Abstract
Background:
Disrupted in Schizophrenia 1 (DISC1) is currently one of the most interesting candidate genes for major mental illness, having been demonstrated to associate with schizophrenia, bipolar disorder, major depression, autism, and Asperger's syndrome. We have previously reported a DISC1 haplotype, HEP3, and an NDE1 spanning tag haplotype to associate to schizophrenia in Finnish schizophrenia families. Because both DISC1 and NDE1 display association in our study sample, we hypothesized that other genes interacting with DISC1 might also have a role in the etiology of schizophrenia.
Methods:
We selected 11 additional genes encoding components of the “DISC1 pathway” and studied these in our study sample of 476 families including 1857 genotyped individuals. We performed single nucleotide polymorphism (SNP) and haplotype association analyses in two independent sets of families. For markers and haplotypes found to be consistently associated in both sets, the overall significance was tested with the combined set of families.
Results:
We identified three SNPs to be associated with schizophrenia in PDE4D (rs1120303, p = .021), PDE4B (rs7412571, p = .018), and NDEL1 (rs17806986, p = .0038). Greater significance was observed with allelic haplotypes of PDE4D (p = .00084), PDE4B (p = .0022 and p = .029), and NDEL1 (p = .0027) that increased or decreased schizophrenia susceptibility.
Conclusions:
Our findings with other converging lines of evidence support the underlying importance of DISC1-related molecular pathways in the etiology of schizophrenia and other major mental illnesses.
Keywords: DISC1, endophenotype, NDEL1, PDE4B, PDE4D, schizophrenia
Disrupted in Schizophrenia 1 (DISC1) is one of the most promising candidate genes for schizophrenia. It was identified as disrupted by a balanced translocation [t(1;11)(q42.1;q14.3)] in a large Scottish family with high prevalence of schizophrenia and other psychiatric disorders (1,2). Independent evidence for DISC1 was found by our work with Finnish cohorts. Initially we found strong and replicated linkage to 1q42 with markers intragenic of DISC1 (3,4). We have since reported an association between allelic haplotypes of DISC1 and schizophrenia (5,6), bipolar disorder (7), autism, and Asperger's syndrome (8). Further studies of the most robust of these haplotypes identified in the original schizophrenia study have implicated that it predisposes to schizophrenia especially in male subjects (6) and associates with poorer visual working memory performance and reduced grey matter volume in the prefrontal cortex (9,10). To date a number of groups have reported association findings between DISC1 and several neuropsychiatric disorders (7,9,11-20) and neurocognitive (21-23) and neuroimaging (9,24) phenotypes. The DISC1 has also been strongly linked to neuronal development on the basis of numerous indirect studies (9,25-27).
The DISC1 is considered a multifunctional “hub” for many protein interactions acting along several pathways (28). Proteins have been recognized as binding to DISC1 through yeast two-hybrid screens or co-immunoprecipitation experiments (26,29-32). A number of these have since been identified as promising candidates for roles in the etiology of schizophrenia and other mental illnesses. We have previously reported an association between NDE1 and schizophrenia in Finnish schizophrenia families (33). Furthermore, NDE1 and NDEL1 have displayed significant interplay with DISC1 that associates with the disorder (34). The PDE4B was initially recognized as a candidate for schizophrenia at a translocation breakpoint between chromosomes 1 and 16 in individuals with schizophrenia (35). Since, several groups have reported association findings between PDE4B and schizophrenia (36-38). The FEZ1 has been reported to associate with schizophrenia in the Japanese population (39). This evidence has led to the concept of the “DISC1 pathway”, a hypothesis that proposes that disruption of the pathways DISC1 is involved in provides risk to mental illness, not just disruption of DISC1 itself (29,39,40).
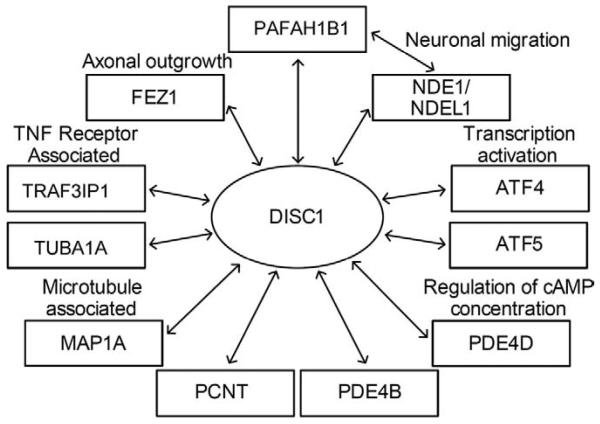
In the present study, we aimed to investigate DISC1 binding partners as potential schizophrenia susceptibility genes in 476 Finnish families ascertained for schizophrenia. We included 11 genes coding for proteins convincingly reported to bind to and interact with DISC1 (18,40). These included NDEL1, PDE4B, and FEZ1 mentioned in the preceding text. Additionally we included PDE4D, PCNT, MAP1A, PAFAH1B1, TUBA1A, TRAF3IP1, ATF4, and ATF5 (Figure 1).
Figure 1.
DISC1 binding partners included in the study with partners grouped by common functions. DISC1, Disrupted in Schizophrenia 1; TNF, tumor necrosis factor; cAMP, cyclic adenosine monophosphate.
Methods and Materials
Sample
In this study we used the same Finnish schizophrenia family sample that has previously been used in DISC1-related linkage and association analyses in Finland (33), plus an additional set of 18 families. The sample collection method has remained un-changed over the years. The individuals with schizophrenia (probands) are identified from three nationwide data sources: the hospital discharge, disability pension, and reimbursed medication registers (41). The first-degree family members of each proband were thereafter identified through the Population Register Centre, enabling the construction of pedigrees. Personal data recorded in the Population Information System are maintained by the Population Register Centre, and local register offices include personal identity code, family relations, and date of birth and death (if applicable). Altogether 33,371 individuals born between 1940 and 1976 and diagnosed with schizophrenia (according to the ICD-8, DSM-III-R, or ICD-10 classifications) between 1969 and 1998 have been identified from these registers (33). Family information has been collected from the Finnish population register, and pedigrees have been constructed. This study now includes 476 nuclear families that include a total 2756 individuals, of which 1857 have been genotyped in this study. The lifetime diagnosis for each case in the study sample has been evaluated according to DSM-IV criteria independently by two psychiatrists (42). Concordance between these two psychiatrists is high (κ values range from 95% to 99%, depending on liability class). However, in the case of a disagreement, a third psychiatrist estimated lifetime diagnosis, and the consensus best-estimate lifetime diagnosis was made. In addition to the proband with schizophrenia, family members with other psychiatric illnesses were also identified. It is therefore possible to define affection with increasingly inclusive liability classes (LC). The LC1 consists of individuals diagnosed with schizophrenia, LC2 adds individuals diagnosed with schizoaffective disorder to the sample, LC3 adds individuals affected with schizophrenia spectrum disorders (schizoid, schizotypal and paranoid personality disorders, schizophreniform, delusional and brief psychotic disorder, and psychosis not otherwise specified) (33), and LC4 adds individuals with bipolar affective disorder or major depression with and without the presence of psychosis. In this study we have restricted our end-state diagnosis phenotype to LC3. It has been shown that in families ascertained for schizophrenia the genetic liability for disorders in both LC2 and LC3 is increased (43), whereas the disorders in LC4—despite having some overlap in genetic risk—display the same liability as the general population background (43-45). According to this LC3 criterion our study includes a total of 886 affected individuals, of which 725 are genotyped.
In this present study, the family sample was randomly split into two non-overlapping sub-samples to be analyzed as exploratory and replication data sets. The exploratory set contained 171 families, and the replication set contained 305 families. The combined sample set included both sample sets. In addition, a sample of 57 anonymous Finnish parent-offspring trios representing a random sample of the Finnish population was used as a control sample.
Neuropsychological Test Methods
For a sub-sample of 186 families, a neuropsychological test battery has been administered. The tests were administered in a fixed order by experienced psychologists or specially trained psychiatric nurses, and the scoring of the tests was performed by experienced psychologists (46). The following variables were used in the analyses. First, from the Wechsler Memory Scale—Revised (WMS-R) (47) we included Verbal and Visual Span forward subtests as measures of auditory and visual attention, respectively. The respective backward condition of these WMS-R subtests was used as measures of verbal and visual working memory. Second, from the California Verbal Learning Test (CVLT) (48), total recall from trials 1–5 were used as a measure of learning. The other included CVLT variables were semantic clustering as a measure of learning strategy and short-delay and long-delay recall. Third, from the Wechsler Adult Intelligence Scale—Revised (WAIS-R) (49), the Vocabulary subtest was included as an estimate of basic ability, and the Digit Symbol subtest was included as a measure of information processing speed. The selected traits are either direct measures of learning and memory—such as short-delay memory, long-delay memory, and verbal learning—or highly relevant to learning process, such as auditory attention, visual working memory, verbal working memory, verbal attention, and semantic clustering. The test scores were normally distributed in our sample.
Genotyping Methods
The SNPs were selected from the international HapMap project database, build #16, phase 1 (50). The linkage disequilibrium (LD) structure for the 11 candidate genes was defined with all the SNPs with a minor allele frequency >5% in the population of European descent (CEU, Utah residents with ancestry from Northern and Western Europe). We used the Haploview program (51) and the solid spine of LD criterion (D′ > .8). The LD blocks with a Hedricks multiallelic (52) D′ ≥ .9 were combined. Haplotype tagging SNPs were prioritized to obtain the optimal coverage of the genes (number of SNPs selected = 91). Additional SNPs from the HapMap and Perlegen (53) databases were selected to provide coverage if genotypes of tagging SNPs were not of high enough quality (number of SNPs selected = 57; total number of SNPs = 148). Genotyping was performed with the Sequenom (San Diego, California) platform according to manufacturer's recommendations (54). The SNPs were later rejected if they had a genotyping success rate <80% (six SNPs). None of the genotyped SNPs showed deviation for Hardy-Weinberg disequilibrium (HWE) (p > .001). After these quality control measures 142 SNPs were included in the analyses (Table 3 in Supplement 1).
Statistical Methods
The SNP genotypes were checked for Mendelian errors with Pedcheck (55). In case of an error in any genotype, all the genotypes for that marker in the relevant family were discarded (Mendelian error rate: .000076/genotype).
Association tests between affection status and gene variants were performed with a two-stage study design described in the preceding text. Such a design has been demonstrated to increase the identification of true positives while also decreasing the rate of false negative associations compared with using the more traditional but overly conservative Bonferroni correction (56,57).
Two-point analyses were performed with the program Pseudomarker (58). This program performs linkage as well as LD analyses on samples of mixed form (families and trios being combined here), correcting for the effect of linkage on the association tests, and is able to deal with cases where parental genotypes are not known (58).
To perform haplotype analyses, we identified haplotype blocks according to the LD structure defined by Haploview, as described in the preceding text. For NDEL1, TRAF3IP1, ATF4, ATF5, MAP1A, TUBA1A, FEZ1, and PCNT, only one haplotype block for each gene was present. The PAFAH1B1, PDE4B, and PDE4D included 2, 10, and 17 LD blocks, respectively. In ATF4, PDE4B, and PDE4D, we were able to tag 0, 8, and 16 blocks, respectively; in other genes, all the blocks were tagged. Generally, the LD structure in our sample correlates well with the LD structure defined with the HapMap CEU population. However, for NDEL1, the LD pattern in the CEU population predicts two LD blocks; meanwhile, according to our sample, one block was predicted. Hedricks D′ between the blocks defined according to CEU population in our sample is 1.00 (compared with .39 in CEU population). Therefore we analyzed these blocks as one.
Haplotype association analyses of end-state diagnosis were performed with the two-stage method described previously. We compared the frequencies of the haplotypes in the affected offspring of the schizophrenia families with that of the founders of the control trios. These frequencies were defined by estimating each individual's most likely haplotypes with Simwalk2, a Markov chain Monte Carlo (MCMC) and simulated annealing program (59). Only haplotype block tagging SNPs were taken into account when constructing the haplotypes. Haplotype association analyses of end-state diagnosis were performed with the χ2 test, testing each possible haplotype against all other haplotypes combined in a 2 × 2 table. A global test was performed in a 2 × n (n = number of alleles) table for the haplotype blocks including one or more haplotypes displaying significant association with affection status in both sample sets. Haplotypes with frequencies <5% in both the case and control samples were combined together to avoid the deviation that these rare haplotypes might have on the result.
The SNPs and haplotypes showing evidence for association with end state diagnosis (p < .05) in our two-stage design were tested for the association with quantitative neurocognitive traits (622 individuals available with quantitative data). These analyses were performed with the QTDT program and the orthogonal model (60,61). Here the haplotypes were re-coded to form a “bi-allelic” marker, so as to test the hypothesized variant against all others combined. We used age, gender, and affection status according to LC3 as covariates in our analysis.
The study has been accepted by the Ministry of Social Affairs and Health (Finland) and institutional review boards. All subjects have provided written informed consent.
Results
We analyzed SNP markers and haplotypes for their association with schizophrenia with a two-stage study design with random sampling of our original 476 Finnish families. Association analyses between affection status and SNPs and haplotypes were first performed in a sample of 171 families. The SNPs and haplotypes displaying evidence for association with p values <.05 proceeded to analyses in a second sample consisting of 305 independent families. Markers and haplotypes passing the replication criteria in the second stage were analyzed with both sample sets combined.
In stage one, 14 SNPs and 19 haplotypes in NDEL1, PDE4B, PDE4D, PAFAH1B1, MAP1A, and TRAF3IP1 met our criteria for progressing to stage two. No markers or haplotypes in FEZ1, PCNT, TUBA1A, ATF4, and ATF5 met this criterion. In stage two, associations with three markers and four haplotypes of NDEL1, PDE4D, and PDE4B were replicated, the same allele showing association with the disorder in both sample sets. These markers and haplotypes were then tested for their overall significance in the combined sample set (Table 1) (Tables 1 and 2 in Supplement 1).
Table 1.
Summary of the Results in the Combined Sample of the SNPs and Haplotypes that Passed Replication in Two Stages
| SNPs |
Haplotypes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Block | SNP | p | F(Affected) | F(Control) | Global p | Alleles | p | F(Affected) | F(Control) |
| NDEL1 | 1 | rs17806986 | .0038 | .27 | .37 | .033 | CGCG | .0027 | .12 | .19 |
| rs1391768 | nt | nt | nt | |||||||
| rs1391766 | nt | nt | nt | |||||||
| rs3817003 | nt | nt | nt | |||||||
| PDE4B | 4 | rs4503327 | nt | nt | nt | .0060 | CCC | .029 | .28 | .21 |
| rs2503222 | nt | nt | nt | |||||||
| rs6588186 | nt | nt | nt | CTT | .0022 | .022 | .058 | |||
| PDE4B | 5 | rs10158178 | nt | nt | nt | nt | nt | |||
| rs7412571 | .018 | .45 | .35 | |||||||
| rs599235 | nt | nt | nt | |||||||
| rs2069278 | nt | nt | nt | |||||||
| PDE4D | 15 | rs13190249 | nt | nt | nt | .0034 | GGACA | .00084 | .40 | .28 |
| rs1120303 | .021 | .13 | .19 | |||||||
| rs921942 | nt | nt | nt | |||||||
| rs10805515 | nt | nt | nt | |||||||
| rs10514862 | nt | nt | nt | |||||||
The p values for the significant single nucleotide polymorphisms (SNPs) and haplotypes are provided with respective allele frequencies (F). The SNPs and haplotypes displaying no evidence for association in stage 1 and stage 2 were not tested (nt).
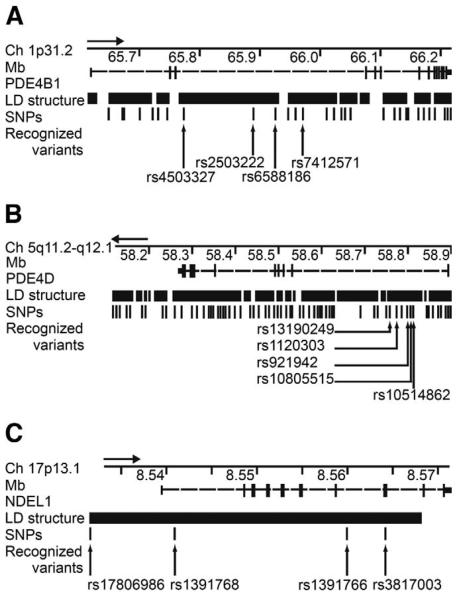
Those SNPs and haplotypes that passed through the two stages are as follows. In PDE4D, rs1120303 showed association with an overall p value of .021 (the minor allele frequency [MAF] in schizophrenia family founders, MAFscz = .13 vs. the minor allele frequency in control family founders, MAFcontrol = .19; Block 15, 65 kb; Minor allele = T) in the combined dataset. The haplotype block in which this SNP is located also provided evidence of association with a haplotype comprising the GGACA alleles of SNPs rs13190249, rs1120303, rs921942, rs10805515, and rs10514862 being significantly over-represented in affected individuals (p = .00084, frequency in affected individuals, Faffected = .40, frequency in control subjects, Fcontrol = .28, global p value .0034). The SNP rs7412571 in PDE4B provided overall association with a p value of .018 (MAFscz = .45, MAFcontrol = .35; Block 5, 76 kb; Minor allele = T). Although the haplotype block in which the rs7412571 SNP is located did not add any further evidence of association, its neighboring block does provide such evidence. Block 4 (104 kb) comprises the SNPs rs4503327, rs2503222, and rs6588186, with two alleles displaying evidence for association in two sets of families and in the combined dataset. The haplotype consisting of the CCC allele was over-represented in affected offspring with an overall p value of .029 (Faffected = .28, Fcontrol = .21), whereas the CTT haplotype of this block was under-represented with overall p value of .0022 (Faffected = .022, Fcontrol = .058). The overall global p value for this block in the combined sample was .0060. In NDEL1, the rs17806986 SNP, located close to the 5′ end of the gene, showed association with an overall p value of .0038 (MAFscz= .27, MAFcontrol = .37; Minor allele = C). The CGCG haplotype of SNPs rs17806986, rs1391768, rs1391766, and rs3817003 extending over the whole gene was under-represented in affected individuals (p = .0027, Faffected = .12, Fcontrol = .19, global p = .033) (Figure 2).
Figure 2.
Schematics show the regions of interest on chromosomes 1 (A),5 (B), and 17 (C), where PDE4B, PDE4D, and NDEL1 are located, respectively, according to UCSC Genome Browser build 16 (released July 2003). Genotyped single nucleotide polymorphisms (SNPs) are indicated with vertical lines. Linkage disequilibrium (LD) structure describes the haplotype blocks that were tested for association. For the SNPs displaying association and SNPs included in the associating haplotype blocks, the rs-numbers are provided. Arrows above the schematics indicate direction from 5′ to 3′.
Previous reports of DISC1 (6) and the DISC1 pathway genes (33,38) have displayed distinct gender differences in their allelic association. We tested for any potential gender differences in allele frequencies of the recognized associating variants in our sample and could detect no differentiating effects between female and male offspring in these Finnish families (Table 2).
Table 2.
Examination of the Differences Between Numbers of Observations of the Recognized Variants and Affected Male and Female Subjects
| F(Allele+) |
F(Allele−) |
F(Haplotype+) |
F(Haplotype−) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Block | SNP | p | M | F | M | F | Haplotype | p | M | F | M | F |
| NDEL1 | 1 | rs17806986 | .57 | .28 | .29 | .72 | .71 | CGCG | .10 | .11 | .13 | .89 | .87 |
| PDE4B | 4 | nt | CCC | .227 | .29 | .28 | .71 | .72 | |||||
| CTT | .47 | .02 | .02 | .98 | .98 | ||||||||
| PDE4B | 5 | rs7412571 | .068 | .56 | .61 | .44 | .39 | nt | |||||
| PDE4D | 15 | rs1120303 | .10 | .15 | .12 | .85 | .88 | GGACA | .10 | .39 | .42 | .61 | .58 |
Affected male and female subjects were divided in groups on the basis of the single nucleotide polymorphism (SNP) and haplotype variants recognized. The testing for deviation in the haplotype and allelic distribution between male and female subjects was performed with the χ2 test. Observations for associated SNP and haplotype are indicated for male (M) and female (F) subjects. F(Allele+) and F(Haplotype+) indicate frequencies of the associated variant, whereas F(Allele−) and F(Haplotype−) indicate frequencies of other than the tested variant. The SNPs and haplotypes displaying no evidence for association in stage 1 or stage 2 were not tested (nt).
Because some of the affected individuals in the sample are siblings, we also tested the identified haplotype variants with only one randomly selected affected offspring/family to account for the possible confounding effect of linkage in the obtained results. Three of the four haplotypes remained significantly associated with the following p values: PDE4D = .0055, NDEL1 = .0018, and PDE4B protective and risk = .0025 and .070, respectively.
Because DISC1 has been previously shown to be associated with visual working memory in these families (10) and with other quantitative neurocognitive traits in other sample sets, we wanted to test these newly identified variants for their association with such traits. In addition to visual working memory, we included several learning- and memory-related variables in the analyses. Due to the reduction of the neuropsychologically tested sample size, we tested all the recognized SNPs and haplotypes only in the combined sample. Of the seven variants, none associated significantly (p < .05) with any of the nine tested traits.
Discussion
We show here that three genes involved in the same intracellular pathways with DISC1 associate significantly with schizophrenia in a Finnish family sample. We identified SNP and haplotype variants in PDE4B, PDE4D, and NDEL1 that were either under-represented or over-represented in families ascertained for schizophrenia. The minor allele of rs7412571 in PDE4B was significantly over-represented in affected individuals. The neighboring haplotype block displayed both an over-represented (“risk”) and an under-represented (“protective”) allele. However, the over-represented haplotype did not remain significantly associated when we tested only one affected offspring/family. This would suggest that the under-represented haplotype is of greater importance. The PDE4D displayed significant association with schizophrenia with an SNP whose minor allele was significantly under-represented in individuals with schizophrenia. A haplotype including the major allele of this same SNP was, consistent with this finding, significantly over-represented in the cases. The minor allele at the NDEL1 rs17806986 SNP displayed significant under-representation in schizophrenia. This SNP was a part of the NDEL1 gene-spanning haplotype that was also significantly under-represented in affected individuals.
Fatemi et al. have recently reported on an association between schizophrenia and the major allele of SNP rs1354064 in PDE4B that is located in the same LD block we detected association with (36). We had not included this SNP in this study, but our SNP rs2503222 tags the variation of this SNP (r2 = .82). Even though we did not see evidence for this SNP alone, our haplotype finding further supports the importance of this locus. Furthermore, a recent report by Pickard et al. (38) also noted a haplotype within PDE4B in the Scottish population to be protective against schizophrenia. Although this haplotype is located in an independent LD block approximately 70 kb toward the 3′ end compared with our haplotype finding, we were able to detect association in the same region as the Scottish haplotype, with the SNP rs7412571, suggesting that the same region might also be involved in the etiology of schizophrenia in the Finnish population. The minor allele of this SNP was over-represented in patients with schizophrenia. This SNP tags (r2 = 1) two SNPs included in the Scottish haplotype. The major alleles of these two SNPs were included in the protective haplotypes in the Scottish study, consistent with our finding. Two previous reports have highlighted protective SNPs recognized close to the 3′ end of the gene, although we did not detect any evidence for association in this region (36,37).
Burdick et al. (34) have reported on an association of NDEL1 with schizophrenia. They recognized an NDEL1 spanning haplotype being under-represented in affected individuals and further reported that the G allele of SNP rs1391768 was over-represented in affected individuals. In contrast, in our sample this G allele is part of a NDEL1 spanning putative protective haplotype.
The NDEL1 has been a strong schizophrenia susceptibility candidate gene (27,62) on the basis of its role in neuronal migration and neuronal outgrowth (63). Furthermore, it is part of the same Dynein signaling pathway as PAFAH1B1, another DISC1 binding partner (64), and RELN (65), a protein that is also implicated in the etiology of schizophrenia (66). Previously, NDEL1 and its homologue NDE1 have been strongly linked to prenatal and early age neuronal development (63,64,67), but recent findings suggest a wider role for NDEL1 (68). According to recent findings, DISC1 has a central role in adult neurogenesis along with NDEL1 in mice. Our finding proposes the involvement of these genes also in the etiology of adult onset disorders. Furthermore, involvement of these genes in the etiology of schizophrenia would support the hypothesis that the vulnerability for developing these disorders might originate during embryonic development.
We are the first to report direct association between PDE4D and schizophrenia. Interestingly PDE4D (5q11.2-5q12.1) was located close to a linkage peak (chromosome 5q12.3, LOD = 2.59) in our previous genome-wide linkage scan when the analysis was conditioned by absence of previously recognized DISC1 risk haplotype HEP3 (33).
Our findings thus support the previous reports especially for PDE4B. Even though our findings are encouraging, naturally further studies are needed to establish the relevance of these genes in the etiology of psychiatric disorders.
It is well-established that patients with schizophrenia have several specific memory deficits (69). Given the evidence supporting the involvement of PDE4B and PDE4D in the etiology of schizophrenia (35-38) and their supposed involvement in memory functions (70,71), it was of interest to investigate whether PDE4D and PDE4B would, in addition to association with schizophrenia, display association with learning- and memory-related quantitative traits. However, no significant association was detected with the recognized variants. Yet this might be due to the reduced sample size in the quantitative trait analyses compared with association tests using affection status.
Because all these genes are shown to biologically interact with DISC1, it would have been interesting to investigate whether these genes demonstrate further combined effects that increase schizophrenia susceptibility within the DISC1 pathways. However, due to the potentially large number of interactions in these pathways, our sample size remains too small for testing this meaningfully (6,33). For the same reason, testing for interaction between the most significant Finnish DISC1 haplotype HEP3 (frequency = .088) and the variants recognized in the current study (frequencies ranging from .058 to .37) remains unreliable, because only .51%–3.3% of the studied individuals would carry combinations of these recognized variants. Yet, as the variants detected here are associated with schizophrenia independent of DISC1, it demonstrates that alterations in components on the DISC1 pathway other than just DISC1 itself might also influence schizophrenia etiology.
Emerging evidence supports a wider role for DISC1 in the development of various psychiatric and neurodevelopmental disorders, with the latest observation being an association with the early-age neurodevelopmental disorders autism and Asperger's syndrome (8). Interestingly, altered expression levels at PDE4B were recently reported within individuals affected with autism (72). The PDE4B has also been shown to associate with major depression (73). Furthermore, PDE4D has been reported to be associated with neuroticism, a psychological trait evidently highly related to major depression and anxiety (74). This would suggest that, like DISC1, the other genes in the DISC1 pathways might play a general role in the development of neurodevelopmental disorders rather than being specific to schizophrenia, potentially being involved in a wider spectrum of psychiatric illness.
We have taken the novel approach of studying candidate genes for schizophrenia on the basis of the known molecular interactions of a previously identified susceptibility gene. This “guided candidate gene approach” is theoretically attractive, because it is plausible that variants in genes involved in the same molecular pathways can cause similar phenotypes. The DISC1 is an excellent starting point on which to anchor such an effort. First, it has been shown to be associated with schizophrenia both in our study sample and others, and there is an abundance of independent multimodal findings implicating this gene in schizophrenia. The DISC1 is known to function as a “hub” for many protein interactions acting along several pathways supporting this approach for this particular gene. With this approach we observed significant association between schizophrenia and two memory-related phosphodiesterase genes (PDE4B and PDE4D) as well as one neurodevelopmentally important peptidase gene (NDEL1). Combined with our previous observation of association with NDE1, this provides strong evidence supporting the DISC1 related pathways in the susceptibility of schizophrenia and should inspire further research into these pathways.
Supplementary Material
Acknowledgments
This work was funded in part by The Centre of Excellence of the Academy of Finland and Biocentrum Helsinki Foundation (LP). The author LT is supported by Sigrid Juselius foundation. The author WH is a long-term EMBO Research Fellow and was also supported by the Finnish Cultural Foundation Piippa Stiina Immonen Grant. The authors AL and AT-H are supported by Academy of Finland post-doctoral fellowships. The author JS is supported by an Academy of Finland research fellowship. The author LP is on the board of Directors of Orion Pharma and Rocla. The authors PL, TP, JE and JL did not report any biomedical financial interests.
We would like to acknowledge Sarah Whittall for help in producing the figures.
Footnotes
The authors declare they have no potential conflicts of interest.
Supplementary data associated with this article can be found, in the online version.
References
- 1.Clair DM, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, et al. Association within a family of balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood DH, Fordyce A, Walker MT, Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: Clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, et al. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet. 2001;10:1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- 4.Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, et al. Genome-wide scan for schizophrenia in the Finnish population: Evidence for a locus on chromosome 7q22. Hum Mol Genet. 2000;9:1049–1057. doi: 10.1093/hmg/9.7.1049. [DOI] [PubMed] [Google Scholar]
- 5.Ekelund J, Hennah W, Hiekkalinna T, Parker A, Meyer J, Lonnqvist J, et al. Replication of 1q42 linkage in Finnish schizophrenia pedigrees. Mol Psychiatry. 2004;9:1037–1041. doi: 10.1038/sj.mp.4001536. [DOI] [PubMed] [Google Scholar]
- 6.Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, Haukka J, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 7.Palo OM, Antila M, Silander K, Hennah W, Kilpinen H, Soronen P, et al. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet. 2007;16:3517–3528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- 8.Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- 9.Cannon TD, Hennah W, van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 10.Hennah W, Tuulio-Henriksson A, Paunio T, Ekelund J, Varilo T, Partonen T, et al. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol Psychiatry. 2005;10:1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- 11.Chen QY, Chen Q, Feng GY, Lindpaintner K, Wang LJ, Chen ZX, et al. Case-control association study of Disrupted-in-Schizophrenia-1 (DISC1) gene and schizophrenia in the Chinese population. J Psychiatr Res. 2007;41:428–434. doi: 10.1016/j.jpsychires.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 12.DeRosse P, Hodgkinson CA, Lencz T, Burdick KE, Kane JM, Goldman D, et al. Disrupted in schizophrenia 1 genotype and positive symptoms in schizophrenia. Biol Psychiatry. 2007;61:1208–1210. doi: 10.1016/j.biopsych.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- 14.Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, Lipsky RH, et al. Disrupted in schizophrenia 1 (DISC1): Association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004;75:862–872. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kockelkorn TT, Arai M, Matsumoto H, Fukuda N, Yamada K, Minabe Y, et al. Association study of polymorphisms in the 5′ upstream region of human DISC1 gene with schizophrenia. Neurosci Lett. 2004;368:41–45. doi: 10.1016/j.neulet.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 16.Maeda K, Nwulia E, Chang J, Balkissoon R, Ishizuka K, Chen H, et al. Differential expression of disrupted-in-schizophrenia (DISC1) in bipolar disorder. Biol Psychiatry. 2006;60:929–935. doi: 10.1016/j.biopsych.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Qu M, Tang F, Yue W, Ruan Y, Lu T, Liu Z, et al. Positive association of the Disrupted-in-Schizophrenia-1 gene (DISC1) with schizophrenia in the Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007;144:266–270. doi: 10.1002/ajmg.b.30322. [DOI] [PubMed] [Google Scholar]
- 18.Thomson PA, Wray NR, Millar JK, Evans KL, Hellard SL, Condie A, et al. Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry. 2005;10:657–668. doi: 10.1038/sj.mp.4001669. 616. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F, Sarginson J, Crombie C, Walker N, Clair D, Shaw D. Genetic association between schizophrenia and the DISC1 gene in the Scottish population. Am J Med Genet B Neuropsychiatr Genet. 2006;141:155–159. doi: 10.1002/ajmg.b.30274. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Tochigi M, Ohashi J, Maeda K, Kato T, Okazaki Y, et al. Association study of the DISC1/TRAX locus with schizophrenia in a Japanese population. Schizophr Res. 2005;79:175–180. doi: 10.1016/j.schres.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 21.Thomson PA, Harris SE, Starr JM, Whalley LJ, Porteous DJ, Deary IJ. Association between genotype at an exonic SNP in DISC1 and normal cognitive aging. Neurosci Lett. 2005;389:41–45. doi: 10.1016/j.neulet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Liu YL, Fann CS, Liu CM, Chen WJ, Wu JY, Hung SI, et al. A single nucleotide polymorphism fine mapping study of chromosome 1q42.1 reveals the vulnerability genes for schizophrenia, GNPAT and DISC1: Association with impairment of sustained attention. Biol Psychiatry. 2006;60:554–562. doi: 10.1016/j.biopsych.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Burdick KE, Hodgkinson CA, Szeszko PR, Lencz T, Ekholm JM, Kane JM, et al. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- 24.Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Austin CP, Ky B, Ma L, Morris JA, Shughrue PJ. Expression of Disrupted-In-Schizophrenia-1, a schizophrenia-associated gene, is prominent in the mouse hippocampus throughout brain development. Neuroscience. 2004;124:3–10. doi: 10.1016/j.neuroscience.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, et al. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- 27.Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- 28.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, et al. Disrupted in Schizophrenia 1 Interactome: Evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 29.Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- 30.Miyoshi K, Asanuma M, Miyazaki I, Diaz-Corrales FJ, Katayama T, Tohyama M, et al. DISC1 localizes to the centrosome by binding to kendrin. Biochem Biophys Res Commun. 2004;317:1195–1199. doi: 10.1016/j.bbrc.2004.03.163. [DOI] [PubMed] [Google Scholar]
- 31.Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: Regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 32.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, et al. Disrupted-in-Schizophrenia-1 (DISC-1): Mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennah W, Tomppo L, Hiekkalinna T, Palo OM, Kilpinen H, Ekelund J, et al. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 34.Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: Evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 36.Fatemi SH, King DP, Reutiman TJ, Folsom TD, Laurence JA, Lee S, et al. PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophr Res. 2008;101:36–49. doi: 10.1016/j.schres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 37.Numata S, Ueno SI, Iga JI, Song H, Nakataki M, Tayoshi S, et al. Positive association of the PDE4B (phosphodiesterase 4B) gene with schizophrenia in the Japanese population. J Psychiatr Res. 2008;43:7–12. doi: 10.1016/j.jpsychires.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Pickard BS, Thomson PA, Christoforou A, Evans KL, Morris SW, Porteous DJ, et al. The PDE4B gene confers sex-specific protection against schizophrenia. Psychiatr Genet. 2007;17:129–133. doi: 10.1097/YPG.0b013e328014492b. [DOI] [PubMed] [Google Scholar]
- 39.Yamada K, Nakamura K, Minabe Y, Iwayama-Shigeno Y, Takao H, Toyota T, et al. Association analysis of FEZ1 variants with schizophrenia in Japanese cohorts. Biol Psychiatry. 2004;56:683–690. doi: 10.1016/j.biopsych.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Hennah W, Thomson P, Peltonen L, Porteous D. Genes and schizophrenia: Beyond schizophrenia: The role of DISC1 in major mental illness. Schizophr Bull. 2006;32:409–416. doi: 10.1093/schbul/sbj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lichtermann D, Hovatta I, Terwilliger JD, Peltonen L, Lonnqvist J. Concordance for sex and the pseudoautosomal gene hypothesis revisited: No evidence of increased sex concordance in a nationwide Finnish sample of siblings with paternally derived schizophrenia. Am J Psychiatry. 1998;155:1365–1375. doi: 10.1176/ajp.155.10.1365. [DOI] [PubMed] [Google Scholar]
- 42.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Press; Washington DC: 1994. [Google Scholar]
- 43.Tienari P, Wynne LC, Laksy K, Moring J, Nieminen P, Sorri A, et al. Genetic boundaries of the schizophrenia spectrum: Evidence from the Finnish Adoptive Family Study of Schizophrenia. Am J Psychiatry. 2003;160:1587–1594. doi: 10.1176/appi.ajp.160.9.1587. [DOI] [PubMed] [Google Scholar]
- 44.Blackwood DH, Pickard BJ, Thomson PA, Evans KL, Porteous DJ, Muir WJ. Are some genetic risk factors common to schizophrenia, bipolar disorder and depression? Evidence from DISC1, GRIK4 and NRG1. Neurotox Res. 2007;11:73–83. doi: 10.1007/BF03033484. [DOI] [PubMed] [Google Scholar]
- 45.Shifman S, Bronstein M, Sternfeld M, Pisante A, Weizman A, Reznik I, et al. COMT: A common susceptibility gene in bipolar disorder and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2004;128:61–64. doi: 10.1002/ajmg.b.30032. [DOI] [PubMed] [Google Scholar]
- 46.Tuulio-Henriksson A, Haukka J, Partonen T, Varilo T, Paunio T, Ekelund J, et al. Heritability and number of quantitative trait loci of neuro-cognitive functions in families with schizophrenia. Am J Med Genet. 2002;114:483–490. doi: 10.1002/ajmg.10480. [DOI] [PubMed] [Google Scholar]
- 47.Wechsler D. Wechsler Memory Scale—Revised (WMS-R) Manual. Psychological Corporation, Harcourt Brace Jovanovich; Cleveland, Ohio: 1987. [Google Scholar]
- 48.Delis DC, Kramer JH, Kaplan E, Ober BA. California Verbal Learning Test: Manual. Research Edition Psychological Corporation, Harcourt Brace & Company; San Antonio, Texas: 1987. [Google Scholar]
- 49.Wechsler D. Wechsler Adult Intelligence Scale—Revised. Psychological Corporation, Harcourt Brace Jovanovich; Cleveland, Ohio: 1981. [Google Scholar]
- 50.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 51.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 52.Hedrick PW. Gametic disequilibrium measures: Proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 54.Jurinke C, van den Boom D, Cantor CR, Koster H. Automated genotyping using the DNA MassArray technology. Methods Mol Biol. 2001;170:103–116. doi: 10.1385/1-59259-234-1:103. [DOI] [PubMed] [Google Scholar]
- 55.O'Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Oord EJ, Sullivan PF. A framework for controlling false discovery rates and minimizing the amount of genotyping in the search for disease mutations. Hum Hered. 2003;56:188–199. doi: 10.1159/000076393. [DOI] [PubMed] [Google Scholar]
- 57.Wen SH, Tzeng JY, Kao JT, Hsiao CK. A two-stage design for multiple testing in large-scale association studies. J Hum Genet. 2006;51:523–532. doi: 10.1007/s10038-006-0393-6. [DOI] [PubMed] [Google Scholar]
- 58.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: Joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet. 2000;66:1310–1327. doi: 10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sobel E, Lange K. Descent graphs in pedigree analysis: Applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 60.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8:545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- 62.Lipska BK, Peters T, Hyde TM, Halim N, Horowitz C, Mitkus S, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 63.Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: Implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Wynshaw-Boris A, Gambello MJ. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- 66.Wedenoja J, Loukola A, Tuulio-Henriksson A, Paunio T, Ekelund J, Silander K, et al. Replication of linkage on chromosome 7q22 and association of the regional Reelin gene with working memory in schizophrenia families. Mol Psychiatry. 2008;13:673–684. doi: 10.1038/sj.mp.4002047. [DOI] [PubMed] [Google Scholar]
- 67.Schaar BT. Cortical development deNUDEd. Neuron. 2004;44:213–214. doi: 10.1016/j.neuron.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aleman A, Hijman R, de Haan EH, Kahn RS. Memory impairment in schizophrenia: A meta-analysis. Am J Psychiatry. 1999;156:1358–1366. doi: 10.1176/ajp.156.9.1358. [DOI] [PubMed] [Google Scholar]
- 70.Davis RL, Cherry J, Dauwalder B, Han PL, Skoulakis E. The cyclic AMP system and Drosophila learning. Mol Cell Biochem. 1995;149–150:271–278. doi: 10.1007/978-1-4615-2015-3_31. [DOI] [PubMed] [Google Scholar]
- 71.Giorgi M, Modica A, Pompili A, Pacitti C, Gasbarri A. The induction of cyclic nucleotide phosphodiesterase 4 gene (PDE4D) impairs memory in a water maze task. Behav Brain Res. 2004;154:99–106. doi: 10.1016/j.bbr.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 72.Braun NN, Reutiman TJ, Lee S, Folsom TD, Fatemi SH. Expression of phosphodiesterase 4 is altered in the brains of subjects with autism. Neuroreport. 2007;18:1841–1844. doi: 10.1097/WNR.0b013e3282f16dca. [DOI] [PubMed] [Google Scholar]
- 73.Numata S, Iga JI, Nakataki M, Tayoshi S, Taniguchi K, Sumitani S, et al. Gene expression and association analyses of the phosphodiesterase 4B (PDE4B) gene in major depressive disorder in the Japanese population [published online ahead of print September 10] Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30852. [DOI] [PubMed] [Google Scholar]
- 74.Shifman S, Bhomra A, Smiley S, Wray NR, James MR, Martin NG, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2008;13:302–312. doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.