Abstract
In the postgenomic era, we need an algorithm to readily translate genes into physiologic principles. The failure to advance biomedicine is due to the false hope raised in the wake of the Human Genome Project (HGP) by the promise of systems biology as a ready means of reconstructing physiology from genes. like the atom in physics, the cell, not the gene, is the smallest completely functional unit of biology. Trying to reassemble gene regulatory networks without accounting for this fundamental feature of evolution will result in a genomic atlas, but not an algorithm for functional genomics. For example, the evolution of the lung can be “deconvoluted” by applying cell-cell communication mechanisms to all aspects of lung biology development, homeostasis, and regeneration/repair. Gene regulatory networks common to these processes predict ontogeny, phylogeny, and the disease-related consequences of failed signaling. This algorithm elucidates characteristics of vertebrate physiology as a cascade of emergent and contingent cellular adaptational responses. By reducing complex physiological traits to gene regulatory networks and arranging them hierarchically in a self-organizing map, like the periodic table of elements in physics, the first principles of physiology will emerge.
Keywords: evolutionary biology, gene regulatory network, cell-cell communication, functional genomics, systems biology
WE NEED AN INTEGRATED MECHANISM FOR EVOLUTION AND PHYSIOLOGY: HOW GENES DETERMINE CELL CROSS TALK
The greatest challenge in the postgenomic era is to effectively integrate functionally relevant genomic data to understand physiological first principles and how they can be used to decode complex traits. This problem is most often addressed stochastically by analyzing large genomic data sets to identify genes that are associated with structural and functional phenotypes; whether they are causal is more often than not indeterminate. This approach is merely an extrapolation from descriptive systematic biology, beginning with Linnaeus's invention of binomial nomenclature.
Binomial Nomenclature
—Carl Linnaeus (1758)
Systems biology can be viewed at several different levels: the level of the gene, the level of the transcript, the level of the protein, the level of the cell, organ, organ system or population; and clearly, evolution could have impacted the process at any one of these levels. There are many such analyses in the literature (12, 20, 41, 42), but they don't provide integrated, functional genomic, evolutionary mechanisms that would lead to further experimentation and ultimately to prediction. Those who have attempted such integrations have used either a “top-down” or bottom-up approach, but selection pressure, intrinsic, extrinsic, or both, must be applied at a level where it can have the desired effect, i.e., at the functional level where the genetic expression is functionally integrated with the phenotype. Based on this precept, we have elected to take a unique “middle-out” approach, focusing on functional nodes defined by ligand-receptor interactions that establish phenotypes during development (1, 25, 36, 39, 40), sustain them physiologically (4, 36), and recapitulate them in injury/repair(4) and regeneration (1). Unlike the former top-down or bottom-up approaches, starting in the middle offers the advantage of minimizing the a priori assumptions by focusing on the gene regulatory networks (GRNs) that generate form and function, particularly those that have “evolved” over space-time (38) using the same ontogenetically/phylogenetically and regeneratively related motifs (1, 4).
Those vertically integrated, cell-to-functional phenotype mechanisms that best represent physiology across species and development, like that of the lung (34–36), are archetypes for the analytic approach we are advocating. Indeed, we have incorporated the role of the external forces of natural selection shaping physiology as a way of understanding the apposition of the lipofibroblast (which recruits and stores neutral lipid) and the epithelial type II cell in the alveolar wall to produce surfactant phospholipid from those neutral lipids, cell types derived from different germ lines (mesoderm, endoderm), which evolve to regulate both surfactant production and alveolar capillary perfusion through a “stretch-regulated” mechanism, from fish to man, as follows: the rise in oxygen in the Phanerozoic era gave rise to the alveolar lipofibroblast, based on the observation that muscle stem cells differentiate into adipocytes in room air, but not in low oxygen environments (7), and the cytoprotective effect of this adaptation (27) against the oxygen in the environment, followed by the production of leptin by these cells (28), led to selection pressure for a stretch-regulated mechanism for the integration of surfactant production and alveolar capillary perfusion, referred to physiologically as ventilation-perfusion matching. We know that the stretch mechanism for parathyroid hormone-related protein (PTHrP) expression is intrinsic to these cells, because in a microgravity environment they contract, resulting in decreased PTHrP expression (30), perhaps a reversion to the evolution of the swim bladder as an adaptation to gravity and feeding (34). Stretch regulation of leptin (29) may have been in response to the overall selection pressure for the transition from water to land, necessitated by Romer's hypothetical drying up of ponds (23), since fat cells are also critical for the evolution of both endothermy (19) and locomotion (6). Such extrinsic factors as oxygen and stretch implicate population genetic mechanisms of evolutionary selection into this cell-cell signaling model for lung evolution.
There have been numerous attempts to “synthesize” biology from its components. Beginning with Darwin, who was a master at seeing the “forest-and-trees” connections within and between species, and his pronouncement of a mechanism for descent with modification, or natural selection, which is clever, but not mechanistic in the context of molecular genetics.
“Descent with modification”
—Charles Darwin (1859).
Darwinian thought then fostered the works of Haeckel, Waddington, Riel, Seilacher, and Gould, to name a few of those who have attempted to further our insights to evolution. And more recently, Morowitz (20) and West (42) have gained notoriety by formulating comprehensive analyses of physiology, but their reductionist/synthetic approaches are similarly descriptive, metabolic pathways and flow patterns in physiology, which are not predictive. The problem with these perspectives is that they reason from existing structures and functions backward in an a posteriori fashion consistent with contemporary biology and medicine, but they do not predict the changes that have occurred over evolutionary space-time, leaving medicine largely nonpredictive and ultimately incomplete in its scope.
The advent of systems biology can be seen as the effort to implement the fruits of reductionism to provide a mechanistic, causal, and explanatory linkage between genotypes and their associated phenotypes by studying the structure and dynamics of convoluted molecular interaction networks that regulate cells and tissues in development and adulthood. Systems biology aims at developing and integrating experimental and mathematical techniques in pursuit of principles that would make the nature of cellular phenotypes more intelligible and their control more deliberate. This effort is catalyzed by the practical need to cure diseases. However, it also reflects a desire for a theoretical framework by which to deconvolute the complexities of the cell and the organism.
A FUNCTIONAL GENOMIC, MIDDLE-OUT APPROACH TO THE EVOLUTION OF COMPLEX PHYSIOLOGICAL TRAITS
“Nothing in Biology Makes Sense Except in the Light of Evolution”
—Theodosius Dobzhansky (1973)
The reductionist genetic approach cannot simply be computed to generate phenotypes (17, 21) because evolution is (largely) not a result of chance, other theories to the contrary (14); rather, it is an “emergent and contingent” process. In our current and future research working environment, we must expand our computational models to encompass a broad, comprehensive, evolutionary approach; as Dobzhansky has famously said, “Nothing in biology makes sense except in the light of evolution” (10).
We have formally proposed using a comparative, functional genomic approach to solving for the evolution of physiological traits that engenders development, homeostasis, and regeneration as a cluster of parallel lines that can be mathematically analyzed as a family of simultaneous equations (34). This perspective provides an empirically feasible and refutable way of systematically integrating such information in its most robust form to retrace its evolutionary origins (see Fig. 1); Among mammals, embryonic lung development is subdivided into two major phases: branching morphogenesis and alveolization. Fortuitously, we have observed that deleting the PTHrP gene results in failed alveolization; the generation of progressively smaller, more plentiful alveoli with thinning walls for gas exchange is the primary vertebrate evolutionary process for the transition from water to air. This, and the fact that PTHrP and its receptor are highly conserved [the PTHrP ortholog tuberoinfundibular protein (TIP39) is expressed as far back in phylogeny as yeast], are stretch-regulated, and form a paracrine signaling pathway mechanistically linking the endodermal and mesodermal germ layers of the embryo to the blood vessels has compelled us to investigate its overall role in lung ontogeny, phylogeny, and evolution and to exploit this key transitional GRN to further our understanding of physiology based on first principles.
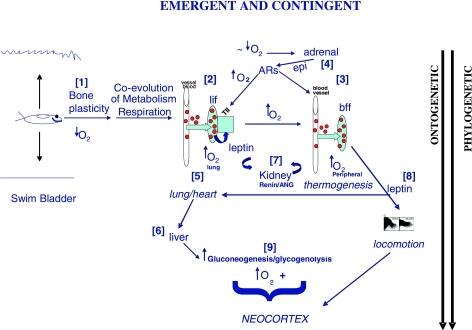
Fig. 1.
Physiological evolution from cells to systems. Selection for lipid metabolism and respiration [1] gave rise to the lung; oxygen induced the lipofibroblast (lif) in the lung [2] and in the peripheral fat cells [3], putting selection pressure on lung/adrenal β-adrenergic signaling [4] to stabilize blood pressure (lung, periphery); coevolution of the lung and heart [5] induced the liver [6]; kidney renin/angiotensin [7] provided further blood pressure stability. Leptin production by fat cells [8] promoted limb and lung development; increased oxygenation and glucose promoted neocortical development [9].
We have demonstrated the utility of this middle-out approach to lung biology both as a basic tool for identifying novel functional genes and to target genes for the treatment of lung disease. For example, we have discovered that leptin is the intermediate for PTHrP signaling to adepithelial lipofibroblasts to stimulate surfactant. Leptin is a pleiotropic metabolic hormone that coordinates the development and evolution of locomotion and metabolism (6), since it stimulates limb development in Xenopus tadpoles. We have now shown that it also stimulates tadpole lung development (37), providing an integrated model for both the ontogeny and phylogeny of these complex physiologic traits. Moreover, the breakdown in the PTHrP-leptin signaling pathway causes chronic lung disease (36), providing novel targets for the diagnosis, prevention, and treatment of chronic lung disease.
This model has the power to transcend the lung based on ontogenetic and phylogenetic principles (see Fig. 1). The effect of leptin on frog lung development is to increase the surface area in tandem with stimulation of surfactant production; the effect of leptin on both the antiatelectatic mechanism and on surfactant protein A, an antimicrobial, points to the evolution of barrier function common to a variety of tissues and organs, e.g., gut, kidney, skin, brain. Viewing these “molecular phenotypes” across ontogenetic and phylogenetic space-time will lead to other such interrelationships, ultimately generating a new paradigm for our understanding of evolutionary biology and physiology (Ref. 34, and see Fig. 2).
Fig. 2.
A periodic table for systems biology. The schematic depicts how developmental cell signaling gene regulatory networks from a variety of tissues could be integrated into a systems biology hologram analogous to the periodic table of the elements.
IT TAKES A PROCESS TO DECIPHER A PROCESS
Evolution is a biologic puzzle. For example, the “solution” for the reassembly of the Dead Sea scrolls was also a puzzle: You are given a box containing the remaining 10,000 fragments of the parchment scrolls. How do you reassemble them based on some mechanism or guiding principle? It takes a process to understand a process, because you need as many equations as variables to solve such complex algebraic problems. The inspiration for the solution to the reassembly of the Dead Sea scroll shards puzzle came from the insight by scientists at the Hebrew University's Koret School for Veterinary Science near Rishon LeZion that the fragments of each scroll found decades earlier in Qumran were parts of one parchment made from one goat skin. Reasoning in the forward direction, from means to ends, these investigators used molecular biology to identify the fragments that were genetically related to the goat skin that the parchment was made from. So a scriptural puzzle had a biological solution. The solution to the evolutionary biological puzzle is even more counterintuitive but must likewise be reasoned from means to ends. The Dead Sea scrolls were reassembled using the DNA signature that was the molecular basis for creating the original parchment. Molecular biology can also be used to decipher physiology, but it must be applied in a way that is compatible with the process being evaluated. The evolution of complex physiological traits was not an acellular, random, statistical event. It was the result of selection pressure for adaptation to the environment, communicated from generation to generation, over eons.
A FOREST-AND-TREES PROBLEM
Perhaps this unique solution to the reassembly of the Dead Sea scrolls is a cipher that may help us overcome the current stagnation in research in biology and medicine, particularly considering all of the technologies to which we now have access. A recent Blue Ribbon Panel of the American Academy of Arts and Sciences charged with determining how to ameliorate the crisis in U.S. funding for biomedical research recommended investing in young scientists and in high-risk, high-reward research (26). But our problem is far more fundamental. It is due to the lack of an effective and accessible algorithm for readily translating genes into phenotypes, or biomolecules into a parchment scroll. The problem is readily apparent compared with the advances in physics over the past 150 years, starting with the Mendeleev version of the periodic table, followed by quantum physics and Einstein's formulation of E = MC2.
“… if all the elements are arranged in order of their atomic weights a periodic repetition of properties is obtained.”
—Demetri Mendeleev (1869)
Ironically, Darwin set us off in search of our evolutionary origins at about the same time that Mendeleev formulated his periodic table of elements. That contrast is now underscored by the publication of the HGP in 2000, which sorely lacks an algorithm like the periodic table to convert genes into phenotypes.
To some extent, the failure to advance biomedicine is due to the false hope raised in the wake of the HGP by the promise of systems biology (12) as a ready means of reconstructing physiology from genes. Like the atom in physics, the cell, not the gene, is the smallest completely functional unit of biology. Trying to reassemble GRNs without accounting for this fundamental feature of evolution will result in a genomic atlas, but not an algorithm for functional genomics. Indeed, the reductionist premise of systems biology is reflective of a recurrent pattern in evolutionary biology, vacillating between genes and phenotypes over its stormy history (11), failing to show that morphogenetic fields exist experimentally, and how they do, in fact, generate structure and function, until only recently (11). The scientific validity of morphogenetic fields has been borne out by contemporary molecular embryology (13, 24, 40), beginning with the breakthrough discovery of homeobox genes (18, 25), demonstrating the homologies across phyla first predicted by Étienne Geoffroy St-Hilaire in the 19th century.
“Nature… tends to repeat the same organs in the same number and in the same relations, and varies to infinity only their form. In accordance with this principle I shall have to draw my conclusions, in determining the bones of the fish's skull, not from a consideration of their form, but from a consideration of their connections.”
—Étienne Geoffroy St-Hilaire (1807)
A PATH THROUGH THE FOREST
This review was composed largely to convey a mechanistic approach to understanding the principles of physiology based on evolutionary precepts, which challenges the prevailing descriptive paradigm. It was motivated by our recent novel insights to the cell-molecular mechanisms of lung evolution, which integrate cell-cell signaling mechanisms common to embryogenesis, homeostasis and regeneration (35).
The evolutionary literature is replete with metaphors that have sustained interest in this esoteric, hermeneutic topic for decades. But such metaphoric thinking has bogged evolutionary biology down in description ever since Darwin first coined the term natural selection (8) to provide a proximate mechanism for evolution.
Concept of Homeostasis
—Walter B. Canon (1932)
There have been many attempts to rationalize the formation of complex physiological systems. For example, Canon formulated the concept that biological systems were designed to “trigger physiological responses to maintain the constancy of the internal environment in face of disturbances of external surroundings,” which he termed homeostasis. He emphasized the need and goal of reassembling the data being amassed for the components of biological systems into the context of whole organism function (5). Weibel, Taylor, and Bolis (41) tested their theory of symmorphosis, the idea that physiology has evolved to optimize biologic function. West, Brown, and Enquist (42) derived a general model for allometry, including a mathematical model demonstrating that metabolism complies with the M3/4 rule. Morowitz (20) has suggested that all of biochemistry can be reduced to hierarchical networks, or “shells.” The significance of all of these observations is that the investigators acknowledge that there are fundamental rules of physiology, but they do not address how and why they have evolved. This review applies the mechanism of cell-cell signaling as the organizing principle for metazoan evolution.
Even to the naïve observer, it is intuitively obvious that there are patterns of size and shape in biology. Darwin was a master at delineating these patterns and defining a process by which they may have evolved through descent with modification, as well as a descriptive mechanism, natural selection. However, such metaphors are grossly inadequate in the age of genomics, and they do not generate testable, refutable hypotheses. Without an understanding of how and why evolution has occurred, we cannot take advantage of the underlying principles, particularly as they might apply to human physiology and medicine. This problem arises over and over again in various ways that are referred to euphemistically as “counterintuitive,” which is an expedient way of dismissing observations that cannot be explained using the prevailing descriptive paradigm. For example, why is it that organ systems have coevolved to link lipid metabolism and respiration (alveolar surfactant and gas exchange), photoreception and circadian rhythms (the pineal as the “third eye”), blood volume control and erythropoiesis, or why ear ossicles evolved from fish jaws. This may be due to the lack of a functionally relevant perspective on the process of evolution.
The term “Paradigm Shift” coined
—Thomas Kuhn (1962)
Alternatively, with the aid of genomics as the basis for biologic analyses, we have reconsidered the process of evolution from a cellular-molecular signaling perspective, because that is where this process emanated from and evolved to (9, 15). Such a Kuhnian paradigm shift would allow us to distinguish forest and trees, and how an understanding of the evolution of structure and function lends itself to the application of genomics to medicine. It seems intuitively obvious that there are fundamental commonalities between ontogeny and phylogeny, given that both start from single cells and form progressively more complex structures through cell-cell interactions mediated by growth factors and their receptors. By systematically focusing on such cell-molecular developmental mechanisms as serial events across vertebrate classes, as is implied by cladograms, it ultimately may be possible to determine the mechanisms of evolution.
The networks of genes that derive from the proposed algorithm generated using a self-organizing map approach (Fig. 2) link specific phenotypes together in ways that seem counterintuitive, offering dynamic new ways of thinking about how the genomic “elements” of physiological systems are recombined through evolution to generate novelty based on cellular principles of phylogeny and development, rather than on static descriptions of structure and function. This is analogous to the periodic table being based on atomic number as an independent “organizing mechanism” for the physical elements. And like the periodic table of elements, which predicts new elements, the biological algorithm will predict novel GRNs. Ultimately, this biological space-time hologram will reveal the underlying rules for the first principles of physiology. Our laboratory has devised several models with which to test this evolutionary cell-molecular concept: the developing rat and mouse, the embryonic chick, and the Xenopus tadpole. These models offer a concerted developmental and phylogenetic approach for determining specific functional GRNs across phyla.
CODA
Just So Stories
—Rudyard Kipling (1902)
Unlike Kipling's Just So Stories about how and why the leopard got its spots, the rhinoceros got its tough skin, or the camel got its hump, the cell-cell signaling model of physiological evolution is not a “just so story.” It is based on mechanisms of cell-molecular embryogenesis, linked to phylogenesis through homeobox genes, for example. It is predictive for chronic disease, as we have shown for the lung (35). And as proof of principle, we have been able to effectively prevent the chronic lung disease of the newborn, bronchopulmonary dysplasia, experimentally, based on principles of lung cell-molecular evolution (35). Furthermore, this model of physiological evolution transcends lung biology, for example, by providing a mechanistic, evolutionary link between the lung and kidney in Goodpasture's syndrome (16). As further proof of principle, persistent failure along this signaling pathway results in the formation of myofibroblasts (2, 3, 32). These interrelationships may ultimately provide an explanation for Potter's syndrome, in which decreased renal function idiopathically leads to oligohydramnios and consequent pulmonary hypoplasia due to lack of fluid distension. So, like the solution to the Dead Sea scrolls, by using common “threads” in the evolutionary fabric of biology, we can solve such complex problems.
Three thousand years of descriptive biology and medicine has brought us to the threshold of predictive molecular medicine. Now, aided by our knowledge of the human genome, we must address the evolutionary origins of human physiology based on phylogenetic and developmental mechanisms. The approach we have proposed may fail to directly identify such first principles because we are missing intermediates from the “molecular fossil record” that failed to optimize survival. But some vestiges of those “failures” were likely incorporated into other existing functional phenotypes or into other molecularly related functional homologies, like those of the lung and kidney, photoreceptors, and circadian rhythms, the lens, and liver enzymes. What this approach does provide is a robust means of formulating refutable hypotheses to determine the ultimate origins and first principles of physiology, forming the basis for predictive medicine (38), rather than merely showing associations between genes and pathology, which is unequivocally a just so story. In this new age of genomics, our reach must exceed our grasp. With this review, we hope to engage you in this new approach to understanding physiology, our “Dead Sea scroll,” by tracing the regulatory pathways affecting our basic operating unit, the cell.
GRANTS
The authors are supported by National Institutes of Health Grants HL-055268 (J. S. Torday) and HL-075405 and HD-051857 (V. K. Rehan).
Acknowledgments
We thank Dr. Karen Chambers for editing this work.
Address for reprint requests and other correspondence: J. S. Torday, Dept. of Pediatrics, The Henry L. Guenther Laboratory for Cell/Molecular Research, Laboratory for Evolutionary Preventive Medicine, Harbor-UCLA Medical Center, Torrance, CA 90502 (e-mail: jtorday@labiomed.org).
REFERENCES
- 1.Barker N The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol 468: 5–15, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Bosch RJ, Rodríguez-Puyol D, Bover J, Rodríguez-Puyol M. Parathyroid hormone-related protein: roles in the glomerulus. Exp Nephrol 7: 212–216, 1999. [DOI] [PubMed] [Google Scholar]
- 3.Bosch RJ, Rojo-Linares P, Torrecillas-Casamayor G, Iglesias-Cruz MC, Rodríguez-Puyol D, Rodríguez-Puyol M. Effects of parathyroid hormone-related protein on human mesangial cells in culture. Am J Physiol Endocrinol Metab 277: E990–E995, 1999. [DOI] [PubMed] [Google Scholar]
- 4.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci 121: 737–746, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Cowley AW Jr. Genomics and homeostasis. Am J Physiol Regul Integr Comp Physiol 284: R611–R627, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Crespi EJ, Denver RJ. Leptin (ob gene) of the South African clawed frog Xenopus laevis. Proc Natl Acad Sci USA 103: 10092–10097, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csete M, Walikonis J, Slawny N, Wei Y, Korsnes S, Doyle JC, Wold B. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. J Cell Physiol 189: 189–196, 2001. [DOI] [PubMed] [Google Scholar]
- 8.Darwin C The Origin of Species. London: John Murray, 1859.
- 9.De Robertis EM Evo-devo: variations on ancestral themes. Cell 132: 185–195, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobzhansky T Nothing in biology makes sense except in the light of evolution. Am Biol Teacher 35: 125–129, 1973. [Google Scholar]
- 11.Gilbert SF, Opitz JM, Raff RA. Resynthesizing evolutionary and developmental biology. Dev Biol 73: 357–372, 1996. [DOI] [PubMed] [Google Scholar]
- 12.Hood L, Rowen L, Galas DJ, Aitchison JD. Systems biology at the Institute for Systems Biology. Brief Funct Genom Proteom 7: 239–248, 2008. [DOI] [PubMed] [Google Scholar]
- 13.Horowitz A, Simons M. Branching morphogenesis. Circ Res 103: 784–795, 2008. [DOI] [PubMed] [Google Scholar]
- 14.Kimura M The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge University Press, 1983.
- 15.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301: 361–363, 2003. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald BA, Sund M, Grant MA, Pfaff KL, Holthaus K, Zon LI, Kalluri R. Zebrafish to humans: evolution of the alpha3-chain of type IV collagen and emergence of the autoimmune epitopes associated with Goodpasture syndrome. Blood 107: 1908–1915, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macklem PT Emergent phenomena and the secrets of life. J Appl Physiol 104: 1844–1846, 2008. [DOI] [PubMed] [Google Scholar]
- 18.McGinnis W, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308: 428–433, 1984. [DOI] [PubMed] [Google Scholar]
- 19.Mezentseva NV, Kumaratilake JS, Newman SA. The brown adipocyte differentiation pathway in birds: an evolutionary road not taken. BMC Biol 6: 17, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morowitz HJ A theory of biochemical organization, metabolic pathways, and evolution. Complexity 4: 39–53, 1999. [Google Scholar]
- 21.Polanyi M Life's irreducible structure. Live mechanisms and information in DNA are boundary conditions with a sequence of boundaries above them. Science 160: 1308–1312, 1968. [DOI] [PubMed] [Google Scholar]
- 22.Rehan VK, Torday JS. Lower parathyroid hormone-related protein content of tracheal aspirates in very low birth weight infants who develop bronchopulmonary dysplasia. Pediatr Res 60: 216–220, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Romer AS Major steps in vertebrate evolution. Science 158: 1629–1637, 1967. [DOI] [PubMed] [Google Scholar]
- 24.Rosenblum ND Developmental biology of the human kidney. Semin Fetal Neonatal Med 13: 125–132, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Scott MP, Weiner AJ. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA 81: 4115–4119, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The American Academy of Arts and Sciences. Advancing Research in Science and Engineering. Cambridge, MA: American Academy of Arts and Sciences, 2008.
- 27.Torday JS, Torday DP, Gutnick J, Qin J, Rehan V. Biologic role of fetal lung fibroblast triglycerides as antioxidants. Pediatr Res 49: 843–849, 2001. [DOI] [PubMed] [Google Scholar]
- 28.Torday JS, Sun H, Wang L, Torres E, Sunday ME, Rubin LP. Leptin mediates the parathyroid hormone-related protein paracrine stimulation of fetal lung maturation. Am J Physiol Lung Cell Mol Physiol 282: L405–L410, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torday JS, Rehan VK. Stretch-stimulated surfactant synthesis is coordinated by the paracrine actions of PTHrP and leptin. Am J Physiol Lung Cell Mol Physiol 283: L130–L135, 2002. [DOI] [PubMed] [Google Scholar]
- 30.Torday JS Parathyroid hormone-related protein is a gravisensor in lung and bone cell biology. Adv Space Res 32: 1569–1576, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Torday JS, Rehan VK. Mechanotransduction determines the structure and function of lung and bone: a theoretical model for the pathophysiology of chronic disease. Cell Biochem Biophys 37: 235–246, 2003. [DOI] [PubMed] [Google Scholar]
- 32.Torday JS, Torres E, Rehan VK. The role of fibroblast transdifferentiation in lung epithelial cell proliferation, differentiation, and repair in vitro. Pediatr Pathol Mol Med 22: 189–207, 2003. [DOI] [PubMed] [Google Scholar]
- 33.Torday JS A periodic table for biology. Scientist 18: 32–33, 2004. [Google Scholar]
- 34.Torday JS, Rehan VK. Deconvoluting lung evolution using functional/comparative genomics. Am J Respir Cell Mol Biol 31: 8–12, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Torday JS, Rehan VK. The evolutionary continuum from lung development to homeostasis and repair. Am J Physiol Lung Cell Mol Physiol 292: L608–L611, 2007. [DOI] [PubMed] [Google Scholar]
- 36.Torday JS, Rehan VK. Developmental cell/molecular biologic approach to the etiology and treatment of bronchopulmonary dysplasia. Pediatr Res 62: 2–7, 2007. [DOI] [PubMed] [Google Scholar]
- 37.Torday JS, Ihida-Stansbury K, Rehan VK. Leptin stimulates Xenopus lung development: evolution in a dish. Evol Dev 11: 218–223, 2009. [DOI] [PubMed] [Google Scholar]
- 38.Torday JS, Rehan VK. Exploiting cellular-developmental evolution as the scientific basis for preventive medicine. Med Hypotheses 72: 596–602, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn 229: 162–175, 2004. [DOI] [PubMed] [Google Scholar]
- 40.Warburton D, Bellusci S, De Langhe S, Del Moral PM, Fleury V, Mailleux A, Tefft D, Unbekandt M, Wang K, Shi W. Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr Res 57: 26R–37R, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Weibel ER, Taylor CR, Bolis L. Principles of Animal Design. Cambridge UK: Cambridge University Press, 1998.
- 42.West GB, Brown JH, Enquist BJ. A general model for the origin of allometric scaling laws in biology. Science 276: 122–126, 1997. [DOI] [PubMed] [Google Scholar]