Abstract
The binding of non-specific human IgM to the surface of infected erythrocytes is important in rosetting, a major virulence factor in the pathogenesis of severe malaria due to Plasmodium falciparum, and IgM binding has also been implicated in placental malaria. Here we have identified the IgM-binding parasite ligand from a virulent P. falciparum strain as PfEMP1 (TM284var1 variant), and localized the region within this PfEMP1 variant that binds IgM (DBL4β domain). We have used this parasite IgM-binding protein to investigate the interaction with human IgM. Interaction studies with domain-swapped antibodies, IgM mutants and anti-IgM mAbs showed that PfEMP1 binds to the Fc portion of the human IgM heavy chain and requires the IgM Cμ4 domain. Polymerization of IgM was shown to be crucial for the interaction because PfEMP1 binding did not occur with mutant monomeric IgM molecules. These results with PfEMP1 protein have physiological relevance because infected erythrocytes from strain TM284 and four other IgM-binding P. falciparum strains showed analogous results to those seen with the DBL4β domain. Detailed investigation of the PfEMP1 binding site on IgM showed that some of the critical amino acids in the IgM Cμ4 domain are equivalent to those regions of IgG and IgA recognised by Fc-binding proteins from bacteria, suggesting that this region of immunoglobulin molecules may be of major functional significance in host-microbe interactions. We have therefore shown that PfEMP1 is an Fc-binding protein of malaria parasites specific for polymeric human IgM, and shows functional similarities with Fc-binding proteins from pathogenic bacteria.
Keywords: Immunoglobulin M, Malaria, Rosetting, Plasmodium falciparum Erythrocyte Membrane Protein 1, Duffy Binding Like domain, Fcα/μR
Introduction
Immunoglobulin M (IgM), the first antibody to be secreted during an immune response, is highly effective at neutralizing and agglutinating pathogens, and also activates the classical complement cascade with 1000-fold increased avidity than IgG (1). This increased avidity is largely due to the pentameric structure of IgM (2). A receptor for IgM (and IgA), the Fcα/μR that is closely related to the polymeric Ig receptor (pIgR) in its ligand binding domain, has recently been identified and shown to be expressed by a subset of B-cells and macrophages, but not on granulocytes, T-cells, or NK cells in the mouse spleen (3). The Fcα/μR mediates endocytosis of IgM coated bacteria and immune complexes and is thought to play a role in antigen processing and presentation during the primary stages of immunity (4,5). Although parasite-specific IgM has been shown to play an important role in limiting parasite replication in rodent models of malaria, its role in human malaria remains largely undetermined (6,7).
Natural IgM, produced by B-1 B-cells of naïve animals, has been identified as a link between innate and adaptive immune responses because of their ability to control the dissemination of both viruses and bacteria (8,9). What role natural IgM plays in immunity to human malaria is less clear, although non-immune IgM is known to bind to the surface of Plasmodium falciparum-infected erythrocytes, and has been shown to correlate with rosetting and severe malaria in both laboratory strains and field isolates (10). Rosetting, the ability of infected erythrocytes to bind uninfected ones, is associated with severe malaria in African children (11,12), and rosettes often contain non-specific human IgM (13-16). Intriguingly, infected erythrocytes implicated in placental adhesion are also able to bind natural non-specific IgM (17,18). Although pathogenic parasite isolates clearly bind IgM, it is unclear what advantage the ability to bind IgM offers a parasite in an infected erythrocyte, although in the case of rosetting phenotypes, it has been suggested that IgM could act as a bridge between infected and uninfected erythrocytes to stabilize rosettes (14,15). However, even in the absence of information concerning their exact biological role, the IgM binding proteins are of considerable interest as immunochemical tools and model systems. A similar situation prevails for the bacterial IgG-binding proteins, staphylococcal protein A and streptococcal protein G, which have been extensively studied (19), but whose biological function is unknown.
The parasite ligands that mediate IgM binding have been shown to be members of the variant erythrocyte surface antigen family P. falciparum erythrocyte membrane protein 1 (PfEMP1), encoded by the var genes (20,21). Every parasite contains 50-60 var genes in its genome, but only one is expressed at the surface of the infected erythrocyte (21). The var gene repertoires of different parasite isolates are largely non-overlapping, resulting in extensive diversity in the PfEMP1 family (22). PfEMP1 molecules are composed of Duffy binding-like (DBL) domains classified into six types (α, β, γ, δ, ε, and X), and cysteine-rich interdomain region domains (CIDR) classified into three types (α, β, and γ) (23,24). Individual var genes differ from each other by the number and type of these domains. A number of different PfEMP1 domains from specific PfEMP1 variants have been shown to bind non-immune IgM, including a CIDR from the FCR3S1.2var1 variant (25), a DBLβ from the TM284S2var1 variant (26), two DBLε domains from FCR3var1CSA and FCR3var2CSA variants (27) and one DBL-X and two DBLε domains from 3D7var2CSA (18). The precise binding sites for IgM within these domains are unknown, and there is no obvious motif shared by these domains that is missing from equivalent domains lacking IgM-binding function. It is also unknown which region of the IgM molecule is bound by PfEMP1, and whether different IgM-binding PfEMP1 variants bind similar or different regions of the IgM molecule.
Here, we identify a new PfEMP1 variant as the IgM-binding ligand from a virulent P. falciparum strain derived from a patient with cerebral malaria, and report experiments analyzing the interaction between IgM and PfEMP1. Using domain-swap antibodies, mutant IgM molecules, and specific mAbs to IgM, we show that PfEMP1 binding requires the Cμ4 domain of the IgM heavy chain, and that IgM polymerization is essential for binding by PfEMP1. In addition, we found that one region of Cμ4 involved in PfEMP1-binding is homologous to the regions in IgG and IgA that are bound by bacterial Fc-binding proteins. Furthermore, using mAbs to the IgM Cμ3 and Cμ4 domains, we show that multiple P. falciparum strains implicated in both severe childhood and pregnancy associated malaria use the same binding site on IgM.
Materials and Methods
Parasite culture and selection
Parasites were cultured in group O erythrocytes in RPMI 1640 medium supplemented with gentamicin, HEPES, glucose and 10% pooled human serum as described previously (28). The parasite lines used were rosetting strains TM284 (29) and HB3R+, and the CSA-binding strains FCR3CSA, 202-CSA and HB3-CSA (17). The TM284 strain used here is the parent of a parasite clone TM284S2 used in a previous study of Ig-binding (26) and the PfEMP1 variant that we have identified (encoded by TM284var1) is distinct from that described by Flick et al., (TM284S2var1). Rosetting parasites were selected by plasmagel flotation (the upper phase contains only non-rosetting parasites and the lower phase contains rosetting parasites and uninfected erythrocytes)(30). After staining an aliquot of parasite suspension with 25 μg/ml ethidium bromide, rosette frequency (percentage of mature infected erythrocytes binding 2 or more uninfected erythrocytes) was assessed by fluorescence microscopy of a wet preparation.
Immunofluorescence assays (IFAs) to detect normal human serum IgM binding to live infected erythrocytes
IFAs were carried out on mature trophozoite or schizont stage parasites grown in medium containing normal human serum as described previously (10), using a mouse mAb to human IgM (Serotec) diluted 1/1000 followed by incubation with Alexa Fluor™ 488 labeled goat anti-mouse IgG (Molecular Probes) diluted 1/500 plus 1μg/ml of 4,6-diamidino-2-phenylindole (DAPI, to stain the parasite nucleic acid and allow identification of infected erythrocytes). Controls were performed using secondary antibody/DAPI only. Parasite cultures were smeared on a slide and viewed under a fluorescent microscope.
Protein extraction and immuno-precipitation assay
At late trophozoite stage, infected erythrocytes were washed twice in incomplete media (supplemented RPMI 1640 as described above, but lacking 10% serum) and half of the culture was treated with trypsin (100 μg/ml) for 10 min at 37°C followed by the addition of Soybean trypsin inhibitor (100 μg/ml) for 10 min at 37°C. Proteins were extracted using a solution of 150 mM NaCl, 5 mM EDTA, 50 mM tris-HCl pH 8.0 and 1% Triton X-100 (NETT) containing one tablet of a cocktail of protease inhibitors (Roche) per 10 ml NETT. After centrifugation (10 min at 13000 rpm, +4°C) the supernatant (Triton X-100 soluble fraction) was removed and used for immunoprecipitation. Pull-down assays were performed using 300 μl of Triton X-100 soluble fraction mixed with 100 μl of 10% slurry of protein L or protein G beads in NETT for 1 hour on a spiramix at +4°C. Beads were washed twice in NETT/1% BSA, once in NETT/350 mM NaCl, twice in NETT and once in tris-saline (150 mM NaCl, 50 mM tris-HCl pH 8.0). Beads were heated at 70°C in loading buffer and pelleted by centrifugation. The supernatant was analyzed on a NuPAGE® Novex 3-8% tris-acetate acrylamide gel (Invitrogen), transferred to PVDF membranes and blocked in PBS containing 5% skimmed milk. Membranes were probed with a 1/1000 dilution of mAb 6H1 (specific for the conserved intracellular portion of PfEMP1 (ref 28, kind gift from S. Rogerson) followed by 1/1000 dilution of HRP-conjugated sheep anti-mouse IgG (Chemicon International) and the signal developed using ECL Plus western blotting detection reagents as described by the manufacturer (Amersham Biosciences).
RNA extraction and var gene reverse-transcriptase PCR
TM284 parasites were divided into isogenic rosetting and non-rosetting sub-populations as described previously (30) and RNA was extracted from late ring stage parasites using Trizol (31). 2 μg of RNA were used for cDNA preparation using Superscript III kit (Invitrogen) according to the manufacturer’s protocol. Briefly, after DNase I treatment for 30 min at room temperature, cDNA was synthesized using random hexamers. Central DBLα domain was amplified by PCR with degenerate primers using AmpliTaq Gold polymerase as described (32). PCR products were cloned into the PCRII vector (Invitrogen) and used to transform competent cells and mini-prep DNA was prepared from white colonies and sequenced as described previously (33).
Sequencing of the TM284var1 gene
A var gene (TM284var1) was identified that comprised the majority of var gene sequences derived from the TM284 rosetting parasites but was not detected in the set of var gene sequences derived from the TM284 non-rosetting parasites. The full-length sequence of TM284var1 was obtained by a PCR-walking strategy using vectorette libraries (30).
Northern blotting
1.5 μg of RNA in formamide loading buffer were loaded on a 1.2% agarose / 1.1% formaldehyde gel, electrophoresed for 2 hours at 150 V in 1x MOPS running buffer, stained with 0.5 μg/ml ethidium bromide for visualisation and transferred overnight onto a nitrocellulose membrane (Roche). Probe fragments were amplified from genomic DNA using AmpliTaq DNA polymerase and cloned into pCRII vector (Invitrogen). Primer sequences were as follows; for the DBL4β probe: Forward 5-tctcgtcagctgaaggatgaatgggattgtaac-3′ and Reverse 5′-acgagtgggcccatgtgaatcatcaatagg-3′. For the exon II probe: Forward 5-aaaaaaccaaagcatctgttggaaatttat-3′ and Reverse 5′- gtgttgtttcgactaggtagtaccac-3′. Inserts were reamplified using Pfx DNA polymerase (Invitrogen), M13 reverse primer and the forward primer of the insert. RNA probes were synthesized from the PCR product using Sp6 RNA polymerase using the DIG Northern starter kit (Roche) according to the manufacturer conditions. Blots were pre-hybridised for 1h in Dig Easy buffer and probed overnight with 2 μl of probe in 7 ml of Dig easy buffer at 58°C for the DBL4β probe or 52°C for the exon II probe. Membranes were washed for 45 min with 0.5x SSC, 0.1% SDS followed by 45 min incubation in 0.25x SSC, 0.1% SDS. Both incubations were performed at 62°C for the DBL4β probe or 55°C for the exon II probe. Membranes were incubated with 1/100 dilution of anti-Dig antibody before chemiluminescent detection with CDP-star (Roche) as described by the supplier.
Expression of TM284var1 domains in COS-7 cells
All PfEMP1 extracellular domains encoded by TM284var1 were PCR amplified and cloned into the pRE4 expression vector using PvuII-ApaI restriction sites as described previously (27,34,35). The amino-acid boundaries of the constructs were as follows: DBL1α (82-422), CIDRβ (455-674), DBL2γ (749-1100), DBL3ε (1082-1499), DBL4β (1481-1952) and DBL5ε (1914-2285). The primers used were: DBL1α 5′-tctcgtcagctgggatatccacccgcaagg-3′ and 5′-acgagtgggccctgttgctacaaattctg-3′; CIDRβ 5′-tctcgtcagctgtcatgtcctgtatttggtgt-3′ and 5′-acgagtgggcccctcattagttacttccaattg-3′; DBL2γ 5′-tctcgtcagctggacaaaaacttgtttcaagtg-3′ and 5′-acgagtgggcccttctttcggtggtgtatcg-3′; DBL3ε 5′-tctcgtcagctgacagactatgatacaaatgc-3′ and 5′-acgagtgggccctggatgatattgtgggtcttc-3′; DBL4β 5′-tctcgtcagctggaagagtcaaatactacag -3′ and 5′-acgagtgggcccagtaacatccgcagtagg-3′; DBL5ε 5′tctcgtcagctggagtgtaacgaacctaacactg-3′ and 5′-acgagtgggcccaatgagacaatcacatttac-3′. PCR conditions were as follow: 3min at 94°C followed by 3 cycles: 94°C: 30 sec, 45°C: 30 sec, 60°C: 1.5 min; 3 cycles: 94°C: 30 sec, 48°C: 30 sec, 60°C: 1.5 min; and 24 cycles: 94°C: 30 sec, 50°C: 30 sec, 60°C: 1.5 min. The pRE4 vector contains the herpes simplex glycoprotein D signal sequence fused to the DBL domain of interest and a transmembrane signal sequence targeting the fusion protein to the surface of transfected COS-7 cells. COS-7 cells were transiently transfected with 1 μg of DNA using 3 μl of Fugene 6 as described by the manufacturer (Roche). Transfection efficiency and ability to bind IgM were assessed by IFA with mAbs DL6 and anti-human IgM respectively, as described before (27).
Bacterial expression of PfEMP1 domains
Bacterial expression of PfEMP1 domains was carried out by modification of the methods of Singh et al., 2003 (36). A PCR product containing a HIS-tagged DBL4β domain was subcloned as a NheI-XhoI fragment into the bacterial expression vector pET28a (Novagen), using the following sets of primers: 5′-ctagctagcgaagagtcaaatactacag-3′ and 5′-ccgctcgagagtaacatccgcagtagg-3′ and transformed into the E. coli BL21 Rosetta strain. Protein expression was induced by isopropyl-β-D-1-thiogalactopyranoside (IPTG) and bacterial pellets lysed in BugBuster (Novagen) containing protease inhibitors (Roche). Inclusion bodies were washed twice in 50 ml of 50 mM Tris-HCl pH7.4 containing 500 mM NaCl, 2M urea, 1 mM EDTA, 0.1% Igepal CA-630 (Sigma) with a final wash in ddH2O. Bacterial pellets were then denatured in 50 ml 8M urea prior to the addition of 5 ml of Talon metal affinity resin (Clontech) and rolled overnight at room temperature to allow binding. Bound protein was washed and eluted with 300 mM imidazole according to manufacturer’s instruction and re-natured into PBS using stepwise dialysis with reducing concentrations of urea at 4°C. Protein concentrations were determined by a BCA™ assay (Pierce) and purity assessed by SDS-PAGE on 12% bis-tris polyacrylamide gels stained with SimplyBlue (Invitrogen) or by immunoblotting with anti-HIS-HRP (Sigma).
Antibodies: domain-swap, point mutants and controls
IgM/IgG and IgM/IgA1 domain-swap, point-mutants, and IgG1 or IgA1 antibodies specific for the hapten NIP (3-iodo-4-hydroxy-5-nitrophenacetyl) were purified from tissue culture supernatants as previously described (37-41). Other antibodies used for investigating binding to PfEMP1 were obtained from commercial sources as follows: human IgA2 and IgM anti-NIP (Serotec); mouse IgMλ (Pharmingen); and IgM Fabμ fragment (Rockland). Detecting antibodies used in ELISAs and IFAs include anti-polyhistidine-HRP mAb (HIS-1, Sigma); goat anti-mouse λ-RPE (Southern Biotech); goat anti-mouse λ-biotin (Southern Biotech); mouse mAb DL6 anti-HSV-1 gD (Santa Cruz Biotechnology); goat anti-mouse IgG Fab-FITC (Sigma); goat F(ab’)2 anti-human IgM Fcμ-RPE (Caltag laboratories); mouse anti-human IgM κ-chain specific (Sigma); goat anti-human IgM μ-chain specific-HRP (Sigma). A panel of anti-IgM μ-chain specific mAbs used in blocking studies (1F11, 4-3, 1G6, 5D7, 196.6b, HB57, 1X11) have been described previously (42,43). The anti-Cμ4 specific mAb 2F11 was kindly provided by Professor Tor Lea, Institute of Immunology, Rikshospitalet, Oslo, Norway.
Construction of IgM mutants
The Cμ4 domain IgM mutants NR445-446HL and PLSP394-397LLPQ were constructed using site-directed mutagenesis (QuickChange IIXL kit; Stratagene) using the following forward and reverse primer sets (5′-catgaggccctgccccacctggtcaaccgagaggac-3′ and 5′-gtcctctcggtgaccaggtggggcagggcctcatg-3′ for IgM NR445-446HL and 5′-gcagagggggcagctcttgccccaggagaagtatgtga-3′ and 5′-tcacatacttctcctggggcaagagctgccccctctgc-3′ for IgM PLSP394-397LLPQ, all from MWG, Ebersberg, Germany) on the wild type pSV2gptVNP IgM plasmid using the polymerase chain reaction (PCR) (37). To verify incorporation of the desired mutations and check for PCR induced errors, the entire μ heavy chain region of all the expression vectors were sequenced. The mutant heavy chain genes were transfected by electroporation into the murine myeloma cell line J558L (European Collection of Cell Cultures), constitutively producing a mouse λ1 light chain and a J-chain, but no Ig heavy chains. The cells were grown in RPMI supplemented with 10% FCS, 100 IU/ml penicillin, and 100 μg/ml streptomycin (Gibco) at 37°C / 5% CO2. Stable transfectants were selected in medium containing 250 μg/ml xanthine and 1 μg/ml mycophenolic acid and IgM positive colonies selected by ELISAs on supernatants using NIP-(15)-BSA (Biosearch Technologies) coated microtiter wells as previously described (41).
Analysis of domain-swap antibody binding to PfEMP1 domains by ELISA
Nunc Maxisorp microtiter plates, coated overnight with 5 μg/ml purified Ab in coating buffer (0.05M sodium carbonate, pH 9.6), were blocked with PBS containing 5% milk powder and 0.1% (v/v) Tween-20. All Abs used to coat wells were of equal concentration as determined by the BCA™ assay and anti-NIP ELISAs with goat anti-mouse λ-biotin and streptavidin-HRP (Serotec). After three washes with PBS, 50 μl of varying concentrations of DBL4β were added in PBS containing 1% milk powder and Tween-20 as above (PBSMT) to duplicate wells and incubated for 2h at room temperature prior to washing as above. In blocking experiments, wells coated with IgM were incubated with varying concentrations of anti-IgM mAbs for 1h prior to the addition of DBL4β. Bound DBL4β was detected using anti-HIS-HRP (Sigma) at 1:1000 in PBSMT and incubated for 1h. After washing as above, reactions were developed by incubating with substrate (Sigma 3,3′,5,5′-tetramethylbenzidine dihydrochloride tablets made up according to manufacturer’s instructions).
Analysis of domain-swap antibody binding to PfEMP1 domains expressed in COS-7 cells by IFA
DBL4β and DBL5ε domains from TM284var1 transiently expressed on the surface of COS-7 cells (as above) were grown on the surface of 12 mm circular coverslips (Raymond A Lamb, UK). Cells were washed in PBS and fixed in ice cold 50:50 methanol acetone solution for 20 min. After washing in PBS, cells were blocked in PBS / 5% FCS for 1h. After washing, 2 ng mAb DL6 to detect the glycoprotein D tag as a marker of transfection efficiency, and 10 ng of Ab in a final volume of 50μl PBS / 1% FCS was spotted onto parafilm and laid on top of each cover slip for 1h in a humid chamber. For blocking experiments, IgM was pre-incubated with varying concentrations of anti-IgM mAb for 1h prior to incubation with DBL transfectants. Cells were washed three times in PBS / 1% FCS prior to the addition of a 1:100 dilution of goat anti-mouse IgG-FITC and goat anti-mouse λ-RPE for 1h in the dark. Cells were washed as above with a final wash in PBS and mounted with Prolong Gold antifade reagent containing DAPI (Invitrogen) and viewed by fluorescence microscopy.
IFAs of live infected erythrocytes with domain-swapped antibodies
To prevent the binding of native IgM from the culture medium, parasites were grown in medium with 10% IgM-depleted human serum, obtained by 3 sequential incubations of human serum with anti-human IgM linked to agarose beads (Sigma). Western blotting with an anti-human IgM antibody confirmed the complete removal of IgM from human serum following this procedure (data not shown). The binding of domain-swapped Ab to infected erythrocytes (Tables 2 and 3) was as described previously with minor modifications (17). Briefly, 50μl aliquots of parasite cultures at 2% hematocrit were washed in PBS and resuspended in PBS containing 1% Ig-free BSA (Sigma)(PBS-BSA). 1.6 μg of Ab were added to each aliquot and cultures incubated on ice for 1 h. Cells were washed twice with PBS-BSA and resuspended in 1:200 dilution of goat anti-mouse λ-FITC (Caltag) in PBS-BSA containing 50 ng DAPI. After a further incubation for 1h and washes as above, cells were resuspended in a 1:1000 dilution of chicken anti-goat Ig-Alexa Fluor 488 (Invitrogen). After another set of incubations and washes, cells were resuspended at 30% hematocrit in PBS-BSA and smeared on slides for immunofluorescence microscopy.
Table 2. Binding of recombinant domain swapped Abs to infected erythrocytes.
| Parasite strain | ||
|---|---|---|
| Antibody | TM284 | FCR3CSA |
| Human IgM | +++ | +++ |
| Human IgG1 | - | - |
| Human IgA1 | - | - |
| Mouse IgM* | - | - |
| Human IgM Fab | - | - |
| γ/Cμ3,4 | ++ | ++ |
| γ/L309C-Cμ4 | +/- | ++ |
| γ/Cμ3 | - | - |
| γ/CμTP | - | - |
| IgM C575S (monomeric IgM) | - | - |
| γ/L309C-μTP (polymeric IgG) | - | - |
| α/Cμ2,3 | - | - |
| α/Cμ2,3,4 | ++ | ++ |
| α/Cμ4 | ++/- | ++ |
Data representing results from two or more experiments are summarized for simplicity. Positive cells showed punctate green fluorescence over the surface of infected erythrocytes as shown in Fig 1A. +++, ≥ 50% of infected erythrocytes positive; ++, 20-40% of infected erythrocytes positive; +, 10-20% of infected erythrocytes positive; -, no infected erythrocytes positive.
Mouse IgM also failed to bind to HB3R+ (rosetting) and HB3CSA, 202CSA (CSA-binding) parasite isolates.
Table 3. Binding of human IgM to infected erythrocytes of various P. falciparum isolates can be blocked by IgM Fc-specific mAbs.
| Parasite Isolate | |||||
|---|---|---|---|---|---|
| mAb | TM284 | FCR3CSA | HB3R+ | 202CSA | HB3CSA |
| No mAb | 23,43,25 | 35,20 | 15,48 | 34,28 | 48 |
| 1X11 (Cμ1) | 28,36,27 | 41,21 | 16,43 | 41,39 | 49 |
| 4-3 (Cμ3) | 2,0,0 | 0,1 | 0,0 | 0,0 | 0 |
| 1F11 (Cμ4) | ND,12,4 | 11,7 | 7,16 | 12,15 | 18 |
| NHS | ND,ND,ND | ND,22 | ND,51 | 32,36 | 45 |
Data represent percentage of parasitized erythrocytes binding human IgM by IFA in the presence or absence of each mAb. NHS = normal human serum control. ND = not determined. Data typical of numerous experiments are shown.
IFAs of live infected erythrocytes with IgM-specific blocking mAbs
Parasites were grown in IgM-depleted human serum as described above. 5 μg of anti-NIP human IgM (Serotec) was incubated for 12 hours at 4°C with 12 μg of the IgM-specific mAbs IX11 (Cμ1-specific), 4-3 (Cμ3-specific) and IF11 (Cμ4-specific). The IgM/blocking mAb mixture was then incubated with live parasites and IFA carried out as described above for the domain-swapped antibodies.
“Pseudo”-rosetting assays
Human erythrocytes were derivatized with NIP and sensitized with varying concentrations of IgM, IgA1 or the α/Cμ4 domain-swap Ab (41). Coating levels for each Ab were found to be equivalent by reactivity with anti-λ light chain-FITC as assessed by flow cytometry (data not shown). COS-7 transfectants were washed twice in PBS, and resuspended at 1 × 106 cells/ml. Rosetting of sensitized erythrocytes to DBL4β transfected COS-7 cells was performed as previously described (41). To investigate the effects of complement at disrupting rosette formation, IgM (20 μg/ml) coated erythrocytes were incubated with C1q (100 μg/ml Sigma) or a 1:200 dilution of serum/plasma in barbitone complement fixation test diluent supplemented with 0.1% gelatin and 0.5% BSA (Veronal buffer; Oxoid, UK). Binding of C1q to IgM-opsonized erythrocytes was confirmed by IFA with anti-C1q conjugated to FITC (Serotec). After 1h incubation erythrocytes in serum, a 1:10 dilution of the anti-Cμ4 mAb 4-3 was added for an additional 1h prior to incubation with the DBL4β transfectants. A pseudo-rosette was defined as a transfected COS-7 cell surrounded by five or more opsonised-erythrocytes.
Results
Identification of PfEMP1 as the IgM-binding ligand from the TM284 strain
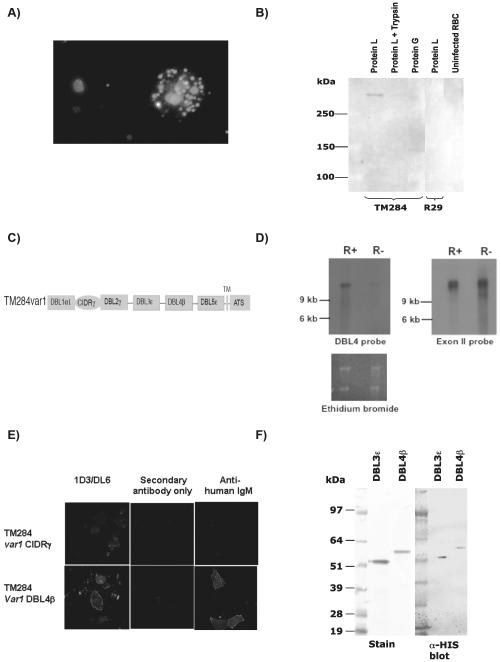
TM284 is a P. falciparum rosetting strain that shows strong binding to human IgM when grown in culture medium containing pooled naïve human serum (10,14)(Fig. 1A). In order to identify whether PfEMP1 was the parasite ligand responsible for IgM binding, we carried out immunoprecipitation experiments using Protein L-sepharose beads on Triton X-100 soluble protein extracts from the TM284 strain (Fig. 1B). A high molecular weight (>250 kDa), trypsin-sensitive protein was immunoprecipitated that could be blotted with mAb 6H1 recognizing the well conserved intracellular domain of PfEMP1 (44). Control immunoprecipitations with protein G (that binds IgG but not IgM) failed to precipitate a similar sized molecule, and no product was detectable with the other controls, including the non-IgM binding IT/R29 parasite strain (10) or uninfected erythrocytes. Taken together, these results indicate that a PfEMP1 variant expressed by TM284 binds human IgM but not IgG.
FIGURE 1.
Characterization of TM284var1 and expression of recombinant DBL domains. (A) IFA on live TM284 infected erythrocytes using mouse anti-human IgM (1/1000) and Alexa Fluor™ 488 conjugated goat anti-mouse secondary antibody (1/500). An IgM-positive rosetting infected erythrocyte shows punctuate surface fluorescence, while a non-rosetting infected erythrocyte is negative. Parasite nuclei stained with DAPI. (B) Protein extracts from TM284 or IT/R29 parasites, in the presence or absence of trypsin, were incubated with either protein-L (to pull out IgM) or protein-G (to pull out IgG) beads. Immunoprecipitated products were run on 3-8% acrylamide tris-acetate gels and transferred to PVDF membranes for blotting with the anti-PfEMP mAb 6H1. The product at >250 kDa represents PfEMP1. (C) Domain structure of the PfEMP1 variant encoded by the TM284var1 gene. (D) Northern-blot of ring-stage rosetting (R+) and non-rosetting (R−) TM284 parasites probed with a DBL4β probe specific for TM284var1 (left panel) or an exon II probe recognising all var genes (right panel). Ethidium bromide stained gel is shown as an indication of loading (lower panel). Bars indicate the position of the 9 Kb and 6 Kb bands of the RNA Millenium™ markers (Ambion). (E) IFA on COS-7 cells transfected with TM284 var1CIDRβ (upper panel) and TM284 var1DBL4β (lower panel). COS-7 cells were incubated with mAbs 1D3 or DL6 to determine transfection efficiency (left hand column), with secondary antibody alone as negative control (middle column) or with mouse mAb to human IgM (right hand column). (F) Western blot analysis of purified DBL domains of TM284var1 expressed in E. coli. PVDF membranes were incubated with peroxidase conjugated anti-HIS as described under ‘Experimental Procedures’. An equivalent SimplyBlue-stained SDS-PAGE 4-15% gradient gel is shown on the left. 2 μg of protein was loaded per lane.
Characterization of the TM284var1 gene
To allow detailed investigation of the molecular interactions between PfEMP1 and human IgM, we cloned and sequenced the var gene encoding the predominant PfEMP1 variant expressed by TM284 rosetting parasites. TM284 was selected to high rosette frequency (>70% of infected erythrocytes in rosettes) and then separated into rosetting and non-rosetting sub-populations (30). RNA was extracted from each sub-population and RT-PCR performed using degenerate primers to the var gene DBLα domain (32). The resulting PCR products were cloned and sequenced, and the predominantly transcribed gene from the rosetting parasites, which was absent from the non-rosetting parasites, was identified as TM284var1. The full-length TM284var1 gene was cloned and sequenced by PCR-walking using vectorette libraries (30). Analysis of the full sequence indicated that TM284var1 encodes a PfEMP1 variant containing 6 extracellular domains (Fig. 1C), plus a transmembrane domain and the intracellular acidic terminal sequence (ATS). The upstream sequence of TM284var1 is of the UpsA type (23), indicating that TM284var1 is a Group A var gene, a subgroup that is associated with rosetting and implicated in the pathogenesis of the most life-threatening forms of malaria (33,45,46). To confirm that TM284var1 is the predominantly transcribed var gene from TM284 rosetting parasites, a northern blot of RNA extracts from TM284 rosetting and non-rosetting sub-populations was probed with an anti-DBL4β probe specific for TM284var1. A strong signal was obtained only in the TM284 rosetting population (Fig. 1D). A probe to the var gene exon 2, encoding the conserved ATS region, that detects many/all var genes was also used and showed that the non-rosetting parasites transcribe other var genes (Fig. 1D).
Additional evidence that TM284var1 encodes the predominant IgM-binding, rosette-mediating PfEMP1 variant comes from a selection experiment in which IgM-binding TM284 infected erythrocytes were FACS-sorted to enrich for IgM positive cells, and then returned to culture. The starting culture was low rosetting and low IgM binding (20%). After FACS sorting for IgM positive cells, and culturing for 2 weeks (necessary to produce enough material for experiments), the culture was 88% rosetting and IgM-binding and a repeat of the RT-PCR confirmed that the predominantly transcribed var gene was TM284var1 (data not shown). Therefore selection for IgM-binding increases rosetting and selects for parasites transcribing the TM284var1 gene.
Identification of DBL4β as the IgM binding domain of the TM284var1 PfEMP1 variant
To identify the IgM-binding domain of TM284var1, each DBL and CIDR domain was cloned into the pRE4 expression vector and expressed at the surface of COS-7 cells (34). Transfection efficiency for each construct was determined by IFA with mAb DL6 specific for the HSV-glycoprotein D epitope expressed C-terminally to each PfEMP1 domain as part of the pRE4 vector (35). IgM-binding was detected by IFA using a mouse mAb to human IgM after incubation of the transfected COS cells in human serum. The level of expression for all domains ranged from 5 to 25% (Table 1). DBL4β was identified as the domain from TM284var1 that binds human IgM (Fig. 1E and Table 1). The DBL4β domain is composed of 291 amino acids including 12 cysteine residues. Phylogenetic comparison with other DBLβ domains showed that TM284var1 DBL4β clusters with domains called “β2” that differ from other DBLβ domains at the nucleotide level and are also characterized by the absence of a PfEMP1 C2 domain, which occurs after β1 type domains and plays an important role in their function (24).
Table 1. TM284var1 domains and their ability to bind human IgM.
| Domain | Transfection efficiency (%)a,b | IgM binding (%)b,c |
|---|---|---|
| DBL1α | 5-10 | 0 |
| CIDRβ | 20-25 | 0 |
| DBL2γ | 15-20 | 0 |
| DBL3ε | 5-10 | 0 |
| DBL4β | 10-15 | 8-12 |
| DBL5ε | 15-20 | 0 |
Transfection efficiency was calculated by IFA using mAb DL6.
Data from at least three experiments for each domain is shown.
IgM binding was determined by IFA using a mouse anti-human IgM mAb.
Bacterial expression of a soluble DBL4β domain
Since the DBL4β domain of TM284var1 uniquely bound human IgM (Table 1), we expressed this domain as a HIS-tagged protein in E. coli to allow further characterization of IgM-PfEMP1 interactions. The recombinant TM284var1 DBL4β domain expressed in E.coli accumulated in inclusion bodies as seen for other DBL domains (36). The protein was extracted and refolded using 8M urea and methods similar to those described previously (36). The final yield of refolded and purified protein from shaken flask cultures was approximately 1 mg/L. The purified DBL4β protein runs as a monomer at approximately 55 kDa on both reducing and non-reducing SDS-PAGE gradient gels, in line with the expected molecular weight of 55.734 kDa determined by ProtParam (http://www.expasy.ch/tools/protparam.html) and confirmed by immunoblotting with anti-HIS-HRP (Fig. 1F). We also expressed in an identical manner two non-IgM-binding DBL domains to act as controls in functional experiments, DBL3ε from the IT/R29var1 variant and DBL5ε (from TM284var1 - ref 27, and data not shown).
The TM284var1 DBL4β domain of PfEMP1 binds to the Fc of IgM
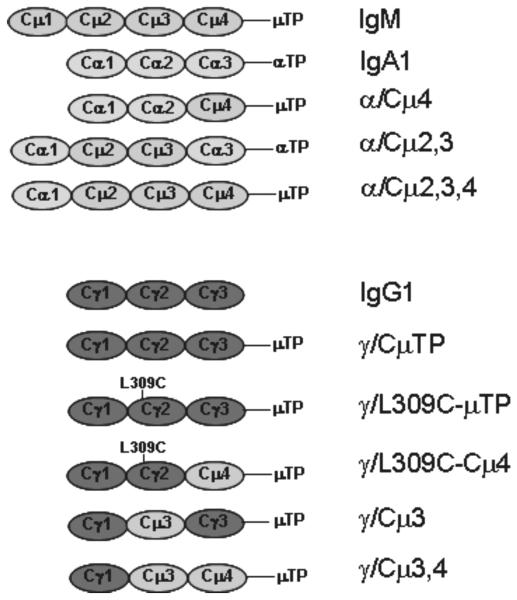
To determine which part of the IgM molecule the TM284var1 DBL4β domain binds, we used two panels of domain-swapped Abs (depicted in Fig. 2), in which homologous domains are exchanged between IgA/IgM (Fig. 2, upper panel) and IgG/IgM (Fig. 2, lower panel). These domain-swapped antibodies consist of mouse light chain and heavy chain variable regions (specific for the hapten NIP) linked to human heavy chain constant regions. The chimaeric domain-swapped antibodies are designated following these examples, α/Cμ4 (constant domain structure Cα1Cα2Cμ4); γ/Cμ3,4 (constant domain structure Cγ1Cμ3Cμ4) and are composed of mixtures of both monomers and polymeric forms. Antibodies with the L309C mutation exist predominantly in higher polymeric forms, including pentamers and hexamers (Fig 5E and refs 37-39).
FIGURE 2.
Overview of antibody heavy chain constructs. Constant heavy chain domains/sequences are shown as blue (μ), yellow (α) or red (γ) ovals. TP = secretory tailpiece. L309C marks a single amino acid replacement of the leucine from IgG to the cysteine from the homologous position in IgM, to create a domain-swap antibody with greater ability to polymerize (38).
FIGURE 5.
Localization of the DBL4β binding site in the Cμ4 domain of IgM. (A) Amino acid alignment of human, mouse and bovine IgM with human IgA1 and IgG1 at two exposed interdomain loop regions within the Cμ4 domain (see Fig. 8B). The IgA1 or IgM sequences are numbered according to the commonly adopted schemes used for IgA1 Bur (59) or IgM (60), whereas Eu numbering is used for IgG (61). Bold residues are conserved between species. Human IgG1 and IgA1 residues involved in contact with protein A and FcαR (CD89) respectively are underlined (41,49). (B) Binding of anti-NIP human IgM and the Cμ4 mutants, NR445-446HL and PLSP394-397LLPQ, were examined for binding to DBL4β by IFA as described for Fig. 3. Neither mutant was seen to bind DBL4β. (C) Binding and detection of IgM point mutants to NIP-BSA immobilized onto microtiter plates. Antibodies were detected with peroxidase conjugated anti-λ of anti-μ as described in ‘Experimental Procedures’. (D) Epitope mapping of mAb binding to the IgM point mutants NR445-446HL and PLSP394-397LLPQ by immunoblotting. (E) Size exclusion chromatograms of recombinant human IgM, γ/L309C-Cμ4 and IgM NR445-446HL antibodies run on a Superdex GL200 column.
The ability of the recombinant TM284var1 DBL4β domain to bind the domain-swapped antibodies was analyzed by ELISA (Fig. 3A and B, left panels). We observed that only those domain-swapped antibodies containing the μ4 constant domain were able to interact with soluble TM284var1 DBL4β with an apparent affinity generally comparable with IgM (Fig. 3). In contrast, no binding was observed with human IgA, human IgG, mouse IgM or domain-swaps lacking the constant μ4 domain. As all these domain-swapped antibodies share similar Fab regions and light chains and are epitope matched for NIP, we can also deduce that the Fc region mediates all of the binding, a result confirmed by the observation that human IgM Fabs failed to bind (see Fig. 4, see below). No binding of any antibody was seen in ELISAs with recombinant IT/R29var1 DBL3ε or TM284var1 DBL5ε domains also expressed in E. coli (data not shown).
FIGURE 3.
Binding of IgA/IgM and IgG/IgM domain-swaps to DBL domains. ELISA analysis of recombinant DBL4β from the TM284var1 variant binding to IgA/IgM (A) or IgG/IgM (B) domain-swap antibodies immobilized in microtitre wells (left panels). No binding was seen with control DBL3ε (R29var1 variant) (30) or DBL5ε (Tm284var1 variant) in the same ELISA. Binding of domain-swaps was also investigated by IFA with COS-7 cells expressing DBL4β or DBL5/3ε (right panels). Positive transfectants were detected with mAb DL6 reactive with the HSV-glycoprotein D tag expressed C-terminally of the DBL domain. DL6 was then detected with a FITC labeled anti-mouse IgG (green). Binding of domain-swap antibodies was detected with a phycoerythrin labeled anti-mouse λ (red) that binds to the common light chain shared by all the domain-swap antibodies. Only transfected cells incubated with IgM, α/Cμ2,3,4, α/Cμ4, γ/L309C-Cμ4 and γ/Cμ3,4 bound DBL4β by IFA and this recapitulated data seen by ELISA. Binding of domain-swap antibodies co-localized with DBL4β seen on the surface of unfixed COS-7 cells (C). No binding of antibodies was seen to control COS-7 cells expressing either DBL3ε or DBL5ε.
FIGURE 4.
The polymerization status of IgM is crucial for binding to DBL domains. Anti-NIP recombinant human IgM and various point mutants deficient in polymerization (IgM C414S and IgM C575S) were examined for binding to DBL4β by ELISA (left panel) and by IFAs (right panel) as described for Fig. 3. A reduced binding was seen for IgM C414S and no binding was seen with IgM C575S. IgM Fab’s or polymeric IgG (IgG L309C-μTP) showed no binding when used at equivalent molar concentrations to IgM in both assays.
To confirm the results from recombinant protein expressed in E. coli, we also transiently expressed TM284var1 DBL4β and DBL5ε (negative control) domains in the pRE4 vector in mammalian COS-7 cells. Only cells expressing DBL4β bound IgM or domain-swaps containing the Cμ4 domain, and failed to bind control IgG or IgA, in a manner that recapitulated our findings with ELISA (Fig. 3A and B, right panels). Confocal microscopy clearly showed that IgM binding co-localizes with the expressed pRE4 tag and DBL4β domain, as witnessed by discrete punctate orange staining on the surface of the COS-7 cell (Fig. 3C), resembling the staining pattern seen with live parasites (Fig. 1A). No binding was seen with the control DBL5ε domain from TM284var1 despite equal transfection efficiencies as determined by the binding of mAb DL6 recognizing the pRE4 tag.
The requirement of the Cμ4 domain for IgM binding was repeated in IFA experiments on live parasites with the rosetting TM284 strain. The parasites were grown in IgM-depleted human serum to prevent the binding of native IgM, and then incubated with the various domain-swapped antibodies. As seen with the TM284var1 DBL4β domain expressed in either E.coli or COS-7 cells (Fig. 3), TM284 live infected erythrocytes only bound those domain-swaps containing the Cμ4 domain (Table 2). Furthermore, binding to a second IgM-binding parasite strain (FCR3CSA) expressing the FCR3var2csa variant (27), known to be implicated in pregnancy malaria (47), was also dependent on the Cμ4 domain of IgM (Table 2). In contrast with the FCR3var2csa variant, the TM284 strain appeared to require contributions made by the Cμ3 domain, since those antibodies uniquely containing the Cμ4 domain did not bind in 2 of the 4 experiments undertaken. Therefore we show that two distinct P. falciparum strains have the ability to bind to the Fc of human IgM via the Cμ4 domain, and in TM284 isolate this binding is mediated by PfEMP1.
Binding of PfEMP1 is dependent on the polymeric nature of IgM
We investigated whether the binding of TM284var1 DBL4β is dependent on the ability of IgM to polymerize. We examined the binding of IgM point mutants with disrupted capability to polymerize and which are secreted from J558L cells principally as monomers (37-40). Two point mutants of IgM, one predominantly monomeric (IgM C575S), and one existing as 70% multimers / 30% monomer (IgM C414S) were significantly reduced in their capacity to bind DBL4β (Fig. 4), in particular the IgM C575S mutant whose tailpiece cysteine (Cys575) is known to make a major contribution to polymerization, as mutation of this residue to serine or alanine leads to secretion of large amounts of monomers (47,48). The monomeric form of IgM (C575S) also failed to bind to live infected erythrocytes by IFA (Table 2), confirming that properties of whole infected erythrocytes are highly similar to those of the individual IgM-binding domain from the TM284var1 PfEMP1 variant.
Polymerization was not, however, the only dictator of whether an antibody binds to PfEMP1, since γL309C-μTP, an IgG molecule in which the tailpiece of IgM had been attached to the C-terminus, and thereby allowing the production of pentameric and hexameric forms of IgG, did not bind to either live parasites or recombinantly expressed TM284var1 DBL4β (Fig. 4 and Table 2). Furthermore, commercial polymeric murine IgM from two separate suppliers failed to bind in any experiment. Similarly, polymeric versions of IgA1 did not bind DBL4β (Fig. 3). Therefore the above data show that the interaction of PfEMP1 with human IgM is dependent on both the primary amino acid sequence in the Cμ4 domain and the polymeric nature of IgM.
Identification of Cμ4 domain residues involved in DBL4β binding by IgM
Since we have shown a clear requirement for the Cμ4 domain of IgM in PfEMP1 binding, we chose to determine if exposed residues in the Cμ4 domain might be directly involved in the interaction with TM284var1 DBL4β. We generated two IgM antibodies, each with amino acid substitutions in either of two exposed loops, comprising Pro394-Pro397 and Pro444-Val447 in the Cμ4 domain. Molecular modeling suggests that one of these loops essentially occupies the analogous position to the Staphylococcus aureus protein A binding site on human IgG, and for the streptococcal IgA-binding proteins (Sir22 and Arp4) on human IgA (49). Since mouse IgM (this study) and bovine IgM do not bind the PfEMP1 variants and parasite strains investigated here (27), we also made amino acid alignments of Cμ4 from different species and discovered that both mouse and bovine IgM differed from human IgM at two locations in the loops of interest (Fig. 5A). Therefore we generated two human IgM mutants, termed PLSP394-397LLPQ and NR445-446HL, which through replacement of Pro-Pro and Asn-Arg at position 394-397 and 445-446, mimicked mouse IgM in these two exposed loops. Both mutations resulted in a complete loss of binding to DBL4β in ELISAs or IFAs (Fig. 5B) when used at equivalent concentrations to human IgM as determined by NIP-BSA specific ELISAs (Fig. 5C). Intriguingly, these regions were also vital for the recognition of the Cμ4-specific mAb 2F11 (Fig. 5D) that also blocked binding of IgM to DBL4β (Fig. 6B and C) and to parasitized red blood cells (data not shown). Size exclusion chromatography revealed that both mutants ran predominantly as monomers when compared with either human IgM or the γ/L309C-Cμ4 mutant (Fig. 5E). Since it could be argued that the lack of binding by the two Cμ4 domain mutants may now arise because they exist principally as monomers, we enriched for pentamers by concentrating numerous FPLC fractions containing the pentameric peak. Again, no binding of PLSP394-397LLPQ and NR445-446HL to DBL4β was observed in IFAs with transfected COS-7 cells.
FIGURE 6.
Epitope mapping of anti-human IgM mAbs and their effect on IgM binding to DBL4β. (A) Domain-swapped and point mutated antibodies were serially diluted from left to right onto nitrocellulose membranes and blotted against a panel of anti-human IgM mAbs from a previous study (42,43). mAbs 4-3, 5D7 and 196.6b bound to the Cμ3 domain, whereas mAbs 1F11 and 1G6 bound to the Cμ4 domain containing antibodies. Binding of antibodies was as for Western blots described in ‘Materials and methods’. (B) ELISA showing blocking of DBL4β binding to IgM by monoclonal antibodies 4-3, 1G6, 1F11, 2F11 and 196.6. IgM was coated onto microtiter plates at 5 μg/ml prior to incubation with varying concentrations of mAb. After washing, a fixed concentration of DBL4β (25 μg/ml) was added and binding detected as described in ‘Experimental Procedures’. (C) Blocking of IgM binding to DBL4β COS-7 cell transfectants by mAb 2F11, in contrast with mAb 1X11 which was unable to block binding at equivalent concentrations (200 ng/ml), as seen in ELISAs. IgM binding was detected by an anti-λ (top panel) or anti-human Fcμ (bottom panel) phycoerythrin labeled antibodies.
Binding of PfEMP1 to IgM can be blocked with monoclonal antibodies to the Cμ4 domain
To confirm the importance of the Cμ4 domain to PfEMP1 binding, we epitope mapped a panel of anti-IgM mAbs (Fig. 6A), previously used to investigate B-cell receptor activation (42,43). All the mAbs recognized human IgM but did not bind IgG or IgA (Fig. 6A). Epitope mapping of a panel of domain-swap antibodies revealed that mAbs 5D7, 4-3 and 196.6b bound to the Cμ3 domain, whereas mAbs 1F11, 1G6 and 2F11 (not shown) bound to the Cμ4 domain. Monoclonal antibodies 1X11 and HB57 bind in the Cμ1 and Cμ2 domains respectively (Fig. 6A), as observed previously and thereby serving as useful negative controls (42,43). Our finding that 5D7, 4-3 and 196.6b bind the Cμ3 domain rather than possessing Cμ4 specificity as previously designated (42,43), suggests that these mAbs may bind epitopes near the C-terminus of the Cμ3 domain, potentially lost during tryptic digestion of IgM in the former analysis.
Preliminary incubation of IgM with the anti-Cμ4 mAbs (mAbs 1F11, 2F11, 1G6) could inhibit IgM binding to DBL4β when used at concentrations as low as 20 ng/ml (Figs. 6B and C). Intriguingly, two anti-Cμ3 mAbs (196.6b and 4-3) could also block binding of human IgM, although higher concentrations of 196.6b were required to achieve this effect than for 4-3, which consistently was the most inhibiting mAb (Figs. 6B and 7C). Both the Cμ3-specific mAb 4-3 and the Cμ4-specific mAb 1F11 significantly blocked binding of IgM to infected erythrocytes from various IgM binding parasite isolates, whereas the Cμ1-specific mAb 1X11 did not (Table 3). That mAb 1F11 was unable to completely inhibit IgM binding to infected erythrocytes when used at similar concentrations to mAb 4-3 fits with the known low affinity for IgM of this particular mAb (42,43). These results indicate underlying similarity in the mechanism of IgM binding by multiple P. falciparum strains.
FIGURE 7.
Development and disruption of ‘pseudo-rosettes’ by Cμ4 specific monoclonal antibodies. (A) Binding of IgM, IgA1 or α/Cμ4 antibodies opsonized onto NIP-coated erythrocytes to DBL4β transfected COS-7 cells assessed by “pseudo”-rosette formation. The results are normalized by expressing specific rosette formation as a percentage of that seen with erythrocytes coated with anti-NIP human IgM (Serotec) at 100 μg/ml. (B) The binding of Ab-coated erythrocytes to DBL4β transfectants (stained with acridine orange) to form rosettes (arrowed) were visualized by white light / fluorescence microscopy and a rosette defined as a DBL4β transfected COS-7 cell surrounded by five or more erythrocytes. (C) IgM coated erythrocytes incubated in the presence of serum or plasma to allow opsonization and activation of components of the classical complement pathway still formed rosettes with DBL4β transfectants. Incubation of erythrocytes opsonized with IgM and complement components to mAb 4-3 totally blocked rosette formation.
Functional analysis of the Cμ4 domain interaction with DBL4β and the role of complement
The ability of the α/Cμ4 domain-swapped antibody to interact with DBL4β transfected COS-7 cells was also assessed by “pseudo”-rosetting assays (Fig. 7). As expected, IgM or the α/Cμ4 domain-swap, when opsonized onto NIP-coated erythrocytes could rosette to DBL4β expressing COS-7 cells with an affinity comparable to that of known Fc-FcR interactions (Figs. 7A and 7B arrowed). A control dimeric IgA1 was unable to form rosettes with DBL4β expressing COS-7 cells as expected. Intriguingly, IgM-opsonized erythrocytes that were incubated in serum or plasma as a source of complement or purified C1q (not shown) were still capable of forming rosettes with DBL4β transfected COS-7 cells (Fig. 7C), allowing us to conclude that the presence of members of the classical pathway, deposited on and around IgM, do not occlude the binding site for DBL4β in the Cμ4 domain. As predicted from our IFA data, the Cμ3 specific blocking mAb 4-3 completely inhibited rosette formation between IgM coated erythrocytes and the DBL4β expressing COS-7 cells, even in the presence of deposited complement components (Fig. 7C).
Discussion
PfEMP1 is a multi-ligand receptor expressed on the surface of infected erythrocytes that is responsible for parasite virulence phenotypes in African children, such as rosetting, and sequestration to the placenta in women (30,50). PfEMP1 is composed of cysteine-rich domains known as DBL domains that play a key role in malaria pathogenesis by mediating adhesion to human cells (51). Here we have identified the PfEMP1 variant transcribed by rosetting parasites from the strain TM284 (encoded by the TM284var1 gene) and shown that a specific domain from this PfEMP1 variant (DBL4β) binds to human IgM. We then used this P. falciparum IgM-binding protein as a tool to investigate the detailed mechanism of interaction between PfEMP1 and human IgM.
In order to analyze regions on IgM critical for the interaction with DBL4β we employed domain-swapped antibodies and point mutants (Fig. 2). As discussed previously (37,38), these mutant proteins are unlikely to have undergone any gross structural aberrations, allowing conclusions to be drawn on the relative contributions of different domains and mutated residues of IgM to the binding of PfEMP1. Our results indicate that DBL4β, whether expressed in E. coli or when presented on the surface of mammalian cell lines, binds to the Fc portion of the IgM heavy chain, and requires the Cμ4 domain (Figs. 3-5). The binding of PfEMP1 to IgM was crucially dependent on the polymeric nature of IgM, since both Fabs and monomeric versions of IgM failed to bind (Fig. 4). This finding is in agreement with previous work showing that IgM monomers do not increase the size of rosettes when added back to IgM-depleted serum (15). Although the affinity of an individual DBL domain for IgM may be low, the pentameric nature of IgM allows for an increased avidity, allowing significant cell to cell interactions as observed in our functional “pseudo”-rosetting assays using Ig-coated erythrocytes and DBL4β expressed in COS-7 cells (Fig. 7).
Importantly, the above results with a single PfEMP1 domain were also mirrored in experiments using the domain-swapped antibodies in IFAs with live infected erythrocytes of strains TM284 (rosetting) and FCR3CSA (CSA-binding) (Table 2). Furthermore, experiments using mAbs specific for different regions of the IgM heavy chain (Fig. 6) showed a similar pattern of inhibition of IgM-binding in 5 distinct parasite strains (2 rosetting and 3 CSA-binding) (Table 3). This suggests that diverse parasite isolates expressing distinct PfEMP1 variants (this study, and ref 27) all bind to the same (or similar) sites on the human IgM molecule. Given the diversity in different PfEMP1 domains implicated in IgM binding (outlined in detail in the introduction), this homogeneity amongst strains in binding to a particular site on IgM is unexpected, and may indicate that this site plays an important role in the host-parasite interaction. It will also be important to determine if the affinity for IgM seen by DBL domains from rosetting (non-CSA-binding) parasites is similar to that for DBL domains from non-rosetting (and CSA-binding) isolates. Future work should also address if there is a common IgM interaction site on these diverse P. falciparum DBL domains. Recent findings suggest that diverse DBLβ-C2 domains from numerous isolates use an equivalent glycan binding region when binding ICAM-1, implying a general model for how DBL domains evolve under dual selection for host receptor binding and immune evasion (52).
We were able to localize some of the amino acid residues within the IgM Cμ4 domain that are of particular importance in PfEMP1 binding (Fig. 5), including the PNRV (residues 444-447) and PLSP (residues 394-397) loops, predicted to lie on the surface of the Cμ4 domain (Fig. 8). Intriguingly, the PNRV loop is homologous to regions on both IgG and IgA responsible for binding to bacterial Fc-binding proteins (49). Since natural selection has shaped the interaction of the Fc region of antibodies with bacterial decoy proteins, the possibility exists that this area on IgM has also been selected for by parasitism (53). Given the overlapping nature of the binding sites in IgA-Fc for FcαRI (CD89) and bacterial IgA binding proteins (41,49,54), it could be postulated that this region is important for IgM function, and that it is in someway beneficial for the infected erythrocyte to block it. Ongoing work in our laboratories has shown that those mAbs that inhibited binding to DBL4β also prevented binding of IgM to the Fcα/μR, suggesting that the binding site for the two ligands lie close to each other. However, the interaction with Fcα/μR does require additional and unique contacts, since the γ/L309C-Cμ4 domain-swap shown to bind to DBL4β in this study, did not bind Fcα/μR; and mouse IgM, shown not to bind DBL4β, was capable of binding Fcα/μR (unpublished data). Furthermore, the extracellular portion of the pIgR (free secretory component; SC) previously shown to bind the Fc of IgM failed to prevent IgM binding to DBL4β. However, free SC did compete out binding of IgM to Fcα/μR, supporting our notion that unique contacts are involved for binding of IgM to either Fcα/μR or the pIgR (40 and unpublished data). Future experiments will address the ability of IgM bound to PfEMP1 to interact with functionally important host receptors for IgM, such as Fcα/μR and the pIgR (3,40).
FIGURE 8.
Molecular models of human IgM highlighting residues interacting with Fc-binding proteins. (A) Model of human IgM (kindly provided by Professor Stephen Perkins, University College, University of London) annotating sites in the Cμ4 domain involved in DBL4β (magenta) binding and sites in the Cμ3 domain important for interaction with C1q (turquoise) (2). (B) Molecular model (PyMol) showing the two sides of IgM-Fc based on the known crystal structure of the analogous IgE-Fc (10OV.pdb)(62). Exposed highlighted residues Pro394-Pro397 (orange) and Pro444-Val447 (red) in the Cμ4 domain have been shown to be involved in the interaction with DBL4β. The C-terminal tailpieces are omitted for clarity.
Further work will also be needed to investigate the effect of IgM binding by PfEMP1 on the ability of IgM to activate complement. The possible involvement of complement in IgM binding and rosette formation is particularly intriguing given the evidence that complement receptor 1 (CR1) is a rosette formation receptor on uninfected erythrocytes (30), and CR1 interacts with P. falciparum-infected cells via its C3b binding sites (55). Existing data suggest that complement activation is not required for rosette formation because rosettes form normally in C3- and C4-deficient human sera (55). However, a role for other complement components in rosetting has not been excluded. C1q is a possible candidate for involvement in rosetting because it is known to bind to the Cμ3 domain of IgM-Fc with high affinity (2), and is also known to interact with CR1 (Fig. 8 and ref 56). We therefore investigated whether C1q influenced the interaction between IgM and PfEMP1 using “pseudo”-rosetting assays in which Ig-coated erythrocytes were incubated with COS-7 cells expressing the IgM-binding PfEMP1 domain DBL4β (Fig. 7). In these pseudo-rosetting assays, IgM would be bound to the erythrocyte in its ‘crab-like’ formation, thus exposing the C1q binding sites in the Cμ3 domain. Despite the presence of saturating levels of complement, it was clear that IgM could still interact with DBL4β (Fig. 7). However, the epitope on the Fc of IgM seen by the blocking mAbs (and presumably DBL4β) is still exposed since these could completely block rosette formation even in the presence of saturating levels of complement. These findings allow us to postulate a number of structural explanations for how complement inactivation might be achieved by the infected erythrocyte for parasite-specific IgM. Recent structural models of the C1 complex have shown that both C1r and C1s, required for activation of the classical pathway, bind in the middle of the cone defined by the C1q stems, where they occupy a large area sitting over the Cμ4 domains of IgM (57). It is therefore possible that DBL4β bound to the Cμ4 domain of IgM, may interfere with complement activation. An alternative, and simpler explanation for the known complement resistance of infected erythrocytes, is that parasite-specific IgM bound to the surface of the parasitized red cell undergoes a conformational change on binding pertinent DBL domains, which re-closes the C1q binding site in the Cμ3 domain, thereby blocking access to C1q. We are currently developing recombinant human IgM molecules with specificity for different DBL domains, including DBL4β, to investigate these possibilities. The tools described in this manuscript will be critical to such analyses.
Rosetting and placental parasite isolates may also bind non-specific IgM to allow masking of critical PfEMP1 domains from the destructive action of specific antibodies; although we were unable to substantiate this hypothesis since DBL4β specific IgG responses were not detectable in a cohort of immune Gambian plasma available to us (58). However, the DBL4β domain described in this work was derived from TM284, a Thai isolate potentially expressing antigenically distinct PfEMP1 epitopes compared to Gambian parasites. It will be interesting to assess the effect of DBL4β specific anti-sera derived in immunization studies on IgM binding. The work shown here focuses on IgM binding by PfEMP1, however the experiments do also shed further light on a recent controversy over whether infected erythrocytes can also bind non-immune IgG. Some previous reports suggest that IgM-binding P. falciparum strains can also bind IgG from normal human serum (14,18,26). However, we have consistently been unable to detect non-immune IgG binding by any P. falciparum rosetting or CSA-binding strain using multiple different detection reagents (10,17). Although only experiments using identical parasite lines used by others will completely rule out non-specific IgG binding, the results shown in this paper indicate that IgM binding is determined both by specific amino acids within IgM and by the polymeric nature of normal IgM molecules. IgG did not bind to infected erythrocytes, even when an artificial polymeric form of IgG was tested (Table 2 and Fig. 4). The experiments that we carried out here, with IgG/IgM domain-swapped antibodies would not have been possible if P. falciparum did bind non-immune IgG. One other controversy in the literature concerns the ability of IgM-binding strains to bind mouse IgM. We reported previously that mouse IgM does bind to the surface of infected erythrocytes (17), however these experiments relied on the use of chloroquine to elute human IgM (derived from the parasite culture medium) from the surface of infected erythrocytes prior to incubation with mouse IgM. In the current study we used infected erythrocytes grown in human IgM-depleted serum, and found that mouse IgM did not bind. It may be that the high concentrations of chloroquine used in our previous study had adverse effects on membrane function and gave misleading results.
In summary, we have demonstrated that PfEMP1 binds to the Cμ4 domain of human IgM and that this mechanism is shared by diverse P. falciparum strains. These findings have allowed us to propose a possible mechanism by which PfEMP1 molecules may bind IgM and simultaneously interact with other serum proteins. Since PfEMP1 from both rosetting and placental isolates bind in the same region of IgM, specific inhibitors of the interaction may prove useful in diverse clinical settings.
Acknowledgements
We would like to thank Professor Tor Lea of the Institute of Immunology, Rikshospitalet, Oslo, Norway, for providing mAb 2F11 and Professor Paul Morgan, Drs Jenny Woof and Melanie Lewis for useful discussions on complement assays. We would also like to thank Mrs Kelly-Ann Vere for help with confocal microscopy and Drs Karen Bunting and Jody Winter for their assistance in expressing various DBL domains in E. coli. We also thank Professor Steve Perkins for kindly providing an α-carbon trace of IgM.
Funding. This study was funded by a Medical Research Council Career Establishment Award (MRC G0300145) and a European Union Marie Curie Excellence Grants, Antibody Immunotherapy for Malaria (MEXT-CT-2003-509670) to RJP. AR, J-PS and JAR were supported by a Wellcome Trust Senior Fellowship, grant number 067431.
Abbreviations
- NIP
3-iodo-4-hydroxy-5-nitrophenacetyl
- mAb
Monoclonal antibody
- HRP
Horse Radish Peroxidase
- ELISA
Enzyme Linked Immunosorbent Assay
- PBS
Phosphate Buffered Saline
- BCA
Bicinchoninic Acid
- IEs
Infected Erythrocytes
- Abs
Antibodies
- DS
Domain-swap
- Pf
Plasmodium falciparum
- DBL
Duffy-Binding-Like
- FITC
Fluorescein isothiocyanate
- PE
Phycoerythrin
- IPTG
Isopropyl-β-D-1-thiogalactopyranoside
- PfEMP1
Plasmodium falciparum Erythrocyte Membrane Protein 1
- DAPI
4′,6-diamidino-2-phenylindole
- DAB
Diaminobenzidine
- IFA
Immunofluorescent Assay
- NIP
3-nitr0-4-hydroxy-5-iodophenylacetate
Footnotes
Competing interests. The authors have declared that no competing interests exist.
References
- 1.Burton DR. Structure and function of antibodies. In: Calabi F, Neuberger MS, editors. Molecular Genetics of Immunoglobulin. Elsevier Science Publishers B.V. (Biomedical Division); 1987. pp. 1–49. [Google Scholar]
- 2.Perkins SJ, Nealis AS, Sutton BJ, J B, Feinstein A. Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation. J. Mol. Biol. 1991;221:1345–1366. doi: 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- 3.Shibuya A, Sakamoto N, Shimizu Y, Shibuya K, Osawa M, Hiroyama T, Eyre HJ, Sutherland GR, Endo Y, Fujita T, et al. Fc alpha/mu receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 2000;1:441–446. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto N, Shibuya K, Shimizu Y, Yotsumoto K, Miyabayashi T, Sakano S, Tsuji T, Nakayama E, Nakauchi H, Shibuya A. A novel Fc receptor for IgA and IgM is expressed on both hematopoietic and non-hematopoietic tissues. Eur. J. Immunol. 2001;31:1310–1316. doi: 10.1002/1521-4141(200105)31:5<1310::AID-IMMU1310>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Cho Y, Usui K, Honda S, Tahara-Hanaoka S, Shibuya K, Shibuya A. Molecular characteristics of IgA and IgM Fc binding to the Fcalpha/muR. Biochem. Biophys. Res. Commun. 2006;345:474–478. doi: 10.1016/j.bbrc.2006.04.084. [DOI] [PubMed] [Google Scholar]
- 6.Harte PG, Cooke A, Playfair JH. Specific monoclonal IgM is a potent adjuvant in murine malaria vaccination. Nature. 1983;302:256–258. doi: 10.1038/302256a0. [DOI] [PubMed] [Google Scholar]
- 7.Couper KN, Phillips RS, Brombacher F, Alexander J. Parasite-specific IgM plays a significant role in the protective immune response to asexual erythrocytic stage Plasmodium chabaudi AS infection. Parasit. Immunol. 2005;27:171–180. doi: 10.1111/j.1365-3024.2005.00760.x. [DOI] [PubMed] [Google Scholar]
- 8.Ochsenbein AF, Fehr F, Lutz C, Suter M, Brombacher F, Hengartner H, Zinkernagel RM. Control of early viral and bacterial distribution and disease by natural antibodies. Science. 1999;286:2156–2159. doi: 10.1126/science.286.5447.2156. [DOI] [PubMed] [Google Scholar]
- 9.Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J. A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. J. Exp. Med. 1998;188:2381–2386. doi: 10.1084/jem.188.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe JA, Shafi J, Kai OK, Marsh K, Raza A. Non-immune IgM, but not IgG binds to the surface of Plasmodium falciparum-infected erythrocytes and correlates with rosetting and severe malaria. Am. J. Trop. Med. Hyg. 2002;66:692–699. doi: 10.4269/ajtmh.2002.66.692. [DOI] [PubMed] [Google Scholar]
- 11.Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet. 1990;336:1457–1460. doi: 10.1016/0140-6736(90)93174-n. [DOI] [PubMed] [Google Scholar]
- 12.Rowe JA, Obeiro J, Newbold CI, Marsh K. Plasmodium falciparum resetting is associated with malaria severity in Kenya. Infect. Immun. 1995;63:2323–2326. doi: 10.1128/iai.63.6.2323-2326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clough B, Atilola FA, Black J, Pasvol G. Plasmodium falciparum: the importance of IgM in the rosetting of parasite-infected erythrocytes. Exp. Parasitol. 1998;89:129–132. doi: 10.1006/expr.1998.4275. [DOI] [PubMed] [Google Scholar]
- 14.Scholander C, Treutiger CJ, Hultenby K, Wahlgren M. Novel fibrillar structure confers adhesive property to malaria-infected erythrocytes. Nat. Med. 1996;2:204–208. doi: 10.1038/nm0296-204. [DOI] [PubMed] [Google Scholar]
- 15.Somner EA, Black J, Pasvol G. Multiple human serum components act as bridging molecules in rosette formation by Plasmodium falciparum-infected erythrocytes. Blood. 2000;95:674–682. [PubMed] [Google Scholar]
- 16.Treutiger CJ, Scholander C, Carlson J, McAdam KP, Raynes JG, Falksveden L, Wahlgren M. Rouleaux-forming serum proteins are involved in the rosetting of Plasmodium falciparum-infected erythrocytes. Exp. Parasitol. 1999;93:215–224. doi: 10.1006/expr.1999.4454. [DOI] [PubMed] [Google Scholar]
- 17.Creasey AM, Staalsoe T, Raza A, Arnot DE, Rowe JA. Nonspecific immunoglobulin M binding and chondroitin sulfate A binding are linked phenotypes of Plasmodium falciparum isolates implicated in malaria during pregnancy. Infect. Immun. 2003;71:4767–4771. doi: 10.1128/IAI.71.8.4767-4771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasti N, Namusoke F, Chêne A, Chen Q, Staalsoe T, Klinkert MQ, Mirembe F, Kironde F, Wahlgren M. Non-immune immunoglobulin binding and multiple adhesion characterize Plasmodium falciparum-infected erythrocytes of placental origin. Proc. Natl. Acad. Sci. U S A. 2006;103:13795–13800. doi: 10.1073/pnas.0601519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nezlin R, Ghetie V. Interactions of immunoglobulins outside the antigen-combining site. Adv. Immunol. 2004;82:155–215. doi: 10.1016/S0065-2776(04)82004-2. [DOI] [PubMed] [Google Scholar]
- 20.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 21.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyes SA, Kraemer SM, Smith JD. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot. Cell. 2007;6:1511–1520. doi: 10.1128/EC.00173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol. Biochem. Parasitol. 2000;110:293–310. doi: 10.1016/s0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, Heddini A, Barragan A, Fernandez V, Pearce SF, Wahlgren M. The semi-conserved head structure of Plasmodium falciparum erythrocyte membrane protein 1 mediates binding to multiple independent host receptors. J. Exp. Med. 2000;192:1–10. doi: 10.1084/jem.192.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flick K, Scholander C, Chen Q, Fernandez V, Pouvelle B, Gysin J, Wahlgren M. Role of nonimmune IgG bound to PfEMP1 in placental malaria. Science. 2001;293:2098–2100. doi: 10.1126/science.1062891. [DOI] [PubMed] [Google Scholar]
- 27.Semblat JP, Raza A, Kyes SA, Rowe JA. Identification of Plasmodium falciparum var1CSA and var2CSA domains that bind IgM natural antibodies. Mol. Biochem. Parasitol. 2006;146:192–197. doi: 10.1016/j.molbiopara.2005.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe JA, Berendt AR, Marsh K, Newbold CI. Plasmodium falciparum: a family of sulphated glycoconjugates disrupts erythrocyte rosettes. Exp. Parasitol. 1994;79:506–516. doi: 10.1006/expr.1994.1111. [DOI] [PubMed] [Google Scholar]
- 29.Carlson J, Wahlgren M. Plasmodium falciparum erythrocyte rosetting is mediated by promiscuous lectin-like interactions. J. Exp. Med. 1992;176:1311–1317. doi: 10.1084/jem.176.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 31.Kyes SA, Pinches R, Newbold CI. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 32.Taylor HM, Kyes SA, Harris D, Kriek N, Newbold CI. A study of var gene transcription in vitro using universal var gene primers. Mol. Biochem. Parasitol. 2000;105:12–23. doi: 10.1016/s0166-6851(99)00159-0. [DOI] [PubMed] [Google Scholar]
- 33.Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, Koné AK, Doumbo OK, Plowe CV, Rowe JA. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol. Biochem. Parasitol. 2006;150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen GH, Wilcox WC, Sodora DL, Long D, Levin JZ, Eisenberg RJ. Expression of herpes simplex virus type 1 glycoprotein D deletion mutants in mammalian cells. J. Virol. 1988;62:1932–1940. doi: 10.1128/jvi.62.6.1932-1940.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh SK, Singh AP, Pandey S, Yazdani SS, Chitnis CE, Sharma A. Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy-binding domains necessary for erythrocyte invasion. Biochem. J. 2003;374:193–198. doi: 10.1042/BJ20030622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sørensen V, Rasmussen IB, Norderhaug L, Natvig I, Michaelsen TE, Sandlie I. Effect of the IgM and IgA secretory tailpieces on polymerization and secretion of IgM and IgG. J. Immunol. 1996;156:2858–2865. [PubMed] [Google Scholar]
- 38.Sørensen V, Sundvold V, Michaelsen TE, Sandlie I. Polymerization of IgA and IgM: roles of Cys309/Cys414 and the secretory tailpiece. J. Immunol. 1999;162:3448–3455. [PubMed] [Google Scholar]
- 39.Sørensen V, Rasmussen IB, Sundvold V, Michaelsen TE, Sandlie I. Structural requirements for incorporation of J chain into human IgM and IgA. Int. Immunol. 2000;12:19–27. doi: 10.1093/intimm/12.1.19. [DOI] [PubMed] [Google Scholar]
- 40.Braathen R, Sorensen V, Brandtzaeg P, Sandlie I, Johansen FE. The carboxyl-terminal domains of IgA and IgM direct isotype-specific polymerization and interaction with the polymeric immunoglobulin receptor. J. Biol. Chem. 2002;277:42755–42762. doi: 10.1074/jbc.M205502200. [DOI] [PubMed] [Google Scholar]
- 41.Pleass RJ, Dunlop JI, Anderson CM, Woof JM. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcα receptor (FcαR) CD89. J. Biol. Chem. 1999;274:23508–23514. doi: 10.1074/jbc.274.33.23508. [DOI] [PubMed] [Google Scholar]
- 42.Rudich SM, Winchester R, Mongini PK. Human B-cell activation. Evidence for diverse signals provided by various monoclonal anti-IgM antibodies. J. Exp. Med. 1985;162:1236–1255. doi: 10.1084/jem.162.4.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudich SM, Mihaesco E, Winchester R, Mongini PK. Analysis of the domain specificity of various murine anti-human IgM monoclonal antibodies differing in human B-lymphocyte signaling activity. Mol. Immunol. 1987;24:809–820. doi: 10.1016/0161-5890(87)90183-0. [DOI] [PubMed] [Google Scholar]
- 44.Duffy MF, Brown GV, Basuki W, Krejany EO, Noviyanti R, Cowman AF, Reeder JC. Transcription of multiple var genes by individual, trophozoite-stage Plasmodium falciparum cells expressing a chondroitin-sulphate A binding phenotype. Mol. Microbiol. 2002;43:1285–1293. doi: 10.1046/j.1365-2958.2002.02822.x. [DOI] [PubMed] [Google Scholar]
- 45.Kaestli M, Cockburn IA, Cortés A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J. Infect. Dis. 2006;193:1567–1574. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, Salanti A, Vestergaard LS, Lusingu JP, Hermsen R, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J. Exp. Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis AC, Collins C, Yoshimura MI, D’Agostaro G, Shulman MJ. Mutations of the mouse mu H chain which prevent polymer assembly. J. Immunol. 1989;143:1352–1357. [PubMed] [Google Scholar]
- 48.Sitia R, Neuberger M, Alberini C, Bet P, Fra A, Valetti G, Williams C, Milstein C. Developmental regulation of IgM secretion: the role of the carboxy-terminal cysteine. Cell. 1990;60:781–790. doi: 10.1016/0092-8674(90)90092-s. [DOI] [PubMed] [Google Scholar]
- 49.Pleass RJ, Areschoug T, Lindahl G, Woof JM. Streptococcal IgA-binding proteins bind in the Calpha 2-Calpha 3 inter-domain region and inhibit binding of IgA to human CD89. J. Biol. Chem. 2001;276:8197–8204. doi: 10.1074/jbc.M009396200. [DOI] [PubMed] [Google Scholar]
- 50.Rowe JA, Kyes SA. The role of Plasmodium falciparum var genes in malaria in pregnancy. Mol. Microbiol. 2004;53:1011–1019. doi: 10.1111/j.1365-2958.2004.04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howell DP, Samudrala R, Smith JD. Disguising itself - insights into Plasmodium falciparum binding and immune evasion from the DBL crystal structure. Mol. Biochem. Parasitol. 2006;148:1–9. doi: 10.1016/j.molbiopara.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Howell DP, Levin EA, Springer AL, Kraemer SM, Phippard DJ, Schief WR, Smith JD. Mapping a common interaction site used by Plasmodium falciparum duffy binding-like domains to bind diverse host receptors. Mol. Micro. 2007;67:78–87. doi: 10.1111/j.1365-2958.2007.06019.x. [DOI] [PubMed] [Google Scholar]
- 53.Abi-Rached L, Dorighi K, Norman PJ, Yawata M, Parham P. Episodes of natural selection shaped the interactions of IgA-Fc with FcalphaRI and bacterial decoy proteins. J. Immunol. 2007;178:7943–7954. doi: 10.4049/jimmunol.178.12.7943. [DOI] [PubMed] [Google Scholar]
- 54.Ramsland PA, Willoughby N, Trist HM, Farrugia W, Hogarth PM, Fraser JD, Wines BD. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc. Natl. Acad. Sci. U S A. 2007;104:15051–15056. doi: 10.1073/pnas.0706028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowe AJ, Rogerson SJ, Raza A, Moulds JM, Kazatchkine MD, Marsh K, Newbold CI, Atkinson JP, Miller LH. Mapping of the region of complement receptor (CR) 1 required for P. falciparum rosetting and demonstration of the importance of CR1 in resetting in field isolates. J. Immunol. 2000;165:6341–6346. doi: 10.4049/jimmunol.165.11.6341. [DOI] [PubMed] [Google Scholar]
- 56.Tas SW, Klickstein LB, Barbashov SF, Nicholson-Weller A. C1q and C4b bind simultaneously to CR1 and additively support erythrocyte adhesion. J. Immunol. 1999;163:5056–5063. [PubMed] [Google Scholar]
- 57.Gaboriaud C, Thielens NM, Gregory LA, Rossi V, Fontecilla-Camps JC, Arlaud GJ. Structure and activation of the C1 complex of complement: unraveling the puzzle. Trend. Immunol. 2004;25:368–373. doi: 10.1016/j.it.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Corran PH, O’Donnell RA, Todd J, Uthaipibull C, Holder AA, Crabb BS, Riley EM. The fine specificity, but not the invasion inhibitory activity, of 19-kilodalton merozoite surface protein 1-specific antibodies is associated with resistance to malarial parasitemia in a cross-sectional survey in The Gambia. Infect. Immun. 2004;72:6185–6189. doi: 10.1128/IAI.72.10.6185-6189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J. Biol. Chem. 1979;254:2865–2874. [PubMed] [Google Scholar]
- 60.Wiersma EJ, Shulman MJ. Assembly of IgM. The role of disulfide bonding and non-covalent interactions. J. Immunol. 1995;154:5265–5272. [PubMed] [Google Scholar]
- 61.Kabat EA, Wu TT, Reid-Miller M, Perry H, Gottesman KS. Sequences of proteins of immunological interest. 4TH Ed. U.S. Department of Health and Human Services, National Institutes of Health; Washington D.C.: 1987. [Google Scholar]
- 62.Wan T, Beavil RL, Fabiane SM, Beavil AJ, Sohi MK, Keown M, Young RJ, Henry AJ, Owens RJ, Gould HJ, Sutton BJ. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nat. Med. 2002;3:681–686. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]