Abstract
BRAF and NRAS are common targets for somatic mutations in benign and malignant neoplasms that arise from melanocytes situated in epithelial structures and lead to constitutive activation of the MAP-kinase pathway1, 2. However, BRAF and NRAS mutations are absent in a number of other melanocytic neoplasms in which the equivalent oncogenic events are currently unknown3. We report frequent somatic mutations in the heterotrimeric G protein alpha subunit, GNAQ, in blue nevi (83%) and ocular melanoma of the uvea (46%). The mutations occur exclusively in codon 209 in the ras-like domain and result in constitutive activation, turning GNAQ into a dominant acting oncogene. Our results demonstrate an alternative route to MAP-kinase activation in melanocytic neoplasia providing new opportunities for therapeutic intervention.
Most melanocytic neoplasms, benign melanocytic nevi as well as melanomas, originate from melanocytes situated within epithelial structures throughout the body, mostly the sun-exposed skin of individuals with light complexion. The majority of nevi and melanomas show oncogenic mutations in signaling components of the MAP kinase pathway, in particular BRAF and NRAS1,2. However, a subset of melanocytic neoplasms does not show mutations in BRAF and NRAS3. One category, uveal melanoma, arises from melanocytes within the choroidal plexus of the eye and is biologically distinct from cutaneous melanoma by characteristic cytogenetic alterations4 and a very strong propensity to metastasize to the liver5. The other category are intradermal melanocytic proliferations, which can be congenital or acquired, and present in diverse ways ranging from discrete bluish moles (blue nevi) to large blue-gray patches affecting the conjunctiva and periorbital skin (nevus of Ota), shoulders (nevus of Ito), and the lower back (Mongolian spot)6. A potential connection between intradermal melanocytic neoplasms and uveal melanoma is suggested by the fact that nevus of Ota is a risk factor for uveal melanoma and by an overlap in some of the histomorphologic features of the two conditions7.
Using a forward genetic screen in mice we previously identified hypermorphic mutations in GNAQ or GNA11as a cause of diffuse skin hyperpigmentation that was due to an increase of intradermal, but not epidermal, melanocytes8. GNAQ and GNA11 encode members of the q class of G-protein alpha subunits involved in mediating signals between G-protein coupled receptors (GPCRs) and downstream effectors. To investigate whether similar pathways were involved in humans, we sequenced the entire coding regions of GNAQ and GNA11 in a broad spectrum of benign and malignant melanocytic neoplasms. We found mutations in GNAQ in 83% of blue nevi (n=29), 50% of “malignant blue nevi” (n=2), and 46% of uveal melanomas (n=48) (Table 1, Supplementary Information, Figure 1a). Nevus of Ota is a condition in which a subtle proliferation of intradermal melanocytes results in hyperpigmentation of the conjunctiva and periorbital skin. To increase the detection limit for mutations in a background of normal cells, we used a mutation-specific assay and found a mutation in one of 14 cases (6%) (Supplementary Information, Figure 1b). No somatic point mutations were found in GNA11.
Table 1.
Frequency of GNAQ mutations in melanocytic neoplasms The number and type of samples analyzed is shown.
| Diagnosis | % Mutant |
N | |
|---|---|---|---|
|
Cutaneous and mucosal
melanomas |
Melanoma on skin without chronic sun-induced damage (non- CSD)22 |
0% | 15 |
| Melanoma on skin with chronic sun-induced damage (CSD)22 | 4% | 27 | |
| Acral melanoma | 0% | 15 | |
| Mucosal melanoma | 0% | 14 | |
| “Malignant blue nevus” | 50% | 2 | |
| Melanoma arising in congenital nevus | 0% | 3 | |
| Spitzoid melanoma | 0% | 2 | |
| Total | 78 | ||
| Nevi | Blue nevus | 83% | 29 |
| Nevus of Ota | 6% | 17 | |
| Congenital nevus | 0% | 7 | |
| Deep penetrating nevus | 0% | 16 | |
| Proliferating nodule in giant congenital nevus | 0% | 7 | |
| Spitz nevus | 0% | 8 | |
| Total | 84 | ||
| Ocular melanomas | Uveal melanoma | 46% | 48 |
| Uveal melanoma cell line | 27% | 15 | |
| Conjunctival melanoma | 0% | 11 | |
| Total | 74 | ||
| Grand Total | 236 |
All mutations in GNAQ were somatically acquired as assessed by sequencing DNA from adjacent tissue, and occurred exclusively at codon 209 (Supplementary Information, Table 1). The glutamine at codon 209 lies within the ras-like domain of GNAQ (corresponding to residue 61 of RAS and is essential for GTP hydrolysis 9. In RAS family members, mutations at this site, and at codon 12, cause loss of GTPase activity with constitutive activation9-11. There is no equivalent of codon 12 in GNAQ. In contrast to the findings in humans, the mutations found in the dark skinned mice occurred at I63, V179, and F335 in the mouse proteins and do not cause constitutive activation8.
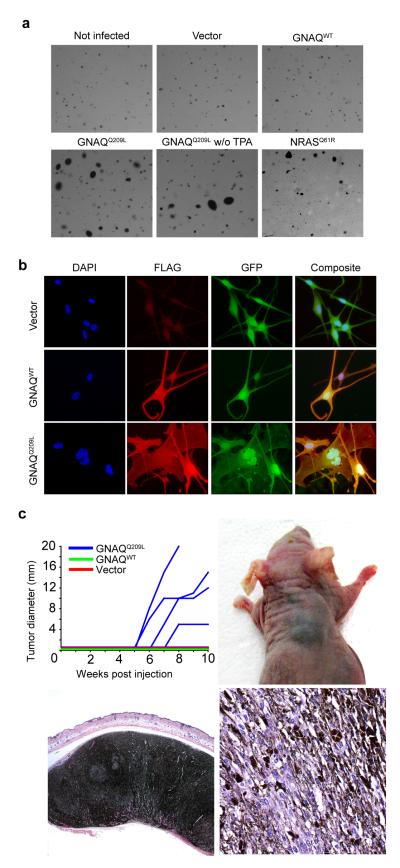
To date no mutations of GNAQ have been described in human neoplasia, but GNAQQ209L has been demonstrated to transform 3T3 cells11. In addition, mutations of the corresponding codon in G alpha S (GNAS) are found in human pituitary and thyroid tumors10, 12. To assess the effect of GNAQQ209L on human melanocytes, we established epitope-tagged lentiviral expression constructs to transfect normal and genetically modified human melanocytes, the latter of which have an extended life span, but still require additional factors (cAMP, TPA) for growth (hTERT/CDK4R24C/p53DD melanocytes13). Stable transfection of GNAQQ209L into primary human melanocytes was insufficient to induce anchorage independent growth (data not shown). In contrast, transfection of GNAQQ209L into hTERT/CDK4R24C/p53DD melanocytes resulted in anchorage independent growth with efficiencies comparable or slightly greater than transfection with NRASQ61R (Figure 1a, Supplementary Information, Table 2). Furthermore, GNAQQ209L but not GNAQwt induced abnormally enlarged nuclei with markedly irregular contours (Figure 1b, Supplementary Information, Table 3). To validate GNAQQ209L as an oncogene in vivo, we performed tumorigenicity studies in nude mice (Figure 1c). GNAQQ209L, but not GNAQwt or vector control transfected melanocytes gave rise to heavily pigmented tumors at the injection site. The tumor morphology resembled closely the spectrum of atypical blue nevus that in humans has been termed animal type melanoma or pigmented epithelioid melanocytoma6.
Figure 1. GNAQQ209L transforms melanocytes.
a, GNAQQ209L induces anchorage independent growth in soft agar of hTERT/CDK4R24C/p53DD melanocytes in a TPA-independent manner with comparable efficiency as NRASQ61R. b, Cells expressing Flag-tagged GNAQQ209L showed enlarged nuclei with irregular contours after 5 days. c, Melan-a cells23 stably transduced with GNAQQ209L, but not with wild-type GNAQ (n=3) or vector control (n=4), induce highly pigmented tumors of spindled and epithelioid melanocytes after 10 weeks in four out of five animals.
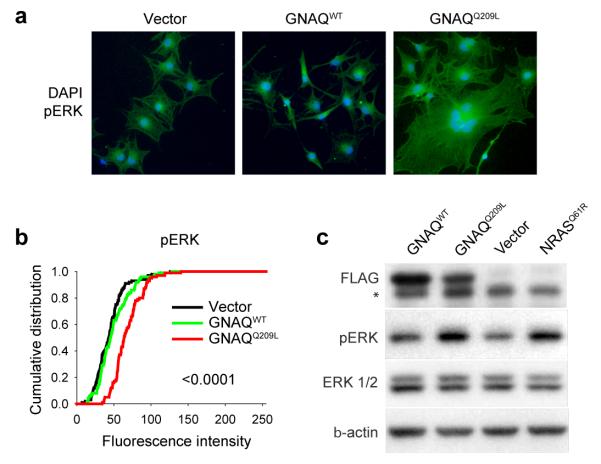
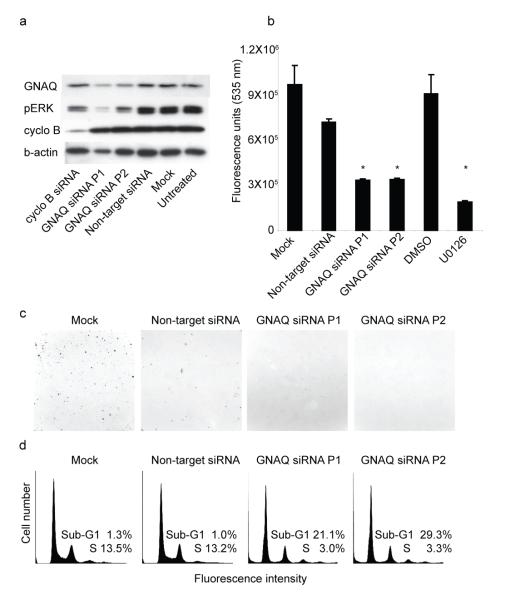
Signaling pathways downstream of GNAQ include activation of protein kinase C family members via the release of diacylglycerol (DAG) by phospholipase Cβ. Consistently, GNAQQ209L-transformed melanocytes grew in soft agar in the absence of TPA, a synthetic DAG analog (Figure 1a, Supplementary Information, Table 2). PKC activation by way of GNAQ activation can activate the MAP-kinase pathway in other cell types14. Uveal melanomas display MAP-kinase activation15, but none of the uveal melanomas we examined in our study showed mutations in BRAF or NRAS (Supplementary Information, Table 4), consistent with other studies3. We therefore tested whether GNAQQ209L would contribute to MAP-kinase pathway activation in human melanocytes and uveal melanoma cells. As shown in Figure 2, GNAQQ209L transfection into hTERT/CDK4R24C/p53DD melanocytes caused increased levels of phospho-ERK compared to control cells transfected with wildtype GNAQ (GNAQWT) or an empty vector (Vector). Similar results were obtained with GNAQQ209L transfection into primary human melanocytes and 293T cells (see Supplementary Information, Figure 2). Conversely, siRNA-mediated knock-down of GNAQ in the uveal melanoma cell line, OMM1.3, which harbors the GNAQ-Q209L mutation, resulted in a decrease of phospho-ERK levels (Figure 3a). In addition, GNAQ knock-down in OMM1.3 cells causes a substantial decrease in cell number (Figure 3b), loss of anchorage independent growth (Figure 3c) and a marked increase in sub-G0/G1 population (Figure 3d) as compared to control cells. Similar results were obtained when another uveal melanoma cell line Mel202 was treated with siRNA against GNAQQ209L (Supplementary Information, Figure 3). Mel202 and OMM1.3 stem from different patients, as confirmed by DNA fingerprinting (data not shown.)
Figure 2. GNAQQ209L induces MAP kinase activation.
a, Increased expression of pERK in hTERT/CDK4R24C/p53DD melanocytes transfected with GNAQQ209L compared to similar melanocytes transfected with GNAQWT or empty vector. b, Cumulative distribution of mean pixel fluorescence intensity per cell obtained from immunofluorescent detection of pERK (p-values: GNAQQ209L vs. vector). c, Western blot showing increased pERK levels in hTERT/CDK4R24C/p53DD melanocytes expressing Flag-tagged GNAQQ209L compared to cells transduced with Flag-tagged GNAQWT or vector control. NRASQ61R transduced melanocytes are shown as a positive control. * The band migrating just below the Flag band is non-specific reactive band in the lysate.
Figure 3. Knockdown of GNAQ in OMM1.3 cells results in MAP-kinase inhibition, reduced growth and apoptosis.
a, Western blot after treatment with 2 pools of siRNAs against GNAQ shows decreased pERK levels compared to control treated cells: cyclophilin B and non-target siRNA. b, After 72 hours, GNAQ knockdown results in marked reduction of cell numbers, similar to the effect of MEK inhibitor U0126. Bars show means and standard error of five replicate assays. * p<0.05, t-test compared to mock or vehicle control, respectively. c, GNAQ knockdown reduces the number of colonies (upper left corner) formed in soft agar. d, Cell cycle profiles showing an increase of the sub-G0/G1 population after GNAQ knockdown.
In summary, our data identify GNAQ as a novel oncogene in human neoplasia. GNAQ operates downstream of several GPCRs that are important in melanocyte homeostasis and neoplasia. GNAQ is involved in endothelin signaling, which is essential for melanocyte survival early during development16. Gq signaling may also contribute to the association observed for melanoma invasion and metastasis with Wnt family members17, since Frizzled receptors can couple to Gq18. Furthermore, Gq signaling is likely to underlie the ability of the metabotropic glutamate receptor gene, GRM1, to cause dermal melanocytic neoplasia and ocular melanoma in transgenic mice19.
The location of neoplastic melanocytes induced by somatic GNAQ mutation in humans is very similar to the location of melanocytes affected by germline GNAQ (and GNA11) mutations in mice8. In both cases, the melanocytic proliferations spare epithelial structures. However, our previous work in mice suggests that these mutations do not interfere with homing of melanocytes to epithelial structures, but instead lead to an increase of the total melanoblast pool8. In humans, it is not known whether the adult dermis contains any residual sparse populations of melanocytes in which GNAQ mutations could occur and induce intradermal melanocytic tumors6, 20. Acquired blue nevi are typically small and well circumscribed, raising the possibility that they arise from an intradermal population of melanocytes that remains to be characterized. By contrast, nevus of Ota involves different structures such as periorbital skin, conjunctiva, neurovascular bundles, ganglia, and uvea, suggesting these GNAQ mutations may arise early in a migrating melanoblast. This later neoplasm shows strong similarities to the murine studies and indicates a specific window for Gq signaling in terms of cell type and developmental time that is conserved across species.
Our findings identify GNAQ as a genetic link between nevus of Ota and uveal melanoma and help explain why nevi of Ota are a risk factor for uveal melanoma7. The risk is small, as only about 1 in 400 nevi of Ota progress to uveal melanoma and future studies will have to elucidate the role of GNAQ mutations in this risk. In our experiments, GNAQ behaves similar to the oncogenes, BRAF and NRAS, in that its mutation is insufficient for full progression to melanoma. This is illustrated best by blue nevi, which are generally stable lesions that rarely become malignant (“malignant blue nevus”)6. Thus MAPK activation appears to be an early event in neoplasms with GNAQ mutations, as it is in neoplasms with BRAF and NRAS mutations2. Further studies are necessary to determine whether the difference in tissue involvement between melanocytic neoplasms with BRAF mutations and those with GNAQ mutations is a functional consequence of the mutations themselves, or indicates differences in the target cell populations in which these mutations occur.
Uveal melanoma is a highly aggressive cancer without any effective treatment options once it metastasizes. Although it only accounts for approximately 5% of all melanomas, it represents the most common intraocular malignancy in the United States and has a five-year disease specific survival rate of approximately 70%21. Our study identifies signaling components downstream of GNAQ as potential targets in this dreadful disease.
Methods Summary
Detailed descriptions of reagents and experiments can be found in the full Methods. GNAQ and GNA11 were sequenced in DNA extracted from archival, paraffin-embedded biopsies under the approval of the institutional review boards at UCSF, Stanford and UBC. Primary, hTERT/CDK4R24C/p53DD, and melan-a melanocytes were stably infected with FG12 lentiviral expression vectors expressing flag-tagged wildtype GNAQ or GNAQQ209L, empty vector or NRASQ61R. Infection efficiencies were estimated by the proportion of cells expressing GFP. To assess anchorage independent growth, melanocytes were plated on soft agar, cultured for 28 days and stained with 0.005% crystal violet. For tumorigenicity experiments, 1 million lentiviral transfected melan-a cells were injected into each nude mouse, housed at UBC according to CCAC guidelines. For immunofluorescence, lentiviral transfected cells were cultured on coverslips for 5 days and incubated with antibodies against pERK (E-4, Santa Cruz Biotechnology), cyclin D1 (M-20, Santa Cruz Biotechnology) and GNAQ (C-19, Santa Cruz biotechnology). OMM1.3 and Mel202 cells were transfected with two different pools of siRNA against GNAQ: pool 1: 5′-CAAUAAGGCUCAUGCACAAUU-3′, 5′-CGACGAGAAUAUCAAUUAUUU-3′, 5′-GCAAGAGUACGUUUAUCAAUU-3′, 5′-UAGUAGCGCUUAGUGAAUAUU-3′; pool 2: 5′-AUGCACAAUUAGUUCGAGAUU-3′, 5′-UAUGAUAGACGACGAGAAUUU-3′, 5′-CAGACAAUGAGAACCGAAUUU-3′, 5′- CGCCACAGACACCGAGAAUUU-3′). For western blot analysis, 5-20 μg of protein was extracted from lentiviral or siRNA transfected cells. Primary antibodies were: pERK, cyclin D1, GNAQ, Cyclophilin B (Abcam), anti-FLAG M2 (Sigma), Anti-ERK ½ pAb (Promega), and β-actin (Sigma). Secondary antibodies were labeled with horseradish peroxidase. Relative cell numbers were quantified by the CyQUANT® Cell Proliferation Assay Kit (Invitrogen). Cell cycle measurements were performed on a FACSCalibur after staining with propidium iodide. Student’s t-test and Fisher’s Exact test were used for statistical comparison.
Full methods
DNA
DNA was obtained from archival paraffin-embedded biopsies under the approval of the institutional review boards at UCSF, Stanford and the University of British Columbia. For each sample, 20 um sections were microdissected, washed in xylene and ethanol and digested with proteinase K. DNA was then extracted with phenol-choloroform-isoamyl-alcohol.
Sequencing
Biopsy DNA was amplified using PCR. For GNAQ exon 5, the primers used were 5′-cccacaccctactttctatcatttac-3′ and 5′-ttttccctaagtttgtaagtagtgc-3′. PCR products were used as templates for sequencing reactions using Big Dye (ABI). Samples identified with mutations in both sequencing directions were replicated at least twice. Mutations 1-3 were verified with RFLP. Mutations 1 and 2 create an Eco0109I restriction site, while mutation 3 produces an AflII restriction site.
Sensitive assay for Q209 mutations in mixed cell populations
The peptide nucleic acid, Actctctgacctttggc-CONH2, was resuspended in 50% DMF and used at a final concentration of 4 uM against 2 ng template DNA in a 25 ul reaction. The reaction conditions were 0.25 mM dNTPs, 6X BSA, 2 U Hotstar Taq, 1× Hotstar Taq buffer, and 0.5 uM each primer, 5′-ttttccctaagtttgtaagtagtgc-3; and 5′-atccattttcttctctctgacc-3′. PCR consisted of 40 cycles of 95 degrees (1 min), 73 degrees (1 min), 57 degrees (45 sec), and 72 degrees (1 min).
Plasmids
A GNAQQ209L cDNA plasmid was obtained from UMR cDNA Resource Center. The wild-type counterpart was generated by site-specific mutagenesis. The coding regions of both constructs were epitope-tagged with an N-terminal Flag-tag and cloned into the lentiviral expression vector FG12. All constructs were sequenced for confirmation.
Cell culture
hTERT/CDK4R24C/p53DD melanocytes were cultured in glutamine containing Ham’s F12 supplemented with 7% FBS, 50 ng/ml TPA, 0.1 mM IBMX, 10 μM Na3VO4, 1 mM dbcAMP. Primary normal melanocytes were cultured in MCDB153 supplemented 20% FBS, 2% chelated FBS, 5 μg/ml L-glutamin, 15 μg/ml cholera toxin, 0.5 ng/ml bFGF, 100 nM ET3 and 1.68 mM SCF. Cell lines OMM1.3, Mel202 and 293T were cultured in RPMI supplemented with 10% fetal bovine serum. Melan-a cells were cultured in glutamine-containing RPMI media supplemented with 10% FCS and 200 nM TPA.
Lentiviral infection
Viral supernatant were generated using 293T cells transfected with 10 μg plasmid and appropriate lentiviral packaging plasmids. Media was changed 16 hr after transfection and the virus was harvested 40 to 56 hr later. hTERT/CDK4R24C/p53DD, melan-a and normal melanocytes were infected and infection efficiencies were estimated by the presence of GFP expressing cells.
Transient transfection
293T cells were seeded in 6-well plates at 1×106 cells per well with RPMI/10% FCS. Transfections were carried out using Lipofectamine 2000 (Invitrogen) and 2 μg plasmid pcDNA™6.2/V5-DEST® Gateway vector (Invitrogen) alone or containing the complete coding region for either GNAQQ209L or GNAQWT, respectively. Cells were lysed 48 hour post-transfection and assayed for protein content.
Tumorigenicity study
Melan-a cells were lentiviral transfected with GNAQWT, GNAQQ209L and empty vector control. Seven days post infection, cells were resuspended in DMEM at 5 million cells per milliliter. Four month old, albino female nude mice (NU/J) were injected with 0.2 ml subcutaneously. Mice were palpated weekly.
Cell proliferation assay
Relative cell numbers were quantified by the CyQUANT® Cell (Invitrogen) Proliferation Assay Kit according to the manufacturer’s protocol using 96-well plates. 7.5×103 cells were transfected and the fluorescent intensity was read after 72 hr.
Immunofluorescence
Human primary and hTERT/CDK4R24C/p53DD melanocytes were cultured on cover slips in 6 well plates and infected with lentiviral vectors containing either GNAQQ209L GNAQWT, or an empty vector as control. Five days after infection, cells were fixed with 4% formaldehyde in PBS, permeabilized with 0.2 % Triton X100 and incubated with 3% BSA. Antibodies against pERK (E-4, Santa Cruz Biotechnology), cyclin D1 (M-20, Santa Cruz Biotechnology) and GNAQ (C-19, Santa Cruz biotechnology) were detected using secondary antibodies labeled with Alexa Fluor 594 and 532 (Molecular Probes). Images were taken at fixed exposures with an Axio Image M1 microscope (Zeiss, Germany). The fluorescence intensities were quantified using ImageJ software and the mean pixel intensities were used for statistical analysis using Microsoft Excel and Data Desk.
Soft agar assay
10×103 human primary melanocytes, hTERT/CDK4R24C/p53DD melanocytes stably expressing GNAQQ209L, GNAQWT, NRASQ61R or vector control and siRNA treated OMM1.3 and Mel202 cells were suspended in full media containing 0.35% agar and plated on a lower layer of 0.5% agar in 6 well plates. After 28 days, cells were stained with 0.005% crystal violet. Colony number and size were quantified using ImageJ software.
Cell cycle analysis
72 hr post transfection with siRNA against GNAQ, OMM1.3 and Mel202 cells were trypsinized, washed with cold PBS and fixed with 70% ethanol. Fixed cells were stained with propidium iodide. Cell cycle measurements were performed on a FACSCalibur (BD Biosciences), with minimum of 20,000 events, and profiles were analyzed using FlowJo and ModFit.
siRNA transfection
OMM1.3 and Mel202 were plated in RPMI/10% FCS in 6 well or 96 well plates at 1.5×105 or 5×103 cells per well, respectively. Two different pools; each comprised of four siRNA duplexes (pool 1: 5′-CAAUAAGGCUCAUGCACAAUU-3′, 5′-CGACGAGAAUAUCAAUUAUUU-3′, 5′-GCAAGAGUACGUUUAUCAAUU-3′, 5′-UAGUAGCGCUUAGUGAAUAUU-3′; pool 2: 5′-AUGCACAAUUAGUUCGAGAUU-3′, 5′-UAUGAUAGACGACGAGAAUUU-3′, 5′-CAGACAAUGAGAACCGAAUUU-3′, 5′-CGCCACAGACACCGAGAAUUU-3′) against GNAQ and ON-TARGETplus controls (all Dharmacon) were transfected in lipofectamine RNAiMax (1μl/pmol siRNA) at 100 nM. siRNA complexes were formed in Optimem. Cells were lysed for analysis 72-96 hours post-transfection. For soft agar assay, 48 post transfection cells were used.
Western blot analysis
Cells were washed twice with ice-cold PBS and lysed in 50 mM Tris—HCl pH 7.8, 1% NP-40, 10% glycerol, 150 mM NaCl, 1 % Sodium deoxycholate, 1% sodium dodecyl sulfate, supplemented with protease inhibitor, phosphatase inhibitor and EDTA (Pierce Biotechnologies). The protein content of the lysates was determined by the BCA Protein Assay Reagent (Pierce Biotechnologies). 5-20 μg of protein were separated by SDS—PAGE and transferred to Immobilon-P membrane (Millipore). Primary antibodies were: pERK (E-4, Santa Cruz Biotechnology), cyclin D1 (M-20, Santa Cruz Biotechnology) and GNAQ (C-19, Santa Cruz biotechnology), Cyclophilin B (Abcam), anti-FLAG M2 (Sigma), Anti-ERK ½ pAb (Promega), and β-actin (Sigma). Secondary antibodies were labeled with horseradish peroxidase.
Statistical analysis
Immunofluorescence data and CyQUANT measurements were analyzed using Student’s t-test. Fisher’s Exact test was used to compare the proportion of atypical cells.
Supplementary Material
Acknowledgements
We thank Dr. William Harbour, Washington University, Saint Louis, for providing additional DNA samples from uveal melanomas and Drs. Pablo Viciana-Rodriguez and Allan Balmain for providing advice. hTERT/CDK4R24C/p53DD melanocytes were a gift from Dr. David Fisher, Dana Farber Cancer Institute13, normal human melanocytes from foreskin were a gift from Dr. Meenhard Herlyn, The Wistar Institute, and OMM1.3 and Mel202 cells were originally from Dr. Bruce Ksander, Schepens Eye Research Institute. The melan-a cells were a gift from Dr. Dorothy Bennett, St George’s University, London23.This work was supported by grants from the National Cancer Institute (P01 CA025874 Project 2 to BCB) the Melanoma Research Alliance, the Canadian Institutes of Health Research (MOP-79511 to CVR), and the National Institutes of Health (GSB).
References
- 1.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Pollock PM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2002;25:25. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 3.Saldanha G, et al. High BRAF mutation frequency does not characterize all melanocytic tumor types. Int J Cancer. 2004;111:705–10. doi: 10.1002/ijc.20325. [DOI] [PubMed] [Google Scholar]
- 4.Horsman DE, White VA. Cytogenetic analysis of uveal melanoma. Consistent occurrence of monosomy 3 and trisomy 8q. Cancer. 1993;71:811–9. doi: 10.1002/1097-0142(19930201)71:3<811::aid-cncr2820710325>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 5.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18:75–84. doi: 10.1016/j.ohc.2004.07.002. viii. [DOI] [PubMed] [Google Scholar]
- 6.Zembowicz A, Mihm MC. Dermal dendritic melanocytic proliferations: an update. Histopathology. 2004;45:433–51. doi: 10.1111/j.1365-2559.2004.01975.x. [DOI] [PubMed] [Google Scholar]
- 7.Singh AD, et al. Lifetime prevalence of uveal melanoma in white patients with oculo(dermal) melanocytosis. Ophthalmology. 1998;105:195–8. doi: 10.1016/s0161-6420(98)92205-9. [DOI] [PubMed] [Google Scholar]
- 8.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS. Effects of G-protein mutations on skin color. Nat Genet. 2004;36:961–8. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science. 1993;262:1895–901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 10.Landis CA, et al. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989;340:692–6. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- 11.Kalinec G, Nazarali AJ, Hermouet S, Xu N, Gutkind JS. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol Cell Biol. 1992;12:4687–93. doi: 10.1128/mcb.12.10.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyons J, et al. Two G protein oncogenes in human endocrine tumors. Science. 1990;249:655–9. doi: 10.1126/science.2116665. [DOI] [PubMed] [Google Scholar]
- 13.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard KB, Hepler JR. Cell signalling diversity of the Gqalpha family of heterotrimeric G proteins. Cell Signal. 2006;18:135–50. doi: 10.1016/j.cellsig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Zuidervaart W, et al. Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer. 2005;92:2032–8. doi: 10.1038/sj.bjc.6602598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin MK, Levorse JM, Ingram RS, Tilghman SM. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402:496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 17.Dissanayake SK, et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282:17259–71. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheldahl LC, Park M, Malbon CC, Moon RT. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr Biol. 1999;9:695–8. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- 19.Marin YE, et al. Stimulation of oncogenic metabotropic glutamate receptor 1 in melanoma cells activates ERK1/2 via PKCepsilon. Cell Signal. 2006;18:1279–86. doi: 10.1016/j.cellsig.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima J, Nogita T, Higaki Y, Horikoshi T, Kawashima M. Dormant melanocytes in the dermis: do dermal melanocytes of acquired dermal melanocytosis exist from birth? Br J Dermatol. 1998;139:349–50. doi: 10.1046/j.1365-2133.1998.02385.x. [DOI] [PubMed] [Google Scholar]
- 21.Chang AE, Karnell LH, Menck HR. The National Cancer Data Base report on cutaneous and noncutaneous melanoma: a summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1998;83:1664–78. doi: 10.1002/(sici)1097-0142(19981015)83:8<1664::aid-cncr23>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 22.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 23.Bennett DC, Cooper PJ, Hart IR. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer. 1987;39:414–8. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.