Abstract
Twist is a basic helix loop helix protein that plays a role both in development and in cancer biogenesis. While characterizing the effects of Twist on breast epithelial cell transformation, we identified E-cadherin as a target gene that is down-regulated by Twist. In this study, we demonstrate that Twist can transcriptionally repress E-cadherin in breast cancer cells. Using transient promoter assays, we show that Twist can down-regulate E-cadherin promoter activity by up to two folds. This is further supported by immunoblot analyses which indicated that over-expression of Twist can decrease E-cadherin protein levels in breast cancer cell lines. Subsequently, chromatin immunoprecipitation was performed on MCF-7/Twist and Hs578 T (high level of endogenous Twist expression), which confirmed Twist binding to the E-cadherin promoter. Finally, the functional relevance of this regulation was verified by quantitative real-time PCR and immunohistochemistry on a cohort of breast cancer samples.
Introduction
Twist is a basic helix loop helix (bHLH) transcription factor essential in embryological morphogenesis. It belongs to the HLH superfamily of proteins which are involved in a variety of regulatory processes in organisms [1]. bHLH proteins dimerize with other members of the family, bind to short conserved sequences called E-boxes [2] in promoter regions and transcriptionally regulate target genes. Twist is a highly conserved protein and was initially identified in Drosophila where it is involved in mesodermal patterning and morphogenetic movement [3]. In mice, Twist is implicated in neural tube closure and null mutants are embryonic lethal and die at E10.5 [4]. In humans, Twist is essential for normal vertebrate development and defects in Twist promote autosomal dominant defects characterized by minor skull and limb anomalies. Twist is mainly expressed in a subset of adult mesodermal cells [5]. Twist also plays a role in cellular determination and in the differentiation of several lineages including myogenesis [6], osteogenesis [7], and neurogenesis [8]. Besides the role of Twist in normal development, Twist is over-expressed in many cancer types such as breast [9], gastric [10] and prostate cancer [11].
E-cadherin is a tumor suppressor transmembrane glycoprotein essential for epithelial cell-cell adhesion [12]. It belongs to the cell adhesion molecule (CAM) family of proteins that are expressed in a wide variety of tissues. Cadherins mediate cell-cell adhesion by interacting via their extracellular domains with other cadherins. E-cadherin switching is indispensable during development where it drives epithelial-mesenchymal transitions (EMT) [13] during gastrulation and organogenesis wherein epithelial cells are converted to mesenchymal cells [14]. The loss or suppression of E-cadherin expression is a critical event signaling loss of the epithelial phenotype and commencement of the invasive program. There is evidence to indicate that defects in E-cadherin lead to development and progression of cancer [15].
A majority of defects in E-cadherin expression have been associated with epigenetic origins such as transcriptional silencing or methylation. Repression by promoter hypermethylation [16], transcriptional repression [17] and mutations [18] are implicated in E-cadherin-mediated cancer progression. The roles of Snail [19] and other factors such as p300 and AML1 have been reported as transcriptional regulators of E-cadherin expression [20].
Earlier we demonstrated that over-expression of Twist occurs in a large number of breast cancers with a concomitant epithelial-mesenchymal transition [9]. This inverse correlation between Twist and E-cadherin expression observed in MCF-7/Twist cells prompted us to decipher the functional role of Twist in regulating E-cadherin expression in breast cancer. We report here that over-expression of Twist causes the transcriptional down-regulation of E-cadherin. We demonstrate by chromatin immunoprecipitation that Twist binds in vivo to the E-cadherin promoter and by promoter-reporter assays that Twist can down-regulate E-cadherin promoter activity. Furthermore, we show by quantitative real-time PCR (qRT-PCR) that Twist expression inversely correlates with E-cadherin expression in breast cancer samples.
Results
Twist downregulates E-cadherin expression in breast cancer cell lines
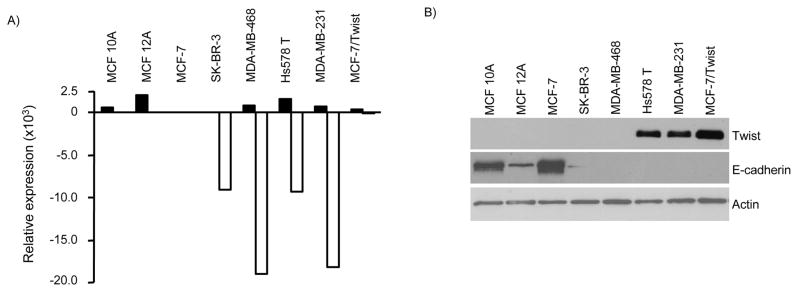
To understand the role of Twist in the biogenesis of breast cancer, we generated MCF-7/Twist - a Twist over-expressing MCF-7 sub-line. Initial characterization of the MCF-7/Twist cell line indicated the occurrence of an EMT with the associated loss of E-cadherin [9]. Microarray analysis showed that E-cadherin levels were down-regulated by 260 fold in MCF-7/Twist cells as compared to MCF-7 cells. To confirm this inverse correlation between Twist and E-cadherin expression, we performed qRT-PCR on a panel of immortalized normal and breast cancer cell lines with varying degrees of tumorigenic and metastatic potential using MCF-7 as a baseline to compare expression levels of Twist and E-cadherin in these cell lines. As seen in Figure 1A, all the cell lines analyzed showed an increase in Twist expression when compared to MCF-7. However, expression of E-cadherin was over 2500 fold less in the tumorigenic lines SK-BR-3, Hs578 T, MDA-MB-231, and MDA-MB-435. MCF-7/Twist cells showed an 826 fold increase of Twist expression and a 225 fold decrease in E-cadherin expression as compared to parental MCF-7 cells.
Figure 1. Twist inversely correlates with E-cadherin expression in breast cancer cells.
A, Quantitative real-time PCR of breast cancer cell lines. Relative expression is calculated with MCF-7 as baseline. An inverse correlation was observed between Twist and E-cadherin mRNA expression as demonstrated by quantitative real-time PCR. B, Immunoblots showing E-cadherin and Twist expression in a panel of normal breast and breast cancer cell lines, which include both invasive and non-invasive phenotypes. Normal cell lines are seen on the left with progressively more invasive cell lines on the right. Antibodies against Twist were made in-house while E-cadherin (BD Biosciences) and β-actin (Sigma-Aldrich) were commercially obtained.
To further confirm the observed correlation between Twist and E-cadherin mRNA levels, immunoblot analysis was performed on a similar panel of normal and breast cancer cell lines. As shown in Figure 1B, immortalized normal breast cells (MCF 10A and MCF 12A) showed no Twist expression but high E-cadherin levels. However there is a significant difference in their E-cadherin levels probably due to genetic heterogeneity. Within the breast cancer cell lines, there was an inverse correlation between Twist and E-cadherin levels in Hs578 T, MDA-MD-231, and MCF-7/Twist cell lines. However in SK-BR-3 and MDA-MB-468 cells, decreased E-cadherin levels were not associated with Twist over-expression. The immunoblot analysis of epithelial breast cancer cell lines further validated our finding that Twist down-regulates E-cadherin expression. However, the levels of Twist mRNA did not correlate with protein levels in MCF 10A and MCF 12A cells. It is possible that the stability of Twist protein in normal immortalized breast cell lines is lower than in cancer cells and needs further investigation.
Twist represses E-cadherin transcription via E-boxes
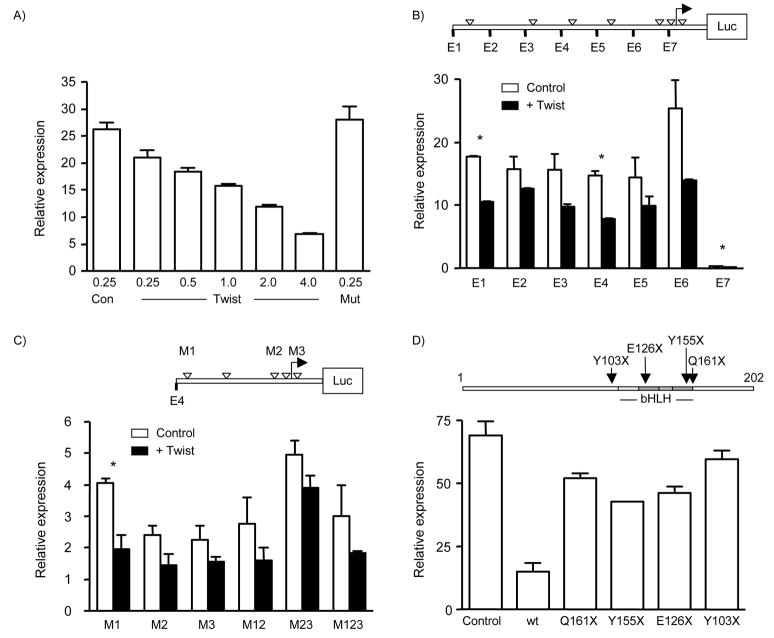
In order to determine if Twist directly regulated E-cadherin at the transcriptional level, we carried out promoter analysis in MCF-7 and MCF-7/Twist cells. We first determined the degree of suppression of the full-length E-cadherin promoter by using increasing amounts of Twist plasmid and found a dose-dependent effect (Figure 2A) which was not seen when a truncated Twist construct (Y103X) was used as the effector plasmid (Figure 2A). Twist mutants were obtained earlier [21] and are shown in Figure 2D.
Figure 2. Twist repression of E-cadherin promoter.
A, Dose dependent repression of E-cadherin promoter. Determination of the degree of suppression of the full-length E-cadherin promoter activity in the presence of increasing amounts of Twist plasmid in transient co-transfection assays. The Y103X mutant Twist construct (Mut) was used to test for specificity of repression. B, Upper panel shows a schematic of the E-cadherin promoter with E boxes shown as hollow triangles. Deletion constructs are indicated at 5′ end with E1–E7. Twist represses E-cadherin transcriptionally. Each of the 6 E-cadherin promoter reporter constructs was repressed 1.2 to 2 fold by Twist. The largest significant repression was seen in construct E4, indicating the importance of the proximal region in E-cadherin regulation by Twist. Statistical significance (P<0.05) is indicated by asterisks. C, Full length Twist is essential for E-cadherin repression. Upper panel shows E-cadherin E4 construct with E-box mutants indicated by M1, M2, and M3. Twist repression was statistically significant only in M1 construct indicating its lack of function in mediating the repression. This indicates that E-box 2 and 3 may play an important role in the down-regulation of E-cadherin by Twist. D, Upper panel shows Twist deletion mutants. Mutations are indicated with amino acids changed to stop codons. Lower panel shows mutant Twist constructs were unable to repress E-cadherin promoter significantly indicating that regions other than the basic helix loop helix domain play a role in the repression of E-cadherin.
Deletion constructs were constructed as described [20] (Figure 2B). Subsequent characterization of the promoter constructs showed that the full-length construct, E1, exhibited significant repression (1.8 fold) in the presence of Twist (P=0.0015) (Figure 2B). However, the maximum repression was observed using constructs E4 and E6 (1.9 folds), indicating the importance of the proximal region in repression of E-cadherin by Twist.
In order to identify which E-boxes are responsible for bringing about this repression, we carried out promoter-reporter assays using different combinations of mutated E-boxes as described in Figure 2C. The down-regulation was significant when the distal E-box was mutated (P=0.047) indicating that the proximal E-boxes 2 and 3 are important in down-regulation of E-cadherin by Twist. All other decreases were non-significant.
Finally, we tested the functional activity of the mutant truncated Twist constructs to repress E-cadherin expression. As shown in Figure 2D, use of wild-type Twist showed the maximum repression as compared to mutant Twist constructs (P=0.014). However, the mutant constructs were unable to completely relieve repression of reporter activity indicating that regions other than the basic helix loop helix domain may play a role in the repression of E-cadherin by Twist. We obtained similar results in MCF-7/Twist cells. However, the repression of promoter activity was 3 magnitudes higher when compared to MCF-7 (data not shown).
Overall, these assays indicate that Twist can transcriptionally repress E-cadherin promoter activity in breast cells.
In vivo binding of Twist to E-boxes in the E-cadherin promoter
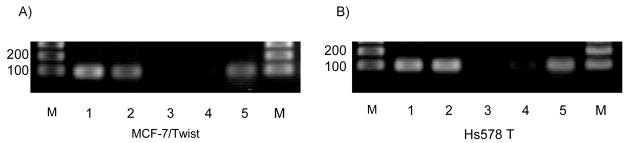
To study mechanistically the role of Twist in down-regulating E-cadherin expression, we examined the in vivo binding of Twist to the E-cadherin promoter. To address this, we carried out chromatin immunoprecipitation (ChIP) assays using MCF-7/Twist cells and Hs578 T, a breast cancer cell line that has an intrinsically high level of Twist. As shown in Figure 3A and 3B (lane 5), the PCR from anti-Twist precipitations resulted in a specific amplification product. Identical amplification products were seen in the positive controls from both total input chromatin and anti-acetyl-histone H3 precipitations (lanes 1 and 2 respectively). Moreover, no amplification was seen in samples that were processed in the absence of chromatin or precipitating antibody (lanes 3 and 4 respectively). We obtained similar results by qRT-PCR (data not shown). These results indicate that Twist binds directly or as part of a complex to the endogenous E-cadherin promoter in vivo.
Figure 3. In vivo binding of Twist protein to the E-cadherin promoter sequence.
ChIP was carried out following established protocols using MCF-7/Twist and Hs578 T cells and analyzed using E-cadherin promoter-specific primers by PCR. Primers amplified a 93 base pair fragment. Identical volumes from the final precipitate were used for the PCR reactions. M-molecular weight marker (bp), lane 1- total input immunoprecipitate, lane 2- acetylated histone H3 immunoprecipitate, lane 3- no chromatin immunoprecipitate, lane 4- no antibody immunoprecipitate and lane 5- Twist immunoprecipitate.
Twist and E-cadherin are inversely correlated in breast cancer patients
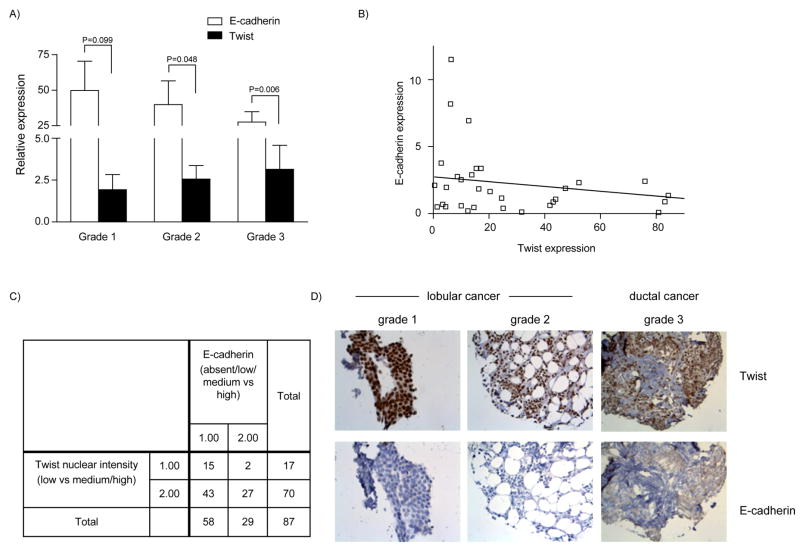
To extend the observed inverse correlation between Twist and E-cadherin expression in breast cancer cell lines to human breast cancers, we quantified Twist and E-cadherin mRNA levels in breast tumors. A total of 31 breast cancers (grade I=6, grade II=12, grade III=13) and four normal breast samples were analyzed by qRT-PCR (Figure 4A). Twist expression in grade III tumors increased 2.9 folds over grade I tumors. In the same samples, E-cadherin expression decreased 1.2 folds as tumor grade increased. For the different tumor grades, the expression levels of Twist and E-cadherin were significantly different in grades 2 and 3 when analyzed by Welch’s t-test. The correlation was not significant when Twist expression levels were compared to E-cadherin by 2-way ANOVA.
Figure 4. Relation between Twist and E-cadherin expression in patient breast cancer samples.
A, mRNA levels of Twist and E-cadherin in breast tumors measured by quantitative real-time PCR. A total of 31 breast tumor (grade I=6, grade II=12, grade III=13) and four normal breast samples were analyzed. Loss of E-cadherin expression was concomitant with increased Twist expression in higher-grade tumors. B, A trend towards an inverse correlation was observed between Twist and E-cadherin expression (r=−0.27, p=0.14) in the breast cancer patient samples. C, Results of immunohistochemical analysis of Twist and E-cadherin expression in eighty-seven human breast cancer samples. The crosstab shows comparison of Twist nuclear staining and its intensity (classified as low vs. medium/high) against E-cadherin expression (absent/low/medium vs. high). D, Representative photomicrographs of human breast tumor samples from lobular and ductal samples that were analyzed for Twist and E-cadherin levels (left panel − 40×, middle and right panel −20×).
When the entire data set of breast tumor samples were analyzed by Pearson’s correlation (Figure 4B), we found a trend towards an inverse correlation between Twist and E-cadherin expression (r=−0.27) although it was not significant (P=0.14). The lack of significance could be due to heterogeneity of the tumor samples studied or the small sample size.
In order to verify the data obtained by qRT-PCR analysis, we immunohistochemically analyzed for Twist and E-cadherin protein levels on a larger number (n=87) of breast cancer samples. As seen in Figure 4C, we compared Twist nuclear intensity staining (low versus medium, high) to E-cadherin staining (absent, low, medium versus high). We found a significant inverse correlation between Twist and E-cadherin expression by Chi-square test (P=0.035) and by Fisher’s exact test (0.045). In Figure 4D, three representative immunohistochemistry pictures of different breast tumor grades showing the inverse correlation between Twist and E-cadherin expression levels are shown.
Discussion
Epithelial-mesenchymal transitions are responsible for the development of the mesoderm from the epithelium during embryo development. The movement of cells that characterizes this process is brought about by a switch in E-cadherin expression, which causes them to lose their ordered epithelial phenotype and become plastic. This is analogous to late-stage tumorigenesis when E-cadherin silencing causes loss of cell-cell adhesion and results in gain of the invasive phenotype.
The complete reverse is true for Twist expression. During gastrulation, Twist is expressed in a subset of epithelial cells causing them to invaginate and eventually form the presumptive mesoderm. Thus Twist expression is obligatory for the occurrence of the EMT during embryo development. Analogously, we have earlier shown that cancer cells over-expressing Twist are motile and invasive and exhibit all the characteristics of having undergone an EMT [9]. This inverse relationship between E-cadherin and Twist expression lead us to further probe into the regulation of E-cadherin by Twist. E-cadherin silencing is thought to be mediated via transcriptional regulation and methylation in various cell types. It has already been shown that the closely related bHLH protein Snail is a repressor of E-cadherin [19].
Following the EMT by MCF-7/Twist cells, we identified E-cadherin as a potential target of Twist. This finding was verified by qRT-PCR and immunoblotting on a series of immortalized breast cell lines and breast cancer cell lines. However, a perfect concordance was not observed between the mRNA and protein levels for Twist and E-cadherin levels. This would indicate that other mechanisms also regulate the expression levels of these two proteins.
The regulatory capacity of Twist is through its ability to bind to E-boxes as homodimers or heterodimers. E-boxes have been shown to be distributed amongst promoter regions of many genes including E-cadherin. The bHLH protein Snail (which is a transcriptional target of Twist) has been shown to regulate E-cadherin expression via E-box 1 [20]. However, our data points to the importance of E-boxes 2 and 3 in the regulation of E-cadherin by Twist and the lack of functionality of E-box 1. As Twist and Snail have different binding affinities for E-boxes, it is likely that over-expression of Twist would facilitate preferential binding to E-box 2 and 3 through a variety of mechanisms such as sequence context and protein binding partners. This is in conformity with published data, which showed that transcriptional regulation forms the major regulatory method modulating E-cadherin expression [17].
The functional importance of the down-regulation of E-cadherin by Twist was verified in human breast cancer samples. Initial experiments using qRT-PCR clearly demonstrated that increased expression of Twist in grade III tumors exhibited decreased E-cadherin levels. The same was true using immunohistochemistry in eighty-seven breast cancer samples. These results paralleled our in vitro data, thus providing further support to our hypothesis that increased Twist expression can promote breast tumor biogenesis and one such mechanism is via regulating E-cadherin expression [9; 17].
Besides Twist, there are several other molecules that play important roles in EMT induction. These include the zinc finger transcriptional repressors Snail and Slug [19; 22], as well as two members of the ZEB family ZEB1 [23] and ZEB2 and the Id family of HLH proteins [24]. Their mechanistic role in EMT and tumorigenesis via the down-regulation of E-cadherin is still being explored [25]. It is possible that many of these regulatory molecules may work in a concerted fashion or in a sequential manner to bring about EMT, thus facilitating the progression of the invasive breast cancer phenotype.
In summary, when seen in the background of high Twist expression in invasive breast cancer cell lines and in high-grade/invasive breast tumors, our results demonstrate that Twist plays a direct role in the EMT mechanism by down-regulating E-cadherin expression and promoting invasive and metastatic phenotype. This would bring into focus the multi-faceted roles of Twist in inducing neoplastic transformation as well as provide alternative pathways to which one can develop novel chemotherapeutic drugs for the treatment of breast cancer.
Materials & Methods
Promoter analysis
As reported previously [20], 6 deletion constructs and 6 mutant constructs were generated from the region spanning 1 kb upstream of the transcription start site of the E-cadherin promoter and cloned into a luciferase reporter vector (Figure 2B and 2C). There are 7 canonical Twist binding E-box sites (CANNTG) in this region. Transient co-transfection assays were carried out using promoter constructs and effector plasmids expressing Twist [9; 26]. All experiments were done in replicates and at least 5 times.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously [27]. A set of PCR primers 5′-CTCCAGCTTGGGTGAAAGAG -3′ and 5′-GGGCTTTTACACTTGGCTGA -3′ was designed for specific amplification of the E-cadherin promoter between the M1 and M2 E-boxes. The primers amplified a 93 bp region from −371 to −279 with respect to the transcription start site.
Twist and E-cadherin levels in human breast cancers
Fresh frozen breast cancer samples controlled for adequate tumor content were obtained from the University Medical Center Utrecht, The Netherlands. RNA was extracted and qRT-PCR performed using iQ SYBR green supermix (BioRad Laboratories, Hercules, CA) on an iCycler5 (Bio-Rad laboratories). Amplification of 36B4, a housekeeping gene, was used for normalizing gene expression values, Ct = Ct (Twist or E-cadherin) − Ct (36B4). Fold difference was calculated by the formula 2 −ΔΔ Ct, where ΔΔCt = ΔCt (Normalized Twist or E-cadherin) − ΔCt (Normalized control samples).
Additional samples were evaluated by immunohistochemistry using in-house prepared Twist antibody and commercial E-cadherin antibody. We stained a total of 87 samples which were grouped according to the staining patterns: Twist low (1.00) vs. medium/high (2.00) and E-cadherin absent/low/medium (1.00) vs. high (2.00).
Statistical analysis
Statistical analysis was done using Prism (GraphPad Software Inc., San Diego, CA). qRT-PCR data was initially screened for outliers by the Grubbs test (α = 0.05). Patient data and promoter assays were analyzed by Students t-test. For correlation, Pearson’s test was used to determine significance and the trend line was determined by non-linear regression. P values below 0.05 were considered significant.
Acknowledgments
We thank UMC Utrecht Biobank and Mrs. M van Blokland for the human breast cancer RNA. We thank Brian Kimble and Ala Lisok for technical help. This work was supported by NIH Grant RO1CA097226.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jan Y, Jan L. Functional Gene Cassettes in Development. PNAS. 1993;90:8305–8307. doi: 10.1073/pnas.90.18.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–83. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 3.Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–53. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–99. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 5.Wang SM, Coljee VW, Pignolo RJ, Rotenberg MO, Cristofalo VJ, Sierra F. Cloning of the human twist gene: its expression is retained in adult mesodermally-derived tissues. Gene. 1997;187:83–92. [PubMed] [Google Scholar]
- 6.Hamamori Y, Wu HY, Sartorelli V, Kedes L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol Cell Biol. 1997;17:6563–73. doi: 10.1128/mcb.17.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MS, Lowe GN, Strong DD, Wergedal JE, Glackin CA. TWIST, a basic helix-loop-helix transcription factor, can regulate the human osteogenic lineage. J Cell Biochem. 1999;75:566–77. doi: 10.1002/(sici)1097-4644(19991215)75:4<566::aid-jcb3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Verzi MP, Anderson JP, Dodou E, Kelly KK, Greene SB, North BJ, Cripps RM, Black BL. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol. 2002;249:174–90. doi: 10.1006/dbio.2002.0753. [DOI] [PubMed] [Google Scholar]
- 9.Mironchik Y, Winnard PT, Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, Burger H, Glackin C, Raman V. Twist Overexpression Induces In vivo Angiogenesis and Correlates with Chromosomal Instability in Breast Cancer. Cancer Res. 2005;65:10801–10809. doi: 10.1158/0008-5472.CAN-05-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–91. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, Wong YC, Wang X. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–62. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 12.Bussemakers MJ, van Bokhoven A, Mees SG, Kemler R, Schalken JA. Molecular cloning and characterization of the human E-cadherin cDNA. Mol Biol Rep. 1993;17:123–8. doi: 10.1007/BF00996219. [DOI] [PubMed] [Google Scholar]
- 13.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–57. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert SF. Developmental Biology. Sinauer Associates; 1997. [Google Scholar]
- 15.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. American Journal of Pathology. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A. 1995;92:7416–9. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji X, Woodard AS, Rimm DL, Fearon ER. Transcriptional defects underlie loss of E-cadherin expression in breast cancer. Cell Growth Differ. 1997;8:773–8. [PubMed] [Google Scholar]
- 18.Berx G, Cleton-Jansen AM, Nollet F, de Leeuw WJ, van de Vijver M, Cornelisse C, van Roy F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. Embo J. 1995;14:6107–15. doi: 10.1002/j.1460-2075.1995.tb00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–9. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 20.Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene. 2005;24:8277–90. doi: 10.1038/sj.onc.1208991. [DOI] [PubMed] [Google Scholar]
- 21.el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–6. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 22.Hajra KM, Chen DYS, Fearon ER. The SLUG Zinc-Finger Protein Represses E-Cadherin in Breast Cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 23.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 24.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 25.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 26.Vesuna F, Winnard P, Jr, Raman V. Enhanced green fluorescent protein as an alternative control reporter to Renilla luciferase. Anal Biochem. 2005;342:345–7. doi: 10.1016/j.ab.2005.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duriseti S, Winnard PT, Jr, Mironchik Y, Vesuna F, Raman A, Raman V. HOXA5 regulates hMLH1 expression in breast cancer cells. Neoplasia. 2006;8:250–8. doi: 10.1593/neo.05766. [DOI] [PMC free article] [PubMed] [Google Scholar]