Abstract
Background: This study examines the impact of dietary fatty acids on regulation of gene expression in mammary epithelial cells before and during puberty. Methods: Diets primarily consisted of n-9 monounsaturated fatty acids (olive oil), n-6 polyunsaturated fatty acids (safflower), saturated acids (butter), and the reference AIN-93G diet (soy oil). The dietary regimen mimics the repetitive nature of fatty acid exposure in Western diets. Diet-induced changes in gene expression were examined in laser capture microdissected mammary ductal epithelial cells at day of weaning and end of puberty. PCNA immunohistochemistry analysis compared proliferation rates between diets. Results: Genes differentially expressed between each test diets and the reference diet were significantly enriched by cell cycle genes. Some of these genes were involved in activation of the cell cycle pathway or the G2/M check point pathway. Although there were some differences in the level of differential expression, all diets showed qualitatively the same pattern of differential expression compared to the reference diet. Cluster analysis identified an expanded set of cell cycle as well as immunity and sterol metabolism related clusters of differentially expressed genes. Conclusion: Fatty acid-enriched diets significantly upregulated proliferation above normal physiological levels during puberty. Higher cellular proliferation during puberty caused by enriched fatty acid diets poses a potential increase risk of mammary cancer in later life. The human homologs of 27 of 62 cell cycle rat genes are included in a human breast cancer cluster of 45 cell cycle genes, further emphasizing the importance of our findings in the rat model.
Keywords: microarray analysis, dietary fat, cell cycle genes
many articles have been written regarding the complexity of breast tissue with regard to both development and mammary cancer. Critical phases of mammary gland development have been identified: 1) the prenatal period when epithelial buds begin to form primitive ducts; 2) puberty, characterized by rapid growth and branching; and 3) pregnancy, characterized by changing the functionality of the gland (19, 44). Previous research from Russo et al. (45) using the rat model has established puberty as a critical window of susceptibility to induction of tumors by environmental agents. This pubertal stage in the rodent model consists of a high prevalence of terminal end buds (TEB) in the mammary gland (45). TEB are characterized by high proliferative indexes, a shortened S-phase, and are increased in prevalence in the 50- to 56-day-old juvenile animal. Juvenile animals receiving a carcinogenic insult with dimethylbenz[a]anthracene (DMBA) at 50 postnatal days (PND50) have enhanced tumor multiplicity and incidence during this time period when TEB are undergoing development to alveolar buds (AB) (46). Sinha et al. (54) established a direct relationship between mammary tumorigenesis induced by DMBA and proliferative (labeling) index at PND50.
Mammary development and proliferation are known to be orchestrated by numerous reproductive hormones as well as transcriptional elements (13, 50). The Sprague-Dawley rodent model has demonstrated remarkably similar changes to the human cyclic ovarian hormone changes seen during the menses, including proliferation indexes, morphologic phenotype, and mRNA expression (16, 51). Transcriptional and hormonal control is necessary to coordinate the development and proliferation needed for normal maturation of the mammary gland. Epidemiologic observations have linked earlier onset of puberty and changes in the morphology of the mammary gland to subsequent enhanced susceptibility to carcinogenesis (9). Hormonal manipulation of the developing gland has been shown to confer some protection against developing mammary tumors in the rodent model (16, 39, 51). The mechanism of these controls during mammary development has not been fully explained. Determining the effects of fatty acids on transcriptional control during development is a subject of this study.
As early as 1975, a relationship between dietary fat intake and breast cancer age-adjusted death rate had been established for human populations (9). Since that time, results on both human and animal studies remain conflicted for a number of reasons (1, 6, 8, 21, 25, 27). Willett (59) reviewed human studies that estimated the diet to be an avoidable causal factor in 10–70% of new breast cancers. Both in vivo and in vitro studies have shown that specific fatty acids have effects on gene expression (11, 12, 42, 56). For example, certain polyunsaturated fatty acids (PUFAs) are often associated with activation of the transcription factor (TF) peroxisome proliferator-activated receptor (alpha, delta, and gamma) (34) and hepatocyte nuclear factor-4 alpha and sterol regulatory element binding protein-1c (33). More recently, effects of soy protein diets on expression of genes regulating metabolism, ion transport, and immune response in laser capture microdissected (LCM) mammary epithelial cells of Sprague-Dawley rats have been reported (56). The expression of proliferation markers has been negatively linked to dietary caloric restriction leading to inhibition of cell cycle progression (26, 32).
Controlled animal studies have sought to link fat consumption with breast cancer incidence with results which remain difficult to interpret. A majority of studies show a link of n-6 PUFA to enhanced susceptibility to carcinogens (18, 24). Ip et al. (28) showed an optimum tumorigenic response with an n-6 diet supplemented with 4% essential fatty acid (28) in the DMBA rat model. Few data exist to support a role of monounsaturated fatty acids in tumorigenesis (23). Some hypothesize that olive oil, while protective against breast cancer, has a mechanism independent of its high monounsaturated fatty acid content (53). In a meta-analysis of rodent studies no conclusive results were obtained regarding inhibitory action of monounsaturated fatty acids in carcinogenesis studies (18). Lasekan showed that olive oil supplemented with a small amount of essential fatty acid when given after DMBA treatment at day 50 resulted in increased tumor promotion (37). The saturated fats found in butter fat have not conclusively been linked to breast cancer in animals (18, 44). There are few data examining the role of these agents in mammary gland differentiation or puberty.
In the present study we altered the consumption and level of dietary fatty acids beginning in the maternal diet of Sprague-Dawley rats and continuing throughout life of the female pups, mimicking the repetitive nature of Western diet exposure. We specifically looked at gene expression profiles from laser-captured mammary gland epithelial cells at day of weaning (PND21) and at the end of puberty (PND50) and identified critical biochemical pathways which may be altered by nutritional modification.
MATERIALS AND METHODS
Dietary exposures and animal breeding.
Female, virgin Sprague-Dawley rats were obtained from Taconic Farms (Germantown, NY) at ∼7 wk of age and placed on one of seven pelleted purified diets [Supplemental (Spl.) Table S11 Research Diets, New Brunswick, NJ)] for 1 mo. Animals were housed individually in a temperature- and humidity-controlled room in the AALAC-approved University of Cincinnati facility under a 12-h light-dark cycle. All rats were provided food and tap water ad libitum, and animals were housed on Sani-chips bedding (P. J. Murphy Forest Products, Montville, NJ). All animal procedures were approved by the University of Cincinnati Institutional Animal Care and Use Committee, and the experiments were performed following the guidelines of the National Institutes of Health (NIH) for the proper and humane use of animals in biomedical research. After 1 mo of diet exposure, female rats were bred with male Sprague-Dawley rats of ∼3 mo of age in stainless steel hanging cages. Mating plugs were noted and recorded as gestational day 1, and females were returned to single housing during the remainder of the gestational period. Males were used to sire only two litters from different diets; dams were used for only one litter. Litters were culled to eight pups within 2nd day after birth (PND2), keeping all female pups and sufficient males to balance. Litters were weighed and monitored throughout the lactational period and for the duration of the study. Pups had access to dam's milk as well as tap water and food throughout lactation. Pups were weaned at PND21 and individually housed on Sani-chips bedding with tap water and appropriate diets ad libitum. Offspring rats were killed by CO2 asphyxiation at approximately PND21 and PND50 with no other treatments.
Diets.
Female rat pups from in-house breeding consumed high-fat (39% of kcal) or low-fat (10% of kcal) versions of one of these diets or the reference diet (16% of kcal from fat). Ten and thirty-nine percent fat by kcal reflect a minimum and high fat content, respectively, consumed in the Western diet. The mothers of the pups consumed the same diet before and throughout gestation and lactation. We evaluated pups at key time points in their lifetime, weaning (PND21) and the end of puberty (PND50), which correspond to important milestones in human mammary development and maturation. Studies detailed here evaluate the effects of AIN-93G-based formulated diets containing primarily one specific dietary fatty acid (olive or safflower oil or butter) on the mammary development in young and pubescent offspring consuming the same diets as their mothers. Diet formulations (Research Diets) appear in Spl. Table S1. Diets reflect a mixture of fatty acids as found in the typical diet. The fatty acids in these diets primarily consisted of n-9 monounsaturated fatty acids (oleic acid-olive oil-HFO, LFO), n-6 polyunsaturated fatty acids (linoleic acid-safflower-HFS, LFS), saturated acids (palmitic-acid-butter-HFB, LFB), or a reference diet (soy oil-AIN-93G or Reference). The diets were controlled for caloric content, vitamins, salts, and protein but varied in fat and carbohydrate content and therefore density. AIN-93G with a soy oil base, a widely used diet for mammary studies, was used here as the reference diet. A basal amount of soy oil was added to each of the diets to provide essential fatty acids lacking in some of the diets.
Tissue preparation and LCM.
The mammary glands were dissected, and the fresh tissue specimens were embedded using Tissue-Tek OCT compound (Sakura Fineteck USA, Torrance, CA), snap-frozen, and stored at −80°C. The frozen mammary tissue was sectioned to 10 μm thickness with a cryostat (Leica Bannockburn, IL) and stored at −80°C until use. The tissue sections were stained with hematoxylin and eosin, and immediately laser capture microdissection was performed using the PixCell II, Arcturus LCM system (Arcturus, Sunnyvale, CA). Mammary ductal epithelial cells were captured at a total of 10–12,000 hits for each sample. RNA was isolated using TRI Reagent according to manufacturer's protocol (#TR118; Molecular Research Center, Cincinnati, OH).
Microarray analysis using Affymetrix GeneChip technology.
The quality of total RNA was first analyzed by Agilent Bioanalyzer 2100. RNA samples with RIN (RNA integrity number) >7.0 are acceptable for microarray analysis (a RIN of >5.0 is acceptable for LCM samples). Total RNA samples of 1 μg or greater were amplified one round, and the amplified RNA (aRNA) was biotinylated using Ambion Biotin-Enhanced Message Amp II kit (#1791; Ambion, Austin, TX), following the manufacturer's protocols. Samples of 10–100 ng including those extracted from LCM samples were amplified two rounds using Ambion's Message Amp II Kit (#1751, Ambion) followed by the Biotin-Enhanced Message Amp II Kit following manufacturer's protocols. Rat Genome 230 2.0 Array GeneChips were hybridized with 15 μg of fragmented aRNA. The hybridization, staining, and washing were carried out using the Affymetrix GeneChip Hybridization Wash and Stain Kit (P/N 900720) following the manufacturer's protocols. The arrays were hybridized for 16 h at 45°C using an Affymetrix Hybridization Oven 640 (P/N 800139). FS450_0001 protocol was used for staining and washing the GeneChips using the Affymetrix Fluidics Station 450 (P/N 00-0079). The GeneChips were scanned with Affymetrix GeneChip Scanner 3000 7G using GCOS software and Affymetrix preset settings.
Data normalization and statistical analysis.
The data were analyzed to identify genes differentially expressed between fatty acid diets and the reference AIN-93G diet, and between the high- and low-fat fatty acid diets. All statistical analyses were performed separately for the two rat age groups (PND21 and PND50). The data were normalized Multichip Average (RMA) protocol (30) and Entrez Gene-specific probe set definitions (14). The statistical analysis was performed using the limma (55) package from the Bioconductor project (22). Initially, the batch effects were factored out by fitting a two-way ANOVA model with the batch designation and the diet type as the explanatory variables and by subtracting the batch effect estimate from each gene expression measurement. Differentially expressed genes were identified by performing separate moderated t-test analysis for each diet compared with the reference AIN-93G diet and for each high-fat vs. corresponding low-fat fatty acid diet using the intensity-based empirical Bayes method (IBMT) (48). We also performed a two-way ANOVA analysis with the high vs. low fat and type of fatty acid as the two factors in the model to assess the overall effect of the high-fat diet adjusted for the effect of the different type of the diet. IBMT-based P values were adjusted for multiple comparisons using the false discovery approach [false discovery rate (FDR)] (7). Genes with statistically significant differences at FDR <0.1 and the magnitude of change at least 1 on the log2 scale (twofold change) were considered differentially expressed. MIAMI-annotated primary and processed microarray data can be downloaded from the support web page http://eh3.uc.edu/supplements/diets and can be interactively queried and analyzed through Genomics Portals (http://GenomicsPortals.org). Data have also been deposited in the Gene Expression Omnibus database under the accession identifier GSE14933.
Functional analysis of differentially expressed genes.
The functional analysis of the differentially expressed genes was performed by assessing the statistical significance of enrichment of different Gene Ontologies (GO) (3) and KEGG pathways (36) using the DAVID functional analysis tool (15), Bioconductor procedures (22), the CLEAN package (20), and the Ingenuity Pathway Analysis (http://www.ingenuity.com).
Cluster analysis.
The cluster analysis was performed to identify genes with a coherent expression pattern across all diets and both age groups (PND21 and PND50). A gene was selected to be clustered if it was differentially expressed between any fatty acid diet and the reference AIN-93G diet with P value <0.01 and the magnitude of at least 1.5-fold. A total of 1,109 genes were selected for clustering based on these criteria. The clustering of log2-average ratios between each diet and the corresponding AIN-93G reference was performed using the context-specific infinite mixture model (38) with expression profiles from different age groups being placed in two different contexts. The functional analysis of resulting hierarchical clustering of gene expression profiles was performed using the CLustering Enrichment ANalysis (CLEAN) framework (20). The significance of functional enrichment obtained in the CLEAN analysis was assessed by comparing it to levels of enrichment obtained by performing the same type of analysis on a set of randomly selected 1,109 genes from our dataset. Results of the cluster analysis can be interactively browsed at the support web page (http://eh3.uc.edu/supplements/diets).
Identifying enriched regulatory motifs.
Computational search for TF binding motifs enriched in promoters of coclustered genes was performed on the human homologs of the rat genes identified in the analysis. Human homologs were established using the National Center for Biotechnology Information's homologene database (49). Only promoter regions with high-level conservation between human, mouse, and rat genomes were considered in the analysis (UCSC Genome Browser conservation score of at least 3,000). For each of the 304 human TFs with at least one position weight matrix (PWM) in the Transfac version 12.1 database, we scored genes as to how likely they were to have such a motif within 500 bases of their transcription start site (TSS). Suppose that θjl is the lth PWM defining the DNA binding motif of length L(θjl) associated with the jth TF and Sijx is any DNA fragment of length L(θjl) in the 1 kbp DNA region around the TSS for the ith gene. The score measuring the likelihood of Sijx being the binding site for the jth TF is calculated as
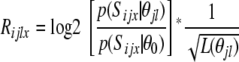 |
(1) |
where p(Sijx θjl) is the probability of Sijx being generated by the product multinomial model with the PWM θjl and p(Sijx θ0) is the probability of Sijx being generated by the background 3rd order Markov chain with the transition matrix θ0 estimated using 1 kb fragments around TSS for all genes in the genome. The gene-specific scores for the ith gene and the jth TF are then calculated as:
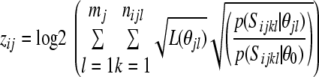 |
(2) |
where the Sijkl is the kth fragment from the promoter of ith gene, which for which Rijlk >1.5, and mj is the number of different PWMs associated with the jth TF. For each TF we selected top 750 targets with the highest score. When fewer than 750 gene promoters had score >0, we selected all such gene promoters. After all rat genes on the microarray were mapped to their respective human genes, most TFs were represented by ∼350 genes. The statistical significance of the overlap between the genes in the cluster and genes with the highest chance of containing a TF binding motif in their promoters was established using the Fisher's exact test.
Analysis of human breast cancer data.
The analysis of human breast cancer data for up- and downregulated genes was performed using the Genomics Portals web tools (http://GenomicsPortals.org) to query the GSE3494Entrez dataset. This dataset was created by downloading the original .CEL files for the GSE3494 dataset (40) from Gene Expression Omnibus (GEO) database (5) and processing them using Entrez Gene-centric definitions (14). The conversions of rat gene identifiers to human gene identifiers were done automatically based on the homologene mappings (49).
Real-time q-PCR.
The cDNA was made from 1 μg of two times amplified RNA from the same samples as used in the microarray analysis, using the High-Capacity cDNA reverse Transcription Kit from Applied Biosystems. TaqMan Gene Expression Assays were purchased from Applied Biosystems [glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Rn99999916_s1, cyclin-dependent kinase inhibitor 3 predicted (Cdkn3_predicted) Rn01414656_m1, polo-like kinase 1 (Plk1) Rn01510931_m1, aurora kinase B (Aurkb) Rn01460656_ m1, cyclin B1 (Ccnab1) Rn00596848_m1, pituitary tumor-transforming 1 (Pttg1) Rn00574373_m1, M-phase phosphoprotein 1 predicted (Mphosph1_predicted) Rn01454431_m1, Ect2 oncogene predicted (Ect2_predicted) Rn01457088_m1, cyclin A2 (Ccna2) Rn01493715_m1, and E2F transcription factor 8 (E2f8) Rn01476919_m1]. The RT-q-PCR was set up according to the manufacturer's protocol and run on the StepOne Plus Real-Time PCR instrument (Applied Biosystems) using the TaqMan Fast Universal PCR Master Mix. Relative differences in q-RT-PCR among samples were determined by the ΔΔCT method. For each sample ΔCT was obtained by averaging CTs from three replicated runs and subtracting the corresponding average GAPDH CT. Statistical significance of differences between ΔCT for AIN-93G samples and fatty acid samples (ΔΔCT) was established using the nonparametric Wilcoxon rank test.
Immunohistochemistry.
Alcoholic-formalin fixed, paraffin-embedded specimens were sectioned on a microtome at 5 μm, and unheated slides were stained by immunohistochemistry for Proliferating Cell Nuclear Antigen (PCNA-Leica Microsystems, Bannockburn, IL); clone PC10 at 1:600 with CC1 mild conditioning. All immunohistochemistry was performed on the Ventana Benchmark (Oro Valley, AZ) automated immunostainer using the Ventana I-View open secondary DAB detection kit. Biotinylated, rat adsorbed horse anti-mouse (Vector Laboratories, Burlingame, CA) was used as the secondary antibody at 1:150. Isotype-specific mouse IgG added in place of the primary antibody was used as a negative control.
RESULTS
We identified differences in gene expression profiles of mammary epithelium induced by the most popular dietary fats found in Western diets (olive oil, safflower oil, and butter) compared with the reference AIN-93G diet (soy oil) (Spl. Fig. S1). Fatty acid composition of test and reference diets is shown in Spl. Table S1. Our most striking finding was the effect of the diets on increased proliferation as evidenced by upregulation of proliferative markers at mRNA and protein level.
Differential gene expression.
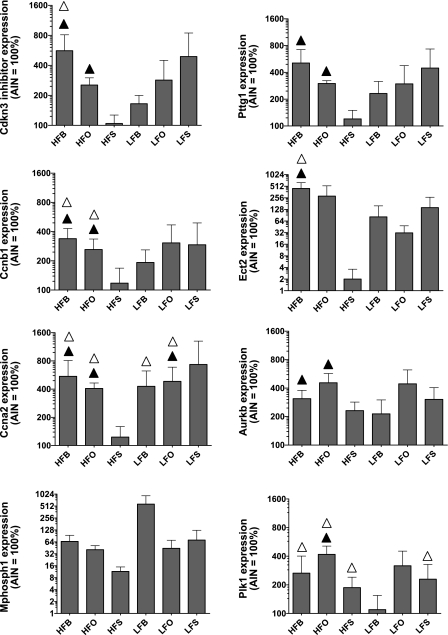
Gene expression levels were examined in LCM-captured epithelial cells using the Affymetrix microarrays. For each age group separately, we identified genes with disregulated expression in at least one of the six fatty acid diets compared with the reference AIN-93G diet. Out of the total of 11,498 genes examined, 44 were differentially expressed (FDR <0.1 and differential expression of at least twofold) between at least one of the test diets and the reference diet at PND21 and 90 gene were differentially expressed at PND50. The combined list contained 113 unique genes differentially expressed either at 21 days, 50 days, or both. Gene expression patterns for all differentially expressed genes are depicted in Spl. Fig. S1 and listed in Spl. Table S2. Differential expression of selected upregulated genes (Ccna2, Ccnb1, Cdkn3, Aurkb, Ect2, Plk1, Pttg1, and Mphosph1) were also examined by the quantitative PCR analysis. Differential expression patterns of all of these selected genes except Mphosph1 were confirmed by q-PCR analysis (Fig. 1).
Fig. 1.
Quantitative PCR validation of differential expression for key gene at day 50. The data are represented as the percent increase over the control diet. Triangles represent statistically significant differential expression at P value <0.1 using the nonparametric Wilcoxon test (▴) and using the simple t-test (▵). Diets: HFB, high fat butter; HFO, high fat olive oil; HFS, high fat safflower oil; LFB, low fat butter; LFO, low fat olive oil; LFS, low fat safflower oil.
We also examined differences in gene expression levels between low- and high-fat diets of the same type. Using the same significance criteria (FDR <0.1 and twofold differential expression), we found none of the 11,498 genes showed differential expression in either simple pair-wise comparisons or in the two-way ANOVA (see materials and methods).
Functional analysis.
Separate functional analyses of the two groups (PND21 and PND50) of differentially expressed genes identified the same GO category, “mitotic cell cycle,” as being the most enriched (FDR <5 × 50−5 at day 21, FDR <4 × 10−17 for day 50, and 8 × 10−18 in the combined list). In the first group, 12 out of 44 genes were cell cycle related, and in the second group 28 out of 90 genes were cell cycle related (Table 1). Although the genes were not selected based on the directionality of differential expression, virtually all cell cycle-related genes showed the concordant differential expression relative to AIN-93G with statistical significance reached in at least one diet at both time points.
Table 1.
Differentially expressed cell cycle genes at PND50 and PND21
| Gene ID | Symbol |
Day 21 |
Day 50
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HFB | HFO | HFS | LFB | LFO | LFS | HFB | HFO | HFS | LFB | LFO | LFS | |||||||||||||||
| Cell Cycle Genes Differentially Expressed at PND50 | ||||||||||||||||||||||||||
| 114494 | Ccna2 | 0.6 | 1.3 | 0.8 | 0.6 | 0.5 | 0.3 | 1.5 | 1.6 | 0.9 | 1.8 | 1.3 | 1.4 | |||||||||||||
| 114592 | Aurkb | 0.8 | 1.5 | 0.9 | 0.6 | 0.4 | 0.4 | 1.6 | 1.8 | 1.1 | 1.8 | 1.3 | 1.7 | |||||||||||||
| 171304 | Kif11 | 0.6 | 0.8 | 0.7 | 0.3 | 0.4 | 0.2 | 1.1 | 1.4 | 0.7 | 1.3 | 1.0 | 1.2 | |||||||||||||
| 171576 | Bub1b | 1.2 | 1.9 | 1.8 | 0.9 | 1.3 | 1.1 | 2.3 | 2.7 | 1.2 | 2.8 | 1.8 | 2.1 | |||||||||||||
| 25203 | Ccnb1 | 0.9 | 1.1 | 0.9 | 0.6 | 0.7 | 0.4 | 1.5 | 1.7 | 0.8 | 1.5 | 1.2 | 1.1 | |||||||||||||
| 25515 | Plk1 | 0.8 | 1.1 | 1.0 | 0.7 | 0.6 | 0.4 | 1.3 | 1.7 | 1.0 | 1.3 | 1.4 | 1.5 | |||||||||||||
| 261730 | Stk6 | 0.8 | 1.0 | 0.6 | 0.7 | 0.5 | 0.7 | 1.4 | 1.8 | 1.0 | 1.6 | 1.6 | 1.2 | |||||||||||||
| 289993 | Cdkn3_predicted | 1.3 | 1.6 | 1.6 | 1.1 | 1.1 | 0.8 | 2.2 | 2.6 | 1.8 | 2.5 | 2.0 | 2.0 | |||||||||||||
| 289997 | Dlg7_predicted | 0.8 | 1.3 | 0.5 | 0.5 | 0.4 | 0.3 | 1.0 | 1.1 | 0.6 | 1.2 | 0.9 | 1.1 | |||||||||||||
| 290326 | Pbk_predicted | 0.6 | 1.7 | 1.0 | 1.1 | 0.7 | 0.5 | 1.5 | 1.6 | 0.7 | 1.6 | 1.2 | 1.3 | |||||||||||||
| 292263 | Fbxo5_predicted | 0.7 | 1.2 | 0.8 | 0.6 | 0.4 | −0.2 | 1.3 | 1.8 | 1.2 | 1.2 | 1.4 | 1.3 | |||||||||||||
| 293502 | Kif22 | 1.0 | 1.2 | 0.9 | 0.9 | 0.9 | 0.5 | 1.9 | 2.1 | 1.1 | 1.7 | 1.2 | 1.5 | |||||||||||||
| 294286 | Kifc1 | 0.7 | 0.7 | 0.7 | 0.3 | 0.4 | 0.3 | 1.3 | 1.5 | 0.9 | 1.5 | 1.2 | 1.4 | |||||||||||||
| 295661 | Spbc25 | 1.4 | 2.1 | 1.3 | 1.1 | 1.0 | 1.0 | 1.6 | 1.8 | 0.9 | 1.6 | 1.0 | 1.4 | |||||||||||||
| 296137 | Bub1_predicted | 0.6 | 1.2 | 0.8 | 0.6 | 0.8 | 0.3 | 1.3 | 1.5 | 0.4 | 1.5 | 0.8 | 0.9 | |||||||||||||
| 296368 | Ube2c_predicted | 0.8 | 1.2 | 1.1 | 0.8 | 0.7 | 0.4 | 1.9 | 2.0 | 1.0 | 1.7 | 1.7 | 1.6 | |||||||||||||
| 297176 | Mad2l1_predicted | −0.1 | 0.3 | 0.2 | −0.1 | 0.1 | 0.0 | 0.9 | 1.2 | 0.3 | 0.8 | 0.9 | 0.9 | |||||||||||||
| 29728 | Mcm4 | 0.8 | 1.2 | 0.9 | 0.5 | 0.7 | 0.3 | 1.4 | 1.5 | 1.0 | 1.1 | 1.2 | 1.4 | |||||||||||||
| 304951 | Cdca1 | 1.0 | 1.4 | 0.9 | 0.8 | 0.6 | 0.5 | 1.5 | 1.6 | 0.7 | 1.8 | 1.3 | 1.2 | |||||||||||||
| 305922 | Lats2_predicted | −0.4 | −0.2 | −0.5 | −0.3 | −0.3 | −0.2 | −0.4 | −1.0 | −0.6 | −0.5 | −0.9 | −0.6 | |||||||||||||
| 308607 | E2f8 | 1.1 | 2.4 | 1.6 | 1.3 | 1.1 | 0.9 | 2.2 | 2.3 | 1.0 | 2.0 | 2.0 | 2.1 | |||||||||||||
| 309523 | Mphosph1_predicted | 1.0 | 2.1 | 1.1 | 0.9 | 1.2 | 0.8 | 2.1 | 2.3 | 1.1 | 2.2 | 1.8 | 1.8 | |||||||||||||
| 315298 | Racgap1_predicted | 0.8 | 0.6 | 0.9 | 0.5 | 0.8 | 0.2 | 1.1 | 1.3 | 0.6 | 1.4 | 1.0 | 0.6 | |||||||||||||
| 315330 | Espl1_predicted | 0.8 | 1.8 | 1.3 | 0.9 | 0.8 | 0.6 | 2.6 | 2.6 | 1.4 | 2.5 | 1.9 | 2.2 | |||||||||||||
| 315740 | Kif23_predicted | 0.5 | 0.7 | 0.4 | 0.4 | 0.4 | 0.1 | 0.9 | 1.1 | 0.4 | 1.1 | 0.8 | 0.6 | |||||||||||||
| 360962 | Tacc3 | 0.6 | 0.9 | 0.8 | 0.4 | 0.4 | 0.2 | 1.5 | 1.6 | 0.9 | 1.6 | 1.2 | 1.3 | |||||||||||||
| 362044 | Cenpe_predicted | 0.3 | 0.3 | 0.5 | 0.0 | 0.2 | −0.2 | 1.4 | 1.3 | 0.7 | 1.5 | 0.6 | 1.3 | |||||||||||||
| 362438 | RGD1562596_predicted | 0.6 | 1.0 | 0.7 | 0.4 | 0.6 | 0.3 | 1.3 | 1.5 | 0.7 | 1.5 | 1.2 | 1.2 | |||||||||||||
| 500545 | Cdca8 | 0.8 | 1.3 | 1.0 | 0.8 | 0.8 | 0.5 | 1.6 | 1.8 | 1.1 | 1.4 | 1.5 | 1.6 | |||||||||||||
| 54237 | Cdc2a | 1.0 | 1.5 | 1.1 | 1.0 | 0.8 | 0.5 | 1.7 | 2.0 | 1.1 | 1.9 | 1.4 | 1.7 | |||||||||||||
| 64193 | Pttg1 | 0.8 | 0.8 | 1.3 | 0.7 | 0.6 | 0.4 | 1.9 | 2.3 | 1.4 | 1.7 | 1.4 | 1.9 | |||||||||||||
| 680089 | LOC680089 | 1.3 | 2.0 | 1.3 | 1.1 | 1.1 | 0.8 | 1.7 | 1.7 | 0.9 | 1.5 | 1.1 | 1.3 | |||||||||||||
| Cell Cycle Genes Differentially Expressed at PND21 | ||||||||||||||||||||||||||
| 114494 | Ccna2 | 0.6 | 1.3 | 0.8 | 0.6 | 0.5 | 0.3 | 1.5 | 1.6 | 0.9 | 1.8 | 1.3 | 1.4 | |||||||||||||
| 114592 | Aurkb | 0.8 | 1.5 | 0.9 | 0.6 | 0.4 | 0.4 | 1.6 | 1.8 | 1.1 | 1.8 | 1.3 | 1.7 | |||||||||||||
| 289997 | Dlg7_predicted | 0.8 | 1.3 | 0.5 | 0.5 | 0.4 | 0.3 | 1.0 | 1.1 | 0.6 | 1.2 | 0.9 | 1.1 | |||||||||||||
| 293502 | Kif22 | 1.0 | 1.2 | 0.9 | 0.9 | 0.9 | 0.5 | 1.9 | 2.1 | 1.1 | 1.7 | 1.2 | 1.5 | |||||||||||||
| 295661 | Spbc25 | 1.4 | 2.1 | 1.3 | 1.1 | 1.0 | 1.0 | 1.6 | 1.8 | 0.9 | 1.6 | 1.0 | 1.4 | |||||||||||||
| 304951 | Cdca1 | 1.0 | 1.4 | 0.9 | 0.8 | 0.6 | 0.5 | 1.5 | 1.6 | 0.7 | 1.8 | 1.3 | 1.2 | |||||||||||||
| 308607 | E2f8 | 1.1 | 2.4 | 1.6 | 1.3 | 1.1 | 0.9 | 2.2 | 2.3 | 1.0 | 2.0 | 2.0 | 2.1 | |||||||||||||
| 309523 | Mphosph1_predicted | 1.0 | 2.1 | 1.1 | 0.9 | 1.2 | 0.8 | 2.1 | 2.3 | 1.1 | 2.2 | 1.8 | 1.8 | |||||||||||||
| 310344 | Plk4_predicted | 1.0 | 1.9 | 1.1 | 1.1 | 0.7 | 0.8 | 1.8 | 1.8 | 0.7 | 1.8 | 1.3 | 1.7 | |||||||||||||
| 315330 | Espl1_predicted | 0.8 | 1.8 | 1.3 | 0.9 | 0.8 | 0.6 | 2.6 | 2.6 | 1.4 | 2.5 | 1.9 | 2.2 | |||||||||||||
| 362519 | Smc2l1_predicted | 0.5 | 1.0 | 1.0 | 0.8 | 0.8 | 0.6 | 0.8 | 0.8 | −0.1 | 1.0 | 0.6 | 0.7 | |||||||||||||
| 399489 | E2f1 | 0.2 | 1.0 | 0.7 | 0.6 | 0.5 | 0.7 | 0.6 | 0.7 | 0.2 | 0.3 | 0.7 | 0.6 | |||||||||||||
| 54237 | Cdc2a | 1.0 | 1.5 | 1.1 | 1.0 | 0.8 | 0.5 | 1.7 | 2.0 | 1.1 | 1.9 | 1.4 | 1.7 | |||||||||||||
| 680089 | LOC680089 | 1.3 | 2.0 | 1.3 | 1.1 | 1.1 | 0.8 | 1.7 | 1.7 | 0.9 | 1.5 | 1.1 | 1.3 | |||||||||||||
PND, postnatal day. Diets: HFB, high fat butter; HFO, high fat olive oil; HFS, high fat safflower oil; LFB, low fat butter; LFO, low fat olive oil; LFS, low fat safflower oil.
In addition to GO we examined enrichment of differentially expressed genes in KEGG pathways and canonical pathways defined by the Ingenuity Pathway Analysis system. Out of all KEGG pathways, cell cycle pathway was most highly enriched by differentially expressed genes (10 out of 90 genes differentially expressed at 50 days, FDR = 1.3 × 10−7). Out of all Ingenuity canonical pathways, G2/M check point pathway was the most significantly (4 out of 90 genes, P value <0.0001). Altogether, based on both GOs and KEGG pathways there were 32 cell cycle-related genes differentially expressed at PND50 and 12 at PND21.
The low- and high-fat butter and the high-fat olive induced statistically significant differential expression at day 50 relative to the reference diet in more cell cycle genes than the other three diets. The other diets (high- and low-fat safflower and low-fat olive) showed some of the same trends but failed to reach the statistical significance in some comparisons (Table 1 and Spl. Fig. S2). Also, the analysis of the PND21 differential gene expression data exhibited upregulation of some of the same genes identified in the 50 days analysis but reached significance of differential expression only in the high-fat olive oil diet. For example, high-fat olive oil reached the statistical significance at PND50 in 32 out of 32 cell-cycle genes significant at PND50 (Spl. Fig. S2, red bars). Although differential expression for high-fat safflower oil reached statistical significance in only 1 out of 32 cell cycle genes at PND50, it had concordant direction of the change compared with the reference AIN-93G diet in all 32 cell cycle genes and was statistically significant at the lower significance level (P value <0.01) in 19 out of 32 cell cycle genes significant at PND50 (Spl. Fig. S2, red bars and green bars).
Cluster analysis.
We used a cluster analysis in combination with functional analysis to define broader patterns of gene expression profiles associated with different diets. For cluster analysis, we selected 1,109 genes with 1.5 differential expression and P values ≤0.01 between at least one of the fatty acid diets and the reference AIN-93G diet at either 21 or 50 days. The cluster analysis was followed by the systematic assessment of functional coherence of all possible clusters (see materials and methods). This analysis yielded three distinct clusters of coexpressed genes highly enriched for genes belonging to relevant functional categories outlined in Spl. Fig. S3 (immunity, sterol metabolism, and cell cycle).
The “cell cycle” cluster consisted of 426 coexpressed genes that included 62 cell cycle-related genes. GO category “cell cycle” showed the highest statistical significance of enrichment (Fisher's FDR = 6 × 10−22). The 62 cell cycle related genes along with their log2 differential expressions are listed in Spl. Table S3. The general expression pattern of these genes followed the expression pattern of cell cycle-related genes identified by the more stringent criteria (Spl. Figs. S1 and S2 and Table 1). In general, they were upregulated for all diets at both time points with PND50 upregulation being stronger.
We performed a computational analysis of promoter regions of genes in this cluster to search for common TF binding motifs. Promoter regions of these genes were enriched (Fisher's test FDR <0.05) for the binding motifs of the NF-Y TF and three E2F complexes (RB-E2F1-DP1, E2F4-DP2, and E2F4-DP1). The involvement of the E2F family of TFs was further assessed by comparing the list of genes in this cluster to E2F targets established by ChIP-chip analysis (60). The 30 gene overlap between the genes in the “cell cycle” cluster and the ChIP chip-derived E2F targets represented highly statistically significant enrichment (Fisher's test P value <10−8). Finally, 9 out of 12 genes upregulated by E2F1 gene in rat fibroblasts in vitro (35) were also among the genes in the “cell cycle” cluster.
The “immunity” cluster consisted of 362 coexpressed genes which included 49 immunity-related genes. The GO category “immune response” showed the highest statistical significance of enrichment (Fisher's FDR = 9 × 10−11). Immunity-related genes along with their log2 differential expressions for various fatty acid diets in comparison to corresponding reference diet are given in Spl. Table S4. Expression levels of these genes indicated downregulation at PND21 but relatively little change or slight downregulation at PND50 (Fig. 2 and Spl. Table S4). Based on the computational analysis, promoters of genes in this cluster were enriched for the binding motifs of the Irf8 TF (Fisher's test FDR <0.05).
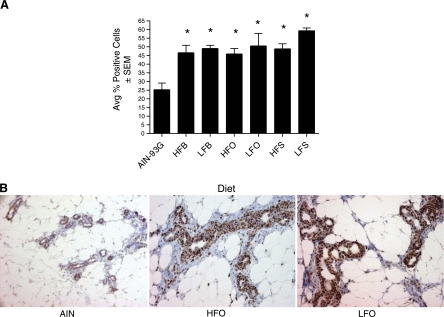
Fig. 2.
PCNA immunohistochemistry of mammary ducts and lobules for pups on different fatty acid diets at postnatal day 50 (PND50). Mammary ducts and lobules stained with PCNA (Leica Clone PC10) and biotinylated horse anti-mouse IgG (Vector) were photographed at ×20 with a Zeiss Axioscope (Carl Zeiss MicroImaging) from the reference diet and low fat (10% of energy as fat) and high fat (39% of energy as fat) olive, safflower, and butter diets. A: for each animal, at least 100 cells from 10 lobular areas were counted with respect to PCNA positivity and negativity, which is expressed as percent, and animals within treatment groups were averaged and compared with AIN-93G. B: examples of PCNA staining in offspring on AIN-93G and HFO and LFO diets for PND50. The percent of PCNA-positive cells in these 3 particular lobular areas were 14.4, 42.7, and 48.2% for AIN-93G HFO and LFO diets, respectively.
The “steroid cluster” consisted of 50 coexpressed genes that included six genes related to steroid biosynthesis (Spl. Table S5). The GO category “sterol biosynthetic process” showed the highest statistical significance of enrichment (Fisher's FDR = 2 × 10−6). These steroid-related genes were upregulated at PND21 and not changed or somewhat downregulated at PND50 (Spl. Fig. S3 and Spl. Table S5). Four out of six sterol-related genes (Sqle, Hmgcr, Dhcr7, Hsd17b7) in this cluster have been shown to be upregulated by estradiol under different physiological conditions (10, 17, 31, 57).
Proliferation by PCNA immunohistochemical staining.
Increased proliferation at PND50 of mammary epithelium of rats on all of the low- and high-fat test diets was confirmed by proliferating cell nuclear antigen (PCNA) antibody staining. The proportion of PCNA positively stained cells was significantly higher (approximately twofold increase, P value <0.05) in the mammary epithelium of rats on all fatty acid diets compared with the reference AIN-93G diet (Fig. 2). Although some of the fatty acid diets seemed to have had a stronger effect on the expression of cell cycle genes than the others, there was no difference in PCNA measurement of cellular proliferation between different test diets.
Proliferation signature in human breast tumors.
We compared 62 cell cycle-related rat genes from our “cell cycle” cluster to a published human breast cancer proliferation cluster of 45 genes (58). Human homologs of 27 of 62 rat cell cycle genes were included in this human breast cancer cluster of 45 genes. The nature of the gene expression pattern identified in the differential gene expression analysis was further characterized by analyzing gene expression profiles of human primary breast tumors (40). We hypothesize that the gene expression patterns in our data are reflective of increased proliferation of mammary epithelium and that the same genes will be associated with the proliferation of human breast tumors. Indeed, 54 of 69 human homologs of rat genes upregulated either at 21 or 50 days according to the high-stringency criteria were positively correlated with the size of the human tumors (Spl. Fig. S4). Conversely, 7 out of 15 human homologs of downregulated rat genes were negatively correlated with the tumor size (Spl. Fig. S5).
DISCUSSION
We investigated the effects of high and low fat-enriched diets on expression of genes in LCM-captured mammary epithelial cells. Our study demonstrated that the exposure to dietary fatty acids prior and during puberty significantly altered the proliferation pathways in the LCM-captured mammary epithelial cells and in some cases the G2M checkpoint pathways compared with the reference AIN-93G diet. Upregulated genes were enriched for proliferation marker genes (e.g., Ccnb1, Ccna2, Plk1, Aurkb, Top2a, Bub1) and genes putatively regulated by E2F family of TFs. Differentially expressed genes were statistically significantly enriched with rat homologs of genes from the previously established human breast cancer proliferation signature. Also, expression profiles of differentially expressed rat genes showed strong correlation with the expression of respective human homologs in human breast tumor expression data. Functional relevance of the upregulation in gene expression of proliferation-related genes was confirmed by the approximately twofold increase in proliferation rates as measured by PCNA immunohistochemistry.
Previous studies have also found that the fat composition of the diet modulates the proliferation of mammary epithelium using PCNA staining (43). The proportion of PCNA-stained mammary ductal epithelial cells in the reference diet in the Olivo and Hilakivi-Clarke study (43) was the same as observed by us (∼250 stained cells per 1,000 cells, Fig. 2). This similarity in proliferation rates between the two studies suggests that this is the baseline mammary ductal proliferation rate for the reference AIN-93G diet in Sprague-Dawley rats at the end of puberty. In that study, there were no differences between the high-fat diets high in n-6 PUFA compared with the low-fat diet high in n-6 PUFA (reference AIN-93G diet). In our study the low- and high-fat test diets showed the same level of increased proliferation compared with the reference AIN-93G diet as measured by PCNA staining (Fig. 2). Some of the fatty acid diets induced statistically significant differential expression in more cell cycle genes compared with the reference AIN-93G diet than others. However, there was no statistically significant differentially expressed genes between high- and low-fat diets, which correlates with similar proliferation rates observed in fatty acid diets at PND50. The fact that our data do not completely correlate with the Olivo and Hilakivi-Clarke study (43) is probably due to significant compositional differences between our test diets and n-6/n-3 PUFAs used in that study (43). Length of diet exposure, rat strain, litter size, and time of sampling are additional differences between the studies which could have contributed to the differences.
A closer inspection of the genes in the cell cycle cluster identified E2F family of TFs as the regulatory effectors of fatty acid diets. Two members of the family (E2F1 and E2F8) were themselves differentially expressed (Spl. Table S3). The DNA binding motifs recognized by E2F TFs were enriched in the promoters of genes from this cell cycle cluster. Also, the cluster was enriched for genes bound by E2F TFs identified in ChIP-chip (60) experiments and for genes upregulated by E2F1 in in vitro experiments (35). E2F TFs have been established as essential regulators of both normal mammary development (2) and tumor progression (41).
Increased proliferation rates have been shown to increase susceptibility to carcinogenesis (47). Other studies have found the significant correlation between diet-induced reduction of mammary proliferation and the reduced sensitivity to chemically induced carcinogenesis in rat (29, 43). Also, in utero dietary exposure to soy protein was shown to be protective against DMBA (4)- and NMU (52)-induced mammary cancers in the same rat model, but the gene expression patterns induced by these dietary regiments are significantly different from our study (56). Future studies will determine the exact mechanisms by which fatty acids are causing the increased proliferation of mammary epithelium and the consequences of such increased proliferation on the increased susceptibility to carcinogen induced tumors during puberty and later in life.
We compared results of our studies to the established human breast cancer proliferation gene expression signature as well as gene expression data from primary human breast tumors. There was a high level of concordance of our cell cycle gene lists with the list of genes defining the human breast cancer proliferation signature (40). Furthermore, when we examined expression levels of human homologs of genes constituting our proliferation signature in primary human tumors we found that they almost perfectly correlate with the tumor size. There are several reasons why these results are relevant in the context of our experimental data analysis. First, the concordance between the “proliferation signature” and the primary tumor data in breast cancers is additional evidence that the pattern we identified is indeed a proliferation signature. The second aspect of this analysis is the connection between the animal model and human cancers. It is not surprising that mammary proliferation in the rat is characterized by the same transcriptional profile as the growth of human breast cancers, but it is still an additional validation of using this model to study human carcinogenesis. The connection with tumor development may be tenuous, however; since the fatty acid diets are capable of upregulating proliferation genes in rats, they might be able to do so as well in human tumors that would otherwise grow at a slower pace. In addition, if these fatty acids are capable of modifying proliferation rates of human mammary epithelium during puberty, they could potentially modify the risk of breast cancer later in life.
GRANTS
This work was supported by NIEHS and NCI Grant U01 ES/CA-12770 and National Human Genome Research Institute (NHGRI) Grant R01HG-003749. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NCI, NHGRI, or NIH.
Supplementary Material
Acknowledgments
We are grateful to Dr. Ronald Jandacek for validating the compositions of different fatty acids in experimental diets and Veronica Ratliff for help in preparing the manuscript. We also thank Think Pink Funds for the purchase of the Applied Biosciences Step One Plus 96 Well RT-PCR System used in these experiments and the Dean of the College of Medicine at University of Cincinnati for some financial support.
This project was carried out as part of the National Institute of Environmental Health Sciences (NIEHS)/National Cancer Institute (NCI) Breast Cancer and the Environment Research Centers, four centers with transdisciplinary research collaborations integrated across biologic, epidemiologic, and community outreach projects.
Address for reprint requests and other correspondence: M. W. Anderson, Dept. of Cancer and Cell Biology, Univ. of Cincinnati, 3125 Eden Ave, Rm. 2312 (PO Box 670521), Cincinnati OH 45267-0521 (e-mail: marshall.anderson@uc.edu).
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Alarcón de la Lastra C, Barranco MD, Motilva V, Herrerias JM. Mediterranean diet and health: biological importance of olive oil. Curr Pharm Des 7: 933–950, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Andrechek ER, Mori S, Rempel RE, Chang JT, Nevins JR. Patterns of cell signaling pathway activation that characterize mammary development. Development 135: 2403–2413, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badger TM, Ronis MJJ, Simmen RCM, Simmen FA. Soy protein isolate and protection against cancer. J Am Coll Nutr 24: 146S-149, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles - database and tools. Nucleic Acids Res D562-D566, 2005. [DOI] [PMC free article] [PubMed]
- 6.Bartsch H, Nair J, Owen RW. Dietary polyunsaturated fatty acids and cancers of the breast and colorectum: emerging evidence for their role as risk modifiers. Carcinogenesis 20: 2209–2218, 1999. [DOI] [PubMed] [Google Scholar]
- 7.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57: 289–300, 1995. [Google Scholar]
- 8.Berry EM Who's afraid of n-6 polyunsaturated fatty acids? Methodological considerations for assessing whether they are harmful. Nutr Metab Cardiovasc Dis 11: 181–188, 2001. [PubMed] [Google Scholar]
- 9.Carroll KK Experimental evidence of dietary factors and hormone-dependent cancers. Cancer Res 35: 3374–3383, 1975. [PubMed] [Google Scholar]
- 10.Chen HJ, Shapiro DJ. Nucleotide sequence and estrogen induction of Xenopus laevis 3-hydroxy- 3-methylglutaryl-coenzyme A reductase. J Biol Chem 265: 4622–4629, 1990. [PubMed] [Google Scholar]
- 11.Clarke SD Polyunsaturated fatty acid regulation of gene transcription: a mechanism to improve energy balance and insulin resistance. Br J Nutr 83, Suppl 1: S59–S66, 2000. [DOI] [PubMed] [Google Scholar]
- 12.Clarke SD, Jump DB. Dietary polyunsaturated fatty acid regulation of gene transcription. Annu Rev Nutr 14: 83–98, 1994. [DOI] [PubMed] [Google Scholar]
- 13.Coletta RD, Jedlicka P, Gutierrez-Hartmann A, Ford HL. Transcriptional control of the cell cycle in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia 9: 39–53, 2004. [DOI] [PubMed] [Google Scholar]
- 14.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F.Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucl Acids Res 33: e175, 2005. [DOI] [PMC free article] [PubMed]
- 15.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3, 2003. [PubMed] [Google Scholar]
- 16.Doisneau-Sixou SF, Sergio CM, Carroll JS, Hui R, Musgrove EA, Sutherland RL. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer 10: 179–186, 2003. [DOI] [PubMed] [Google Scholar]
- 17.Duan WR, Parmer TG, Albarracin CT, Zhong L, Gibori G. PRAP, a prolactin receptor associated protein: its gene expression and regulation in the corpus luteum. Endocrinology 138: 3216–3221, 1997. [DOI] [PubMed] [Google Scholar]
- 18.Fay MP, Freedman LS, Clifford CK, Midthune DN. Effect of different types and amounts of fat on the development of mammary tumors in rodents: a review. Cancer Res 57: 3979–3988, 1997. [PubMed] [Google Scholar]
- 19.Fenton SE Endocrine-disrupting compounds and mammary gland development: early exposure and later life consequences. Endocrinology 147: S18–S24, 2006. [DOI] [PubMed] [Google Scholar]
- 20.Freudenberg MJ, Joshi KV, Medvedovic M. CLEAN: CLustering Enrichment ANalysis. Under Review 2008. [DOI] [PMC free article] [PubMed]
- 21.Gago-Dominguez M, Yuan JM, Sun CL, Lee HP, Yu MC. Opposing effects of dietary n-3 and n-6 fatty acids on mammary carcinogenesis: the Singapore Chinese Health Study. Br J Cancer 89: 1686–1692, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerber M Olive oil, monounsaturated fatty acids and cancer. Cancer Lett 114: 91–92, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Hilakivi-Clarke L, Clarke R, Onojafe I, Raygada M, Cho E, Lippman M. A maternal diet high in n - 6 polyunsaturated fats alters mammary gland development, puberty onset, and breast cancer risk among female rat offspring. Proc Natl Acad Sci USA 94: 9372–9377, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes MD, Hunter DJ, Colditz GA, Stampfer MJ, Hankinson SE, Speizer FE, Rosner B, Willett WC. Association of dietary intake of fat and fatty acids with risk of breast cancer. JAMA 281: 914–920, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med 54: 131–152, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Hursting SD, Thornquist M, Henderson MM. Types of dietary fat and the incidence of cancer at five sites. Prev Med 19: 242–253, 1990. [DOI] [PubMed] [Google Scholar]
- 28.Ip C, Carter CA, Ip MM. Requirement of essential fatty acid for mammary tumorigenesis in the rat. Cancer Res 45: 1997–2001, 1985. [PubMed] [Google Scholar]
- 29.Ip C, Singh M, Thompson HJ, Scimeca JA. Conjugated linoleic acid suppresses mammary carcinogenesis and proliferative activity of the mammary gland in the rat. Cancer Res 54: 1212–1215, 1994. [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Ivanga M, Labrie Y, Calvo E, Belleau P, Martel C, Luu-The V, Morissette J, Labrie F, Durocher F. Temporal analysis of E2 transcriptional induction of PTP and MKP and downregulation of IGF-I pathway key components in the mouse uterus. Physiol Genomics 29: 13–23, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Zhu Z, Thompson HJ. Effect of energy restriction on cell cycle machinery in 1-methyl-1-nitrosourea-induced mammary carcinomas in rats. Cancer Res 63: 1228–1234, 2003. [PubMed] [Google Scholar]
- 33.Jump DB, Clarke SD. Regulation of gene expression by dietary fat. Annu Rev Nutr 19: 63–90, 1999. [DOI] [PubMed] [Google Scholar]
- 34.Jump DB, Clarke SD, Thelen A, Liimatta M, Ren B, Badin MV. Dietary fat, genes, and human health. Adv Exp Med Biol 422: 167–176, 1997. [DOI] [PubMed] [Google Scholar]
- 35.Kalma Y, Marash L, Lamed Y, Ginsberg D. Expression analysis using DNA microarrays demonstrates that E2F-1 up-regulates expression of DNA replication genes including replication protein A2. Oncogene 20: 1379–1387, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480-D484, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasekan JB, Clayton MK, Gendron-Fitzpatrick A, Ney DM. Dietary olive and safflower oils in promotion of DMBA-induced mammary tumorigenesis in rats. Nutr Cancer 13: 153–163, 1990. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Sivaganesan S, Yeung KY, Guo J, Bumgarner RE, Medvedovic M. Context-specific infinite mixtures for clustering gene expression profiles across diverse microarray dataset. Bioinformatics 22: 1737–1744, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medina D, Peterson LE, Moraes R, Gay J. Short-term exposure to estrogen and progesterone induces partial protection against N-nitroso-N-methylurea-induced mammary tumorigenesis in Wistar–Furth rats. Cancer Lett 169: 1–6, 2001. [DOI] [PubMed] [Google Scholar]
- 40.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. From The Cover: an expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 102: 13550–13555, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niida A, Smith A, Imoto S, Tsutsumi S, Aburatani H, Zhang M, Akiyama T. Integrative bioinformatics analysis of transcriptional regulatory programs in breast cancer cells. BMC Bioinformatics 9: 404, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ntambi JM, Bene H. Polyunsaturated fatty acid regulation of gene expression. J Mol Neurosci 16: 273–278, 2001. [DOI] [PubMed] [Google Scholar]
- 43.Olivo SE, Hilakivi-Clarke L. Opposing effects of prepubertal low- and high-fat n-3 polyunsaturated fatty acid diets on rat mammary tumorigenesis. Carcinogenesis 26: 1563–1572, 2005. [DOI] [PubMed] [Google Scholar]
- 44.Russo I Morphology and development of the mammary gland. In: Integument and Mammary Glands, edited by Jones TC, Mohr U, and Hunt RD. Berlin: Springer-Verlag, 1989, p. 233–266.
- 45.Russo J, Gusterson BA, Rogers AE, Russo IH, Wellings SR, van Zwieten MJ. Comparative study of human and rat mammary tumorigenesis. Lab Invest 62: 244–278, 1990. [PubMed] [Google Scholar]
- 46.Russo J, Tay LK, Russo IH. Differentiation of the mammary gland and susceptibility to carcinogenesis. Breast Cancer Res Treat 2: 5–73, 1982. [DOI] [PubMed] [Google Scholar]
- 47.Russo J, Russo IH. Influence of differentiation and cell kinetics on the susceptibility of the rat mammary gland to carcinogenesis. Cancer Res 40: 2677–2687, 1980. [PubMed] [Google Scholar]
- 48.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics 7: 538, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucl Acids Res 37: D5–D15, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schedin P, Mitrenga T, Kaeck M. Estrous cycle regulation of mammary epithelial cell proliferation, differentiation, and death in the Sprague-Dawley rat: a model for investigating the role of estrous cycling in mammary carcinogenesis. J Mammary Gland Biol Neoplasia 5: 211–225, 2000. [DOI] [PubMed] [Google Scholar]
- 51.Silberstein GB, Van HK, Hrabeta-Robinson E, Compton J. Estrogen-triggered delays in mammary gland gene expression during the estrous cycle: evidence for a novel timing system. J Endocrinol 190: 225–239, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Simmen RCM, Eason RR, Till SR, Chatman J, Velarde MC, Geng Y, Korourian S, Badger TM. Inhibition of NMU-induced mammary tumorigenesis by dietary soy. Cancer Lett 224: 45–52, 2005. [DOI] [PubMed] [Google Scholar]
- 53.Simonsen NR, Fernandez-Crehuet Navajas J, Martin-Moreno JM, Strain JJ, Huttunen JK, Martin BC, Thamm M, Kardinaal AF, van't Veer P, Kok FJ, Kohlmeier L. Tissue stores of individual monounsaturated fatty acids and breast cancer: the EURAMIC study. European Community Multicenter Study on Antioxidants, Myocardial Infarction, and Breast Cancer. Am J Clin Nutr 68: 134–141, 1998. [DOI] [PubMed] [Google Scholar]
- 54.Sinha DK, Pazik JE, Dao TL. Progression of rat mammary development with age and its relationship to carcinogenesis by a chemical carcinogen. Int J Cancer 31: 321–327, 1983. [DOI] [PubMed] [Google Scholar]
- 55.Smyth GK Limma: linear models for microarray data. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor, edited by Gentleman R, Carey V, Dudoit S, Irizarry R, and Huber W. New York: Springer, 2005, p. 397–420.
- 56.Su Y, Simmen FA, Xiao R, Simmen RC. Expression profiling of rat mammary epithelial cells reveals candidate signaling pathways in dietary protection from mammary tumors. Physiol Genomics 30: 8–16, 2007. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe H, Suzuki A, Kobayashi M, Lubahn DB, Handa H, Iguchi T. Similarities and differences in uterine gene expression patterns caused by treatment with physiological and non-physiological estrogens. J Mol Endocrinol 31: 487–497, 2003. [DOI] [PubMed] [Google Scholar]
- 58.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer 6: 99–106, 2006. [DOI] [PubMed] [Google Scholar]
- 59.Willett WC Goals for nutrition in the year 2000. CA Cancer J Clin 49: 331–352, 1999. [DOI] [PubMed] [Google Scholar]
- 60.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 17: 1550–1561, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.