-40). Samples or serial dilutions of purified WNV NS1 for the standard curve were added to plates coated with 10NS1 and incubated for 2 hr at room temperature. Plates were washed, and incubated serially with 55 μl of biotinylated 3NS1 (5 μg/ml, for 1.5 hr) and streptavidin-conjugated horseradish peroxidase (2 μg/ml, Invitrogen) for 1.5 hr, and developed as described above. In the assay, the net value of optical density reading at 450 nm (OD450) was defined after subtracting OD450 value for negative control. The concentration of NS1 was calculated after comparison with a standard curve, which was performed on each ELISA plate.
NS1-Antbody Immune Complex Dissociated Capture ELISA
To release NS1 from immune complexes, plasma samples were incubated with an alkaline solution. A plasma sample (55 μl) or soluble WNV NS1 (for the standard curve) was mixed with 6 μl of virus-inactivating solution (0.25% NP-40 and 150 mM NaCl). After a 30 min incubation at room temperature, the samples were mixed with 61 μl of dissociation solution (1 M Tris-base (pH10.5),2% TritonX-100, and 150 mMNaCl), incubated for 1.5 hr at 37°C, and then neutralized with 20 μl of 2N HCl. After an additional 1 hr incubation at 37°C and addition of 100 μl of 150 mM NaCl, the mixture (~240 μl) was added to microtiter wells containing the 10NS1 capture antibody. After an overnight incubation at 4°C, the plate was washed, incubated with 55μl ofbiotinylated 3NS1 (5 μg/ml) and processed as described above.
Statistical Analysis
All data were analyzed with Prism software (Graph-Pad Software). Statistical significance was determined using a two-tailed paired or unpaired Student’s t-test.
RESULTS
Development of the NS1 Capture ELISA
To create a specific and sensitive antigen capture ELISA that measured levels of soluble NS1 of WNV, we screened a panel of 22 previously generated mAbs against WNV NS1 [Chung et al., 2006]. The criteria for selecting two mAbs in the capture ELISA included strength of binding to NS1, lack of competitive binding, and an absence of cross-reactivity with other flavivirus NS1 proteins. After iterative testing, 10NS1 and 3NS1 were selected as the capture and detection antibodies, respectively. These two mAbs bound with high avidity to soluble NS1 (data not shown), and did not cross-react with DENV-2 NS1 or interfere with each other in competitive binding assays [Chung et al., 2006]. In addition, both mAbs bound to conformationally sensitive epitopes (data not shown).
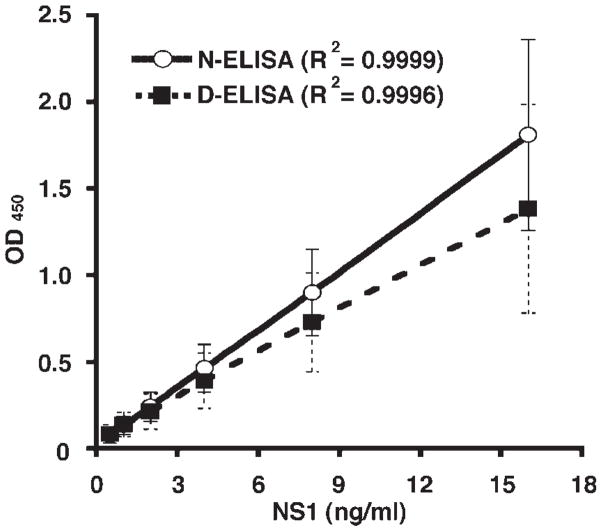
To determine the limit of detection of the capture ELISA, we serially diluted recombinant NS1 that was generated in insect cells. Purified NS1 was diluted in 10% FBS or mouse plasma to determine the sensitivity of the assay. We observed little inhibitory effect of mouse plasma up through the maximum concentration (22%) tested, results that agree with that seen with human serum and a DENV NS1 capture ELISA [Young et al., 2000]. The limit of detection of WNV NS1 capture ELISA was 0.5 ng/ml and a dose-dependent linear response was observed in the range of 0.5–16 ng/ml (Fig. 1); higher concentrations resulted in a plateau in the signal, although dilution of these samples increased the assay dynamic range (data not shown). Similar results were observed with NS1 generated from BHK21 hamster cells that stably propagate a WNV replicon (data not shown). Thus, the two mAb ELISA using 10NS1 and 3NS1 show ~20-fold greater sensitivity than previously developed capture ELISA against DENV [Young et al., 2000] or WNV NS1 [Macdonald et al., 2005].
Fig. 1.
WNV NS1 capture ELISA. NS1 capture ELISA dose–response curve with different concentrations of untreated (N-ELISA; solid line) or alkaline-treated (D-ELISA; dashed lines) purified NS1. Untreated or alkaline-treated NS1 was diluted serially and added to 10NS1-coated plates, washed, and detected with biotinylated 3NS1. The graph represents the average results of four independent experiments performed in duplicate. Error bars indicate standard deviations. The R2 values of the linear regressions are indicated.
NS1 Capture ELISA Performance
To test whether the NS1 capture ELISA reliably detected secreted NS1 from WNV-infected mammalian cells, we assayed BHK cell supernatants after infection. Supernatants were harvested at 6, 12, 24, and 48 hr after infection at different doses (MOI of 0.1, 0.5, 2.5, or 12.5) with a lineage I (New York 2000) virulent WNV isolate. Supernatants were clarified by low-speed centrifugation and NS1 was measured by capture ELISA after serial dilution. At 6 hr, which corresponds to a time when the viral RNA is transitioning from translation to replication [Lindenbach and Rice, 2001], we could not detect secreted NS1 in any of the samples (Fig. 2A). By 12 hr, ~2–100 ng/ml of NS1 was detected in supernatants of cells infected at MOI of 0.5–12.5; as expected, the levels of soluble NS1 directly correlated with the inoculating MOI. At 48 hr after infection, high levels of NS1 were detected in all samples, ranging from 5,000 to 8,000 ng/ml. Although lower levels were measured, the capture ELISA also detected NS1 from a genetically divergent lineage II WNV strain (Fig. 2A). The lower amounts of NS1 observed in supernatants from cells infected with the lineage II WNV strain may reflect a slightly decreased avidity of the detection mAb, 3NS1, for lineage II NS1 [Chung et al., 2006] or an inherently reduced replication rate of this attenuated strain [Pierson et al., 2005].
Fig. 2.
Detection of NS1 from WNV-infected BHK and primary cells. A: Quantitation of NS1 from WNV-infected BHK cells. BHK cells were infected with lineage I (Lin I, New York 1999) or lineage II (Lin II, 956) WNV strains at an MOI of 0.1, 0.5, 1, 2.5, or 12.5. The culture media were collected at 6, 12, 24, and 48 hr after infection and measured by NS1 capture ELISA. The graph shows the average results of two independent experiments performed in duplicate. Note the break in the y-axis. B: Detection of secreted NS1 from WNV-infected mouse primary cells. Primary cortical neurons (neuron), BM-DC (DC), and BM-Mφ (Mφ) were infected with WNV (Lin I) at an MOI of 0.01 and extensively washed. At 72 hr after infection, NS1 was quantitated by capture ELISA. In parallel, infectious virus in culture fluid was measured by plaque assay on BHK cells. Each dot and black bar represents viral titers and NS1 concentrations at each point. The graph shows the average results of two independent experiments. Error bars indicate standard deviations. The concentration of NS1 was calculated after comparison with a standard curve, which was performed on each ELISA plate.
To test whether the NS1 capture ELISA could readily detect secreted NS1 from primary cells, we generated and infected mouse primary cortical neurons, BM-Mφ, and dendritic cells (BM-DC) at an MOI of 0.01. At 72 hr after infection, the levels of NS1 reached ~40 and 110 ng/ml in supernatants from cortical neurons and BM-DC, respectively. In contrast, the BM-Mφ which are less permissive as evidenced by the lower viral yield, did not secrete NS1 or secreted NS1 at levels below the sensitivity of detection of our assay (Fig. 2B). Thus, the level of NS1 in the supernatant of infected primary cells was directly related to virus production.
NS1 is a conserved glycoprotein among flaviviruses (44% amino acid identity and 65% amino acid homology). Not surprisingly, some anti-NS1 antibodies have exhibited cross-reactivity with other flaviviruses [Falconar and Young, 1991; Puttikhunt et al., 2003; Chung et al., 2006; Clark et al., 2007]. To establish the specificity of the NS1 capture ELISA, we tested the cross-reactivity of 3NS1 and 10NS1 with clinically relevant flaviviruses that co-circulate in Western Hemisphere: YFV, SLEV and DENV. Notably, 3NS1 and 10NS1, showed no immunoreactivity with cells infected with YFV, SLE, or DENV-2 (Fig. 3), and did not detect secreted forms of NS1 from these viruses (data not shown). In addition, 10NS1 did not recognize insect cells infected with the closely related JEV and MVEV (R. Hall, personal communication). Against the clinically relevant flaviviruses that we examined, the two mAb capture ELISA appears specific for WNV NS1.
Fig. 3.
Flow cytometry analysis of cross-reactivity of 3NS1 and 10NS1 mAb against flaviviruses. BHK cells were infected with DENV, YFV, SLEV, and WNV at an MOI of 0.2. Immunoreactivity of 3NS1 and 10NS1 mAb was analyzed in saponin-permeablized cells. The cross-reactive 4G2 mAb against flavivirus E was used as a positive control and uninfected BHK cells were used as negative control. Arrows indicate that the binding of mAbs to NS1. The data is representative of two independent experiments.
Detection of NS1 in Plasma Samples
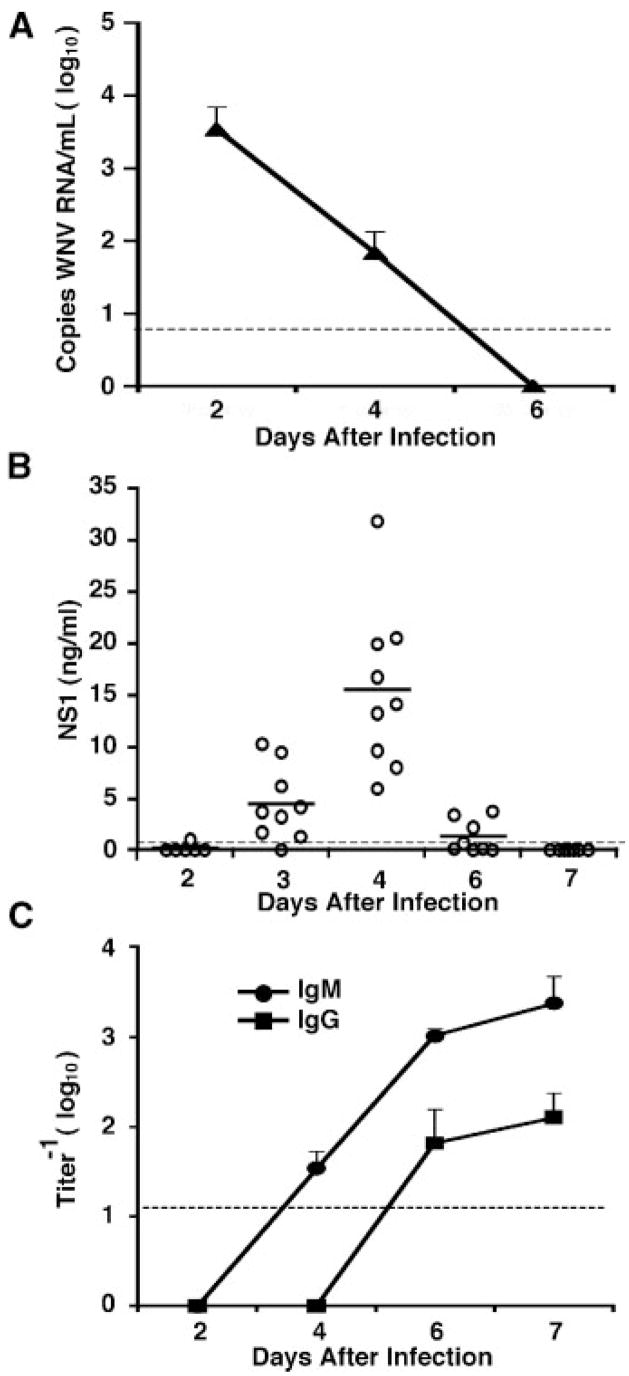
To begin to examine the applicability of the NS1 capture ELISA as a diagnostic tool, plasma was collected from mice (days 0, 2, 3, 4, 6, and 7) after infection with 2 × 105 PFU of WNV. Although viral RNA was detected in plasma by fluorogenic quantitative RT-PCR at day 2 after infection, it rapidly decreased to levels of below detection by day 6 (Fig. 4A). By day 6, viral RNA in all mice (5 of 5) was undetectable. Soluble NS1 was reliably detected beginning at day 3 after infection, with peak NS1 concentrations at day 4 after infection (Fig. 4B). At day 3, NS1 was detected in 89% (8 of 9) of plasma samples but by day 6, only 38% (3 of 8) of plasma samples were positive for NS1 antigen. By day 6 after infection, mice showed evidence of systemic infection as judged by a 16% reduction in body weight (data not shown). Unfortunately, the capture ELISA did not detect NS1 at day 7 after infection on untreated plasma. The decrease of detectable NS1 in plasma was coincident with the production of IgM against WNV NS1 (Fig. 4C). Based on this, we speculated that the development specific anti-NS1 antibody decreased the levels of free NS1 and reduced the sensitivity of capture ELISA. To test this, we initially evaluated the effect of NS1-immune complex formation on the sensitivity of the assay using a polyclonal anti-NS1 antibody and purified NS1. NS1-antibody immune complexes markedly reduced the sensitivity of the capture ELISA (Fig. 5). Thus, to improve the capture ELISA, we attempted to dissociate antibody-NS1 antigen immune complexes using acid treatment (0.2N HCl) and heat-exposure (37°C for 90 min), as has been described for HIV and DENV [Miles et al., 1993; Koraka et al., 2003]. Unfortunately, this treatment did not increase the sensitivity of the assay at these late time points, possibly because of the acid liability of flavivirus NS1 [Crooks et al., 1994].
Fig. 4.
Viremia, quantitation of NS1 in plasma, and titers of IgM and IgG against WNV NS1. Five-week-old wild type mice were infected with 2 × 105 PFU of WNV, and serum or plasma was collected at the indicated days. A: WNV RNA levels in the serum were determined by quantitative RT-PCR. The data reflects 5 mice per time point. B: After infection with 2 × 105 PFU of WNV, plasma samples were collected at days 2, 3, 4, 6, and 7. The level of NS1 in plasma was analyzed by capture ELISA without dissociation of immune complexes. Each circle represents a sample from an individual mouse. Solid dashes denote the average NS1 concentration at each time point. The concentration of NS1 was calculated after comparison with a standard curve, which was performed on each ELISA plate. C: Titers of anti-NS1 specific IgM and IgG were determined after incubation of plasma with purified WNV NS1 adsorbed to microtiter plates. In a subset of samples, low levels of anti-NS1 IgM were detected at day 3 after infection (data not shown). The data reflects at least 4 mice per time point. The dotted line represents the limit of sensitivity of the assay. Error bars indicate standard deviations.
Fig. 5.
Release of NS1 from NS1-antibody immune complexes. After preparation of NS1-antibody immune complexes, soluble NS1 was detected by capture ELISA with (D-ELISA; filled bar) or without (N-ELISA; open bar) dissociation of immune complexes. The difference in the level of NS1 between the D-ELISA and N-ELISA was statistically significant (asterisks) at two different serum dilutions (1:2,000, P = 0.0002; 1:100, P = 0.002). Error bars indicate standard deviations. The dashed line indicates the limit of sensitivity.
We developed an alternate strategy to dissociate NS1-Ab immune complex using an alkaline solution. NS1 immune complex dissociation and capture ELISA sensitivity was noticeably increased after incubation in a solution of 0.5 M Tris-base pH 10.5 and 1% Triton X-100 with mild heat treatment (37°C for 1.5 hr), with subsequent neutralization with 2N HCl (Fig. 5). Alkaline treatment of purified NS1 did not significantly affect the limit of detection, the linearity, or the sensitivity of the assay (Fig. 1). Titration experiments with known concentrations of soluble NS1 indicated that ~30% of the NS1 originally bound in immune complexes was dissociated in a form that maintained epitope integrity (Fig. 5, and data not shown). Thus, dissociation of NS1-antibody immune complexes increased the sensitivity of NS1 capture ELISA.
Enhanced Detection of Soluble NS1 in Plasma After Disruption of Immune Complexes
Using our alkaline treatment to dissociate NS1-antibody immune complexes, we re-tested plasma samples for NS1 levels. After dissociating immune complexes, 90% (9 of 10) and 38% (3 of 8) of samples at days 6 and 7, respectively, showed positive signal in our assay compared to 38% and 0% of samples in the absence of treatment. Moreover, the levels of secreted NS1 in plasma after immune complex dissociation were significantly increased (Fig. 6). Overall, dissociation of antibody–antigen complexes augmented the sensitivity of our NS1 capture ELISA at late time points.
Fig. 6.
Comparison of detection of WNV NS1 levels in plasma samples with or without dissociation of immune complexes. Wild type mice were infected with 2 × 105 PFU of WNV and plasma samples were collected at days 3, 4, 6, and 7. The level of NS1 in plasma was measured by capture ELISA with (D-ELISA; filled circle) or without (N-ELISA; open circle) dissociation of immune complexes. The increase in the level of NS1 after dissociation of immune complex was significant at the following days: day 4, P < 0.0001; day 6, P = 0.02; and day 7, P = 0.03. Each circle and line represents one pair of samples from an individual mouse with or without immune complex dissociation. The solid dashed line indicates the limit of sensitivity. Asterisks indicate time points at which difference are statistically significant. The number of mice for each time point ranged from 6 to 10 from 2 to 3 independent experiments. The concentration of NS1 was calculated after comparison with a standard curve, which was performed on each ELISA plate.
DISCUSSION
Many flaviviruses cause human disease with extensive morbidity and mortality. Recent studies have shown that antibody-based therapeutics may be useful treatments for flavivirus infections, such as WNV [Roehrig et al., 2001; Diamond, 2005]. Because therapeutic intervention is expected to have a relatively narrow window for benefit, we set out to develop a rapid, sensitive, and specific ELISA-based tool that could be used early during the course of WNV infection, prior to the development of a serological response. We generated a dual mAb capture ELISA for detection and quantitation of secreted WNV NS1 protein. This assay was highly sensitive and detected as little as 0.5 ng/ml of soluble WNV in the native form. By developing a simple protocol to release NS1 from antibody immune complexes, we extended the utility of the capture ELISA to later time points, after the induction of a specific antibody response. Overall, an NS1 capture ELISA could be useful for rapidly and specifically identifying individuals infected with WNV infection early during the course of infection.
The testing of several combinations of capture and detection mAbs from a large panel produced a highly sensitive assay. 3NS1 recognizes a determinant in the N-terminal region of NS1 whereas 10NS1 binds a distinct, non-overlapping epitope in the middle of the protein [Chung et al., 2006]. For reasons that remain unclear, a parallel NS1 capture ELISA comprised of 3NS1 or 10NS1 and a polyclonal anti-NS1 antibody showed less sensitivity (data not shown). An NS1 antibody-based capture ELISA comprised of polyclonal and a monoclonal or a single monoclonal antibody also showed less sensitivity [Young et al., 2000; Macdonald et al., 2005]. Although further biophysical analysis is warranted, we speculate that polyclonal antibodies may contain moderate and lower avidity antibodies with higher off-rates that decrease the sensitivity of the assay.
NS1 is a conserved glycoprotein and serological studies have shown significant cross-reactivity of anti-NS1 antibodies between flaviviruses. Indeed, in a previously published WNV NS1 ELISA, the capture mAb recognized a highly conserved epitope, and thus could be used more broadly to diagnose flavivirus infection [Macdonald et al., 2005]. Nonetheless, because specific and accurate diagnosis of WNV infection may prompt a decision to initiate expensive therapies against WNV early during the course of infection, in regions where multiple flaviviruses co-circulate, it will be important to have a rapid, WNV-specific diagnostic assay. The 3NS1 and 10NS1 mAbs used in the NS1 capture ELISA showed no appreciable cross-reactivity against YFV, SLEV, and DENV, and also did not bind to NS1 of the closely related JEV and MVEV (R. Hall, personal communication).
The capture ELISA reliably measured WNV NS1 in plasma at day 3 after infection prior to the development of an IgM response. Under standard analysis conditions, NS1 antigenemia peaked at day 4 and then declined by day 6, coincident with production of anti-NS1 IgM. The sensitivity of the capture ELISA under standard conditions was reduced by the formation of NS1-immune complexes, which prevented the detection of free, soluble NS1. Historically, treatment of serum or plasma with an acidic solution has been described for dissociating viral antigen–antibody immune complexes for early diagnosis of HIV, DENV, and hepatitis B and C viruses [Miles et al., 1993; Panakitsuwan et al., 1997; Troisi and Hollinger, 1997; Weber et al., 2001; Koraka et al., 2003]. Although previous studies with DENV demonstrated that NS1 immune-complex dissociation under acid conditions augmented the sensitivity of detection by immunoblot assay, this protocol did not work well for WNV NS1 (data not shown), likely because of its acid lability [Crooks et al., 1994] and our selection of mAbs that recognized conformationally sensitive epitopes. As such, treatment of plasma with an alkaline solution and a non-ionic detergent partially dissociated NS1 immune complexes and improved the sensitivity of the capture ELISA. Disruption of immune complexes in plasma samples extended the time window for measurement of antigenemia, to a point that exceeded detection of viral RNA by quantitative RT-PCR.
The NS1 capture ELISA can be used to accurately measure the kinetics and amplitude of WNV infection in primary and transformed cells. We reproducibly detected significant levels of NS1 within 12 hr of infection, which corresponds to the completion of the first replication cycle [Lindenbach and Rice, 2001; Brinton, 2002]. High (~5 μg/ml) and significant (~100 ng/ml) levels of NS1 were detected in supernatants from BHK and primary neuron or DC, respectively. Interestingly, we did not detect soluble NS1 in the supernatant of macrophages infected with WNV; although further metabolic labeling studies are necessary, these cells may inefficiently secrete NS1 or secrete NS1 at levels that are below detection by our assay. Regardless, the capture ELISA can be used to screen cell types and conditions that optimize production of NS1 for biochemical and biophysical studies. Alternatively, comparison of the levels of NS1 in supernatants of cells treated with candidate small molecule inhibitors can be used for inexpensive, high-throughput 96-well plate screening of chemical antagonists of WNV infection [Nouiery et al., 2007].
An expanding WNV epidemic necessitates the development of new diagnostics that are rapid and simple for detection of early WNV infection. An NS1 capture ELISA that uses mAbs specific to WNV NS1 may allow rapid distinction of WNV infections from other endemic flaviviruses; this may be especially important in parts of the world (e.g., Asia and South America) where DENV infections occur with high (>90%) prevalence or populations have been vaccinated against other flaviviruses (JEV, YFV, and TBEV), making serological tests difficult to interpret [Mackenzie et al., 2004]. Although more testing is necessary, the NS1 capture ELISA, when combined with immune complex dissociation, shows promise for rapid and specific diagnosis of early WNV infection.
Acknowledgments
NIH; Grant number: U01 AI061373; Grant sponsor: Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research; Grant number: U54 AI057160; Grant sponsor: Pediatric Dengue Vaccine Initiative; Grant sponsor: Chonbuk National University (partial support to K.M.C.); Grant number: NP-2007–17.
The authors thank members of their laboratories for experimental advice, A. Barrett for providing the SLEV strain, and R. Hall for sharing preliminary results.
Footnotes
The authors declare no financial conflict of interest.
References
- Agrawal AG, Petersen LR. Human immunoglobulin as a treatment for West Nile virus infection. J Infect Dis. 2003;188:1–4. doi: 10.1086/376871. [DOI] [PubMed] [Google Scholar]
- Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol. 2002;40:376–381. doi: 10.1128/JCM.40.2.376-381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nathan D, Lustig S, Tam G, Robinzon S, Segal S, Rager-Zisman B. Prophylactic and therapeutic efficacy of human intravenous immunoglobulin in treating west nile virus infection in mice. J Infect Dis. 2003;188:5–12. doi: 10.1086/376870. [DOI] [PubMed] [Google Scholar]
- Brinton MA. The molecular biology of West Nile Virus: A new invader of the western hemisphere. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Busch MP, Caglioti S, Robertson EF, McAuley JD, Tobler LH, Kamel H, Linnen JM, Shyamala V, Tomasulo P, Kleinman SH. Screening the blood supply for West Nile virus RNA by nucleic acid amplification testing. N Engl J Med. 2005a;353:460–467. doi: 10.1056/NEJMoa044029. [DOI] [PubMed] [Google Scholar]
- Busch MP, Tobler LH, Saldanha J, Caglioti S, Shyamala V, Linnen JM, Gallarda J, Phelps B, Smith RI, Drebot M, Kleinman SH. Analytical and clinical sensitivity of West Nile virus RNA screening and supplemental assays available in 2003. Transfusion. 2005b;45:492–499. doi: 10.1111/j.0041-1132.2005.04382.x. [DOI] [PubMed] [Google Scholar]
- Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. Antibodies against West Nile virus non-structural (NS)-1 protein prevent lethal infection through Fc gamma receptor-dependent and independent mechanisms. J Virol. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DC, Lobigs M, Lee E, Howard MJ, Clark K, Blitvich BJ, Hall RA. In situ reactions of monoclonal antibodies with a viable mutant of Murray Valley encephalitis virus reveal an absence of dimeric NS1 protein. J Gen Virol. 2007;88:1175–1183. doi: 10.1099/vir.0.82609-0. [DOI] [PubMed] [Google Scholar]
- Crooks AJ, Lee JM, Easterbrook LM, Timofeev AV, Stephenson JR. The NS1 protein of tick-borne encephalitis virus forms multimeric species upon secretion from the host cell. J Gen Virol. 1994;75:3453–3460. doi: 10.1099/0022-1317-75-12-3453. [DOI] [PubMed] [Google Scholar]
- Diamond MS. Development of effective therapies against West Nile virus infection. Expert Rev Anti Infect Ther. 2005;3:931–944. doi: 10.1586/14787210.3.6.931. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J Virol. 2000a;74:7814–7823. doi: 10.1128/jvi.74.17.7814-7823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Roberts T, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000b;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Shrestha B, Marri A, Mahan D, Engle M. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol. 2003a;77:2578–2586. doi: 10.1128/JVI.77.4.2578-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Sitati E, Friend L, Shrestha B, Higgs S, Engle M. Induced IgM protects against lethal West Nile Virus infection. J Exp Med. 2003b;198:1–11. doi: 10.1084/jem.20031223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, Dupuis AP, III, Ngo K, Nicholas D, Kauffman E, Jones SA, Young D, Maffei J, Shi PY, Bernard K, Kramer L. Partial genetic characterization of West Nile Virus strains, New York State, 2000. Emerg Inf Dis. 2001;7:650–653. doi: 10.3201/eid0704.010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle M, Diamond MS. Antibody prophylaxis and therapy against West Nile Virus infection in wild type and immunodeficient mice. J Virol. 2003;77:12941–12949. doi: 10.1128/JVI.77.24.12941-12949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconar AK, Young PR. Production of dimer-specific and dengue virus group cross-reactive mouse monoclonal antibodies to the dengue 2 virus non-structural glycoprotein NS1. J Gen Virol. 1991;72:961–965. doi: 10.1099/0022-1317-72-4-961. [DOI] [PubMed] [Google Scholar]
- Gould LH, Sui J, Foellmer H, Oliphant T, Wang T, Ledizet M, Murakami A, Noonan K, Lambeth C, Kar K, Anderson JF, de Silva AM, Diamond MS, Koski RA, Marasco WA, Fikrig E. Protective and therapeutic capacity of human single chain Fv-Fc fusion proteins against West Nile virus. J Virol. 2005;79:14606–14613. doi: 10.1128/JVI.79.23.14606-14613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Nisbet DJ, Pham KB, Pyke AT, Smith GA, Khromykh AA. DNA vaccine coding for the full-length infectious Kunjin virus RNA protects mice against the New York strain of West Nile virus. Proc Natl Acad Sci USA. 2003;100:10460–10464. doi: 10.1073/pnas.1834270100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubalek Z, Halouzka J. West Nile fever—A reemerging mosquito-borne viral disease in Europe. Emerg Inf Dis. 1999;5:643–650. doi: 10.3201/eid0505.990505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG, Winger QA, Olsen AL, Day CW, Sidwell RW, Morrey JD. Treatment of West Nile virus-infected mice with reactive immunoglobulin reduces fetal titers and increases dam survival. Antiviral Res. 2005;65:79–85. doi: 10.1016/j.antiviral.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol. 1999;73:10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Lin E, Zhang B, Luster AD, Tollett J, Samuel MA, Engle M, Diamond MS. Neuronal CXCL10 directs CD8+ T cell recruitment and control of West Nile virus encephalitis. J Virol. 2005;79:11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraka P, Suharti C, Setiati TE, Mairuhu AT, Van Gorp E, Hack CE, Juffrie M, Sutaryo J, Van Der Meer GM, Groen J, Osterhaus AD. Kinetics of dengue virus-specific serum immunoglobulin classes and subclasses correlate with clinical outcome of infection. J Clin Microbiol. 2001;39:4332–4338. doi: 10.1128/JCM.39.12.4332-4338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraka P, Burghoorn-Maas CP, Falconar A, Setiati TE, Djamiatun K, Groen J, Osterhaus AD. Detection of immune-complex-dissociated nonstructural-1 antigen in patients with acute dengue virus infections. J Clin Microbiol. 2003;41:4154–4159. doi: 10.1128/JCM.41.9.4154-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer LD, Chandler LJ. Phylogenetic analysis of the envelope gene of St. Louis encephalitis virus. Arch Virol. 2001;146:2341–2355. doi: 10.1007/s007050170007. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. Rapid detection of west nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol. 2000;38:4066–4071. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis. 2002;186:1165–1168. doi: 10.1086/343813. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Flaviviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 991–1041. [Google Scholar]
- Macdonald J, Tonry J, Hall RA, Williams B, Palacios G, Ashok MS, Jabado O, Clark D, Tesh RB, Briese T, Lipkin WI. NS1 protein secretion during the acute phase of West Nile virus infection. J Virol. 2005;79:13924–13933. doi: 10.1128/JVI.79.22.13924-13933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Martin DA, Noga A, Kosoy O, Johnson AJ, Petersen LR, Lanciotti RS. Evaluation of a diagnostic algorithm using immunoglobulin M enzyme-linked immunosorbent assay to differentiate human West Nile Virus and St. Louis Encephalitis virus infections during the 2002 West Nile Virus epidemic in the United States. Clin Diagn Lab Immunol. 2004;11:1130–1133. doi: 10.1128/CDLI.11.6.1130-1133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PW. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989;169:354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles SA, Balden E, Magpantay L, Wei L, Leiblein A, Hofheinz D, Toedter G, Stiehm ER, Bryson Y. Rapid serologic testing with immune-complex-dissociated HIV p24 antigen for early detection of HIV infection in neonates. Southern California Pediatric AIDS Consortium. N Engl J Med. 1993;328:297–302. doi: 10.1056/NEJM199302043280501. [DOI] [PubMed] [Google Scholar]
- Morrey JD, Siddharthan V, Olsen AL, Roper GY, Wang H, Baldwin TJ, Koenig S, Johnson S, Nordstrom JL, Diamond MS. Humanized monoclonal antibody against West Nile virus E protein administered after neuronal infection protects against lethal encephalitis in hamsters. J Infect Dis. 2006;194:1300–1308. doi: 10.1086/508293. [DOI] [PubMed] [Google Scholar]
- Muylaert IR, Chambers TJ, Galler R, Rice CM. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: Effects on virus replication and mouse neurovirulence. Virology. 1996;222:159–168. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- Nouiery AO, Olivo PD, Slomczynska U, Zhou Y, Buscher B, Geiss B, Engle M, Roth RM, Chung KM, Samuel MA, Diamond MS. The identification of novel small molecule inhibitors of West Nile virus infection. J Virol. 2007;81:11828–11839. doi: 10.1128/JVI.01358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oceguera LF, III, Patiris PJ, Chiles RE, Busch MP, Tobler LH, Hanson CV. Flavivirus serology by Western blot analysis. Am J Trop Med Hyg. 2007;77:159–163. [PubMed] [Google Scholar]
- Oliphant T, Engle M, Nybakken G, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panakitsuwan S, Yoshihara N, Hashimoto N, Miyamura K, Chotpitayasunondh T. Early diagnosis of vertical HIV infection in infants by rapid detection of immune complex-dissociated HIV p24 antigen. AIDS Patient Care STDS. 1997;11:429–433. doi: 10.1089/apc.1997.11.429. [DOI] [PubMed] [Google Scholar]
- Petersen LR, Marfin AA, Gubler DJ. West Nile virus. JAMA. 2003;290:524–528. doi: 10.1001/jama.290.4.524. [DOI] [PubMed] [Google Scholar]
- Pierson TC, Diamond MS, Ahmed AA, Valentine LE, Davis CW, Samuel MA, Hanna SL, Puffer BA, Doms RW. An infectious West Nile Virus that expresses a GFP reporter gene. Virology. 2005;334:28–40. doi: 10.1016/j.virol.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Puttikhunt C, Kasinrerk W, Srisa-ad S, Duangchinda T, Silakate W, Moonsom S, Sittisombut N, Malasit P. Production of anti-dengue NS1 monoclonal antibodies by DNA immunization. J Virol Methods. 2003;109:55–61. doi: 10.1016/s0166-0934(03)00045-4. [DOI] [PubMed] [Google Scholar]
- Ratterree MS, Gutierrez RA, Travassos da Rosa AP, Dille BJ, Beasley DW, Bohm RP, Desai SM, Didier PJ, Bikenmeyer LG, Dawson GJ, Leary TP, Schochetman G, Phillippi-Falkenstein K, Arroyo J, Barrett AD, Tesh RB. Experimental infection of rhesus macaques with West Nile virus: Level and duration of viremia and kinetics of the antibody response after infection. J Infect Dis. 2004;189:669–676. doi: 10.1086/381461. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Staudinger LA, Hunt AR, Mathews JH, Blair CD. Antibody prophylaxis and therapy for flaviviral encephalitis infections. Ann NY Acad Sci. 2001:286–297. doi: 10.1111/j.1749-6632.2001.tb02704.x. [DOI] [PubMed] [Google Scholar]
- Russell PK, Nisalak A. Dengue virus neutralization by the plaque reduction neutralization test. J Immunol. 1967;99:291–294. [PubMed] [Google Scholar]
- Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BRG, Silverman RH, Gale M, Diamond MS. PKR and RNAse L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejvar JJ, Haddad MB, Tierney BC, Campbell GL, Marfin AA, Van Gerpen JA, Fleischauer A, Leis AA, Stokic DS, Petersen LR. Neurologic manifestations and outcome of West Nile virus infection. JAMA. 2003;290:511–515. doi: 10.1001/jama.290.4.511. [DOI] [PubMed] [Google Scholar]
- Tardei G, Ruta S, Chitu V, Rossi C, Tsai TF, Cernescu C. Evaluation of immunoglobulin M (IgM) and IgG enzyme immunoassays in serologic diagnosis of West Nile Virus infection. J Clin Microbiol. 2000;38:2232–2239. doi: 10.1128/jcm.38.6.2232-2239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, Geuijen C, Goudsmit J, Bakker AQ, Korimbocus J, Kramer RA, Clijsters-van der Horst M, de Jong M, Jongeneelen M, Thijsse S, Smit R, Visser TJ, Bijl N, Marissen WE, Loeb M, Kelvin DJ, Preiser W, ter Meulen J, de Kruif J. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile Virus. J Virol. 2006;80:6982–6992. doi: 10.1128/JVI.00551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisi CL, Hollinger FB. Detection of antibodies to hepatitis C virus in seronegative patients using an immune complex dissociation assay. J Viral Hepat. 1997;4:383–386. doi: 10.1046/j.1365-2893.1997.00068.x. [DOI] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Weber B, Melchior W, Gehrke R, Doerr HW, Berger A, Rabenau H. Hepatitis B virus markers in anti-HBc only positive individuals. J Med Virol. 2001;64:312–319. doi: 10.1002/jmv.1052. [DOI] [PubMed] [Google Scholar]
- Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89:423–437. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- Winkler G, Randolph VB, Cleaves GR, Ryan TE, Stollar V. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology. 1988;162:187–196. doi: 10.1016/0042-6822(88)90408-4. [DOI] [PubMed] [Google Scholar]
- Winkler G, Maxwell SE, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989;171:302–305. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- Yamada K, Takasaki T, Nawa M, Kurane I. Virus isolation as one of the diagnostic methods for dengue virus infection. J Clin Virol. 2002;24:203–209. doi: 10.1016/s1386-6532(01)00250-5. [DOI] [PubMed] [Google Scholar]
- Young PR, Hilditch PA, Bletchly C, Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J Clin Microbiol. 2000;38:1053–1057. doi: 10.1128/jcm.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]