Abstract
The yeast Sir2/3/4 protein complex forms a repressive heterochromatin-like structure that confers transcriptional silencing. The complex nucleates at silencers and then spreads distally by a process that requires the NAD+-dependent deacetylase activity of Sir2 and the affinity of Sir3/4 for deacetylated histone tails. A byproduct of the Sir2 reaction, O-acetyl-ADP-ribose (OAADPr), is thought to aid spreading by binding one of the Sir proteins. We have used a protein chimera approach to reexamine the essential roles of Sir2 in silencing. We show that a Sir3 chimera bearing Hos3, an unrelated NAD+-independent histone deacetylase, substitutes for Sir2 in transcriptional repression. Sir3-Hos3 requires silencers and Sir4, consistent with the chimera operating within the Sir pathway. Furthermore, the chimera silences in strains lacking all five OAADPr-producing deacetylases, indicating that the OAADPr is not required. Silencing by an analogous Hos3 hybrid bearing the Sir2 targeting motifs shows that the motifs operate independently of the Sir2 reaction. Together, these data demonstrate that protein deacetylation is the only necessary function of Sir2 in the creation of silenced chromatin.
Keywords: yeast transcriptional silencing, sirtuin, Sir2 histone deacetylase, Hos3, O-acetyl-ADP-ribose (OAADPr)
Introduction
Transcriptional silencing is a form of transcriptional control that regulates large chromosomal domains rather than individual genes. It involves specialized chromatin structures that propagate over kilobases in budding yeast and tens of megabases in humans. The extent of propagation can vary from one cell to the next, yielding variegated patterns of gene expression that are inherited epigenetically.
Transcriptional silencing is typified by silent chromatin in yeast Saccharomyces cerevisiae. The heterochromatin-like structure, built from a complex of Sir2, Sir3 and Sir4, controls the expression of genes in subtelomeric regions and the auxiliary mating-type loci, HML and HMR (Rusché et al., 2003). Assembly of silent chromatin at these locations is directed by cis-acting regulatory sequences known as silencers that bind the multifunctional factors Rap1, Abf1, and ORC directly, and Sir1 via ORC. Sir3 and Sir4 possess an affinity for these factors, as well as the deacetylated N-terminal tails of histone H3 and H4 (Moretti and Shore, 2001; Carmen et al., 2001; Liou et al., 2005). Sir2 belongs to a large evolutionarily conserved family of nicotinamide adenine dinucleotide NAD+-dependent protein deacetylases known as the sirtuins (Sauve et al., 2006). The enzyme acts preferentially on the acetylated lysine 16 (K16) of histone H4 in vitro (Imai et al., 2000). Deacetylation of this site by Sir2 is necessary for silencing in vivo (Johnson et al., 1990; Suka et al., 2002; Kimura et al., 2002).
A prevailing view holds that silent chromatin assembles in two operationally defined steps: nucleation and spreading. The Sir complex first nucleates at silencers by associating with silencer-bound proteins. Spreading of the complex from silencers requires Sir2 enzymatic activity, which creates additional binding sites for Sir3 and Sir4 by removing acetyl groups from histone tails of adjacent nucleosomes (Luo et al., 2002; Hoppe et al., 2002; Rusché et al., 2002; figure 1A). Through iterative cycles of Sir2 deacetylation followed by Sir3/Sir4 binding, silent chromatin assembles processively to exert transcriptional repression on promoters located kilobases away from silencers.
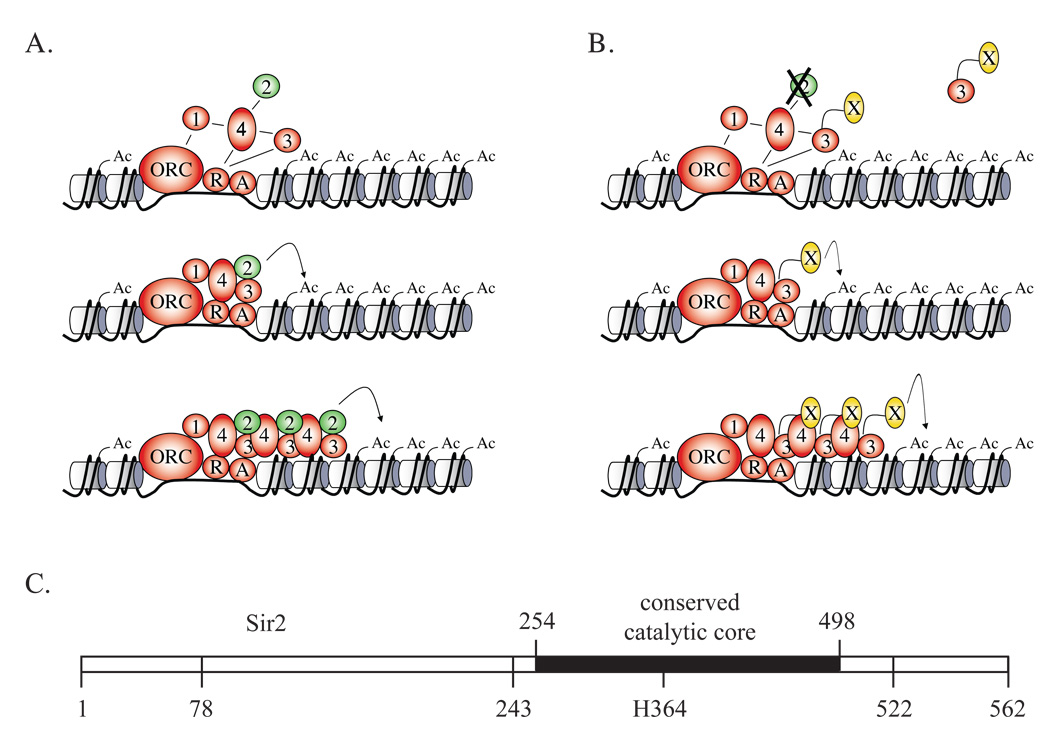
Figure 1. Experimental Strategy.
A) Nucleation and spreading of silencing proteins (adapted from Rusché et al., 2002 with permission). Nucleation (top panel) occurs when the Sir2/3/4 complex associates with silencer-bound factors. Lines between the Sir proteins (numbered) and silencer-bound factors (R = Rap1, A = Abf1) depict the network of protein-protein interactions (see references in (Rusché et al., 2003). Removal of acetyl groups (Ac) from histone tails by Sir2 facilitates binding of additional Sir2/3/4 complexes. Interactions between individual Sir2/3/4 complexes may also facilitate spreading. B) Nucleation and spreading of silencing proteins by tethering Sir3 to a heterologous deacetylase (labeled X). C) The domain structure of Sir2 and the locations of truncation endpoints used in this study. Histidine H364 is required for enzymatic activity (Imai et al., 2000). The truncation point at position 243 corresponds to the N-terminus of Sir2-Af1, a Sir2 family member whose structure has been solved (Min et al., 2001). Amino acids 1–77 are omitted in the LexA-Sir278–562 chimera.
In each reaction cycle Sir2 converts NAD+ to nicotinamide and 2’-O-acetyl-ADP-ribose (OAADPr) while deacetylating a single lysine residue (Tanner et al., 2000; Tanny and Moazed, 2001). Recent work by Moazed and colleagues has shown that OAADPr promotes association of Sir3 with preformed Sir2/Sir4 pairs (Liou et al., 2005). The binding event yields Sir complexes with super-stochiometric amounts of Sir3, which produce fibers 15–20 nm in diameter when combined with yeast oligonucleosomes (Onishi et al., 2007). On this basis it was hypothesized that OAADPr acts as a small molecule mediator of silent chromatin assembly. Understanding the precise physiological role for OAADPr, however, remains an open question.
Two large families of evolutionarily conserved NAD+-independent histone deacetylases yield free acetate instead of OAADPr during deacetylation. Yeast Hda1 is the founding member of one that includes yeast Hos1 and Hos2. Yeast Rpd3 is founding member of another that includes yeast Hos3 gene. Enzymes of both classes reside within large macromolecular complexes that, in general, must be intact for deacetylase activity (Carmen et al., 1999). Whereas some deacetylases are targeted to promoters, others seem to act by a diffusible non-specific mechanism (Millar and Grunstein, 2006). None are currently thought to reside within extended chromatin structures like Sir2 within silent chromatin.
In this study we sought to clarify the roles of Sir2 and OAADPr in Sir-mediated transcriptional repression. We reasoned that if histone deacetylation is the only critical Sir2 function in creating extended silent domains, then other histone deacetylases should substitute for Sir2 if targeted appropriately. Traditional targeting assays in which a protein of interest is tethered to DNA via a heterologous DNA binding domain would not likely be sufficient. This scenario provides localized deacetylation activity but not an activity that would spread from a nucleation point. To overcome this limitation, we fused histone deacetylases to a silencing factor that both binds at and spreads from silencers (Figure 1B). We show here that chimeras containing Sir3 and either truncated forms of Sir2 or the NAD+-independent deacetylase Hos3, can silence genes at a distance. The data indicate that Sir2 and the OAADPr it generates can be bypassed in forming silenced chromosomal domains.
Results
Restoration of silencing by fusing a nonfunctional Sir2 mutant to Sir3
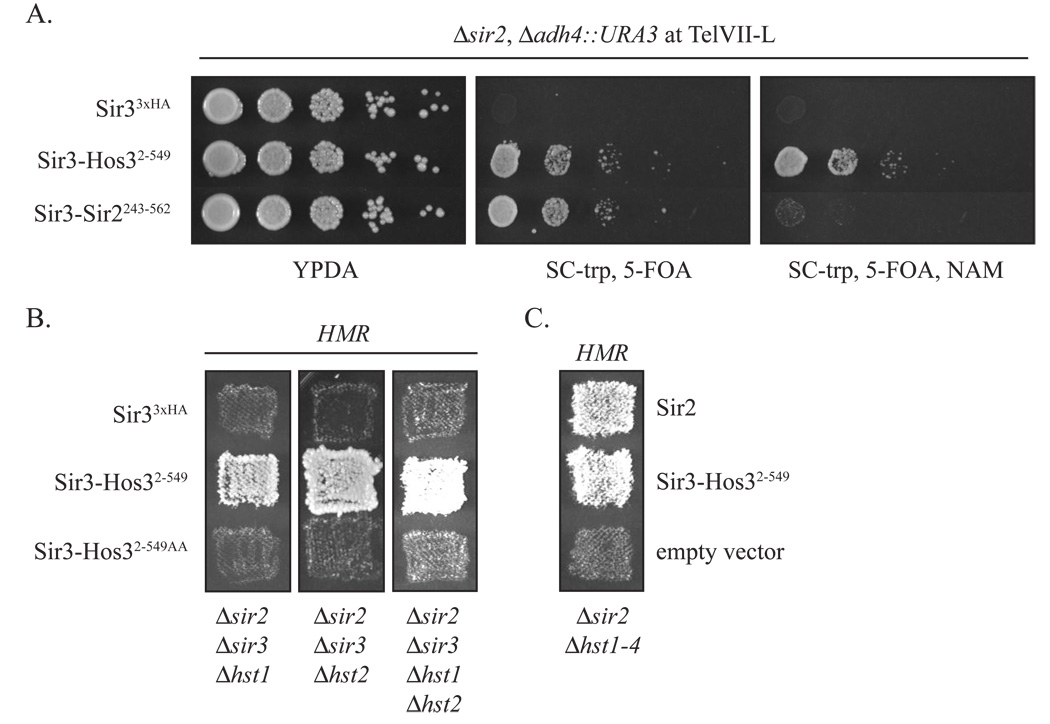
The deacetylase activity of Sir2 lies within a conserved core domain at the center of the polypeptide (Imai et al., 2000; Figure 1C). Flanking amino and carboxyl terminal domains, which are required for silencing in vivo, target the enzyme to sites of action (Sherman et al., 1999; Cuperus et al., 2000; Cockell et al., 2000; Mead et al., 2007; Hickman et al., 2007). We sought a sir2 allele with a nucleation and spreading defect and began with a truncation that removed the first 242 amino acids, yielding Sir2243–562. Here, telomeric silencing was measured with a URA3 reporter gene embedded at TelVII-L. Expression of the gene kills cells exposed to the toxic metabolite 5-fluoro orotic acid (5-FOA). Silencing of HMR and HML was measured with a traditional patch-mating assay. Derepression of the HMR a1 gene in MATα cells creates a pseudo-diploid state that blocks mating and subsequent growth of mated cells on SD indicator plates. Similarly, derepression of the α genes at HML blocks mating of MATa strains. Figures 2A and 2B show that full length Sir2 restored telomeric and mating-type silencing in sir2 null mutants. Sir2243–562, on the other hand, did not. In further studies we tethered the Sir2 fragment to a silencer and found that it still did not yield silencing (Figure S1). These results indicate that Sir2243–562 does not support the spread of silent chromatin, even when linked directly to DNA. A likely explanation is that Sir2243–562 cannot form stable ternary complexes with Sir3 and Sir4.
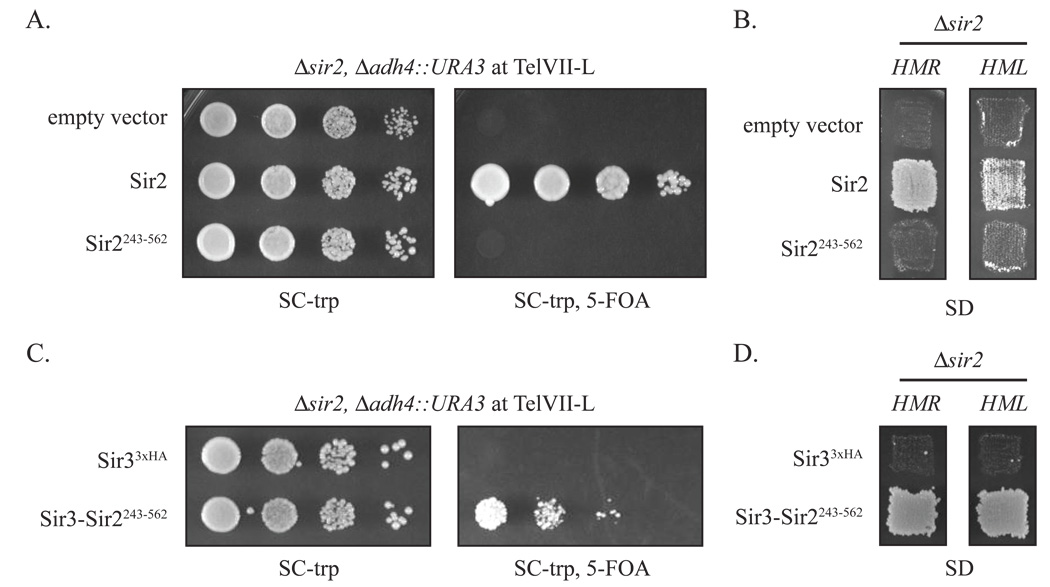
Figure 2. Silencing by Sir3-Sir2243–562.
A) Sir2243–562 does not support telomeric silencing. Repression of a URA3 reporter at Tel-VIIL was measured by growth on 5-FOA in strain CCC1 (MATα Δsir2 Δadh4::URA3) transformed with empty vector (pRS414), a full length Sir2 expression vector (pCC7) or a vector expressing Sir2243–562 (pCC8). SC-trp plates provided loading and growth controls. B) Sir2243–562 does not support mating-type silencing. HMR silencing in strain CCC1 and HML silencing in strain GCY16 (MATa Δsir2) were measured by growth on SD plates after patch mating to tester strains K125 (MATa) and K126 (MATα), respectively. The same plasmids as in (A) were used. C) Silencing of a telomeric URA3 reporter gene by Sir3-Sir2243–562. Strain CCC1 was transformed with plasmids that express Sir33xHA (pSIR3-HA TRP1) or Sir3-Sir2243–562 (pCC4). D) Silencing of the mating-type loci by Sir3-Sir2243–562. Strains CCC1 and GCY16 were transformed with plasmids described in (A) and patch mated to K125 and K126, respectively.
We fused the Sir2243–562 to Sir3 in an attempt to restore silencing. We anticipated that a Sir3-Sir2 chimera would associate with silencers and combine the activities necessary for spreading into a single polypeptide: 1) deacetylation of histone tails by the Sir2 fragment, and 2) binding of the deacetylated tails by Sir3. Figures 2C and 2D show that Sir3-Sir2243–562 silenced the URA3 marked telomere and both HM loci in sir2 null reporter strains. This result indicates that the silencing defect of Sir2243–562 is not loss of enzymatic function. Rather, it suggests Sir2243–562 does not associate with other proteins necessary for nucleation and spreading. The experiment serves as a proof of principle that Sir3 can deliver a deacetylase for silencing at sites distant from a silencer.
Restoration of silencing by fusing Sir3 to a heterologous histone deacetylase
If the only role for Sir2 in silent chromatin is to deacetylate histones, then fusion of Sir3 to other histone deacetylases might also restore silencing. To explore this notion, we linked Sir3 to yeast Hos3, an NAD+-independent deacetylase that is distinct because it requires no other yeast proteins for activity (Carmen et al., 1999). The chimera contained most of Hos3 (residues 2–549) but omitted the C-terminal 148 amino acids. Remarkably, a Sir3-Hos32–549 silenced both HMR and HML, as well as the telomeric reporter gene (Figures 3A and 3B). We conclude that a heterologous deacetylase can substitute for Sir2 in silencing if it is bound to a protein that brings it to appropriate sites of action. A corollary to this finding is that the byproduct of the Sir2 reaction, OAADPr, need not be generated locally for silencing by Sir3-deacetylase hybrids.
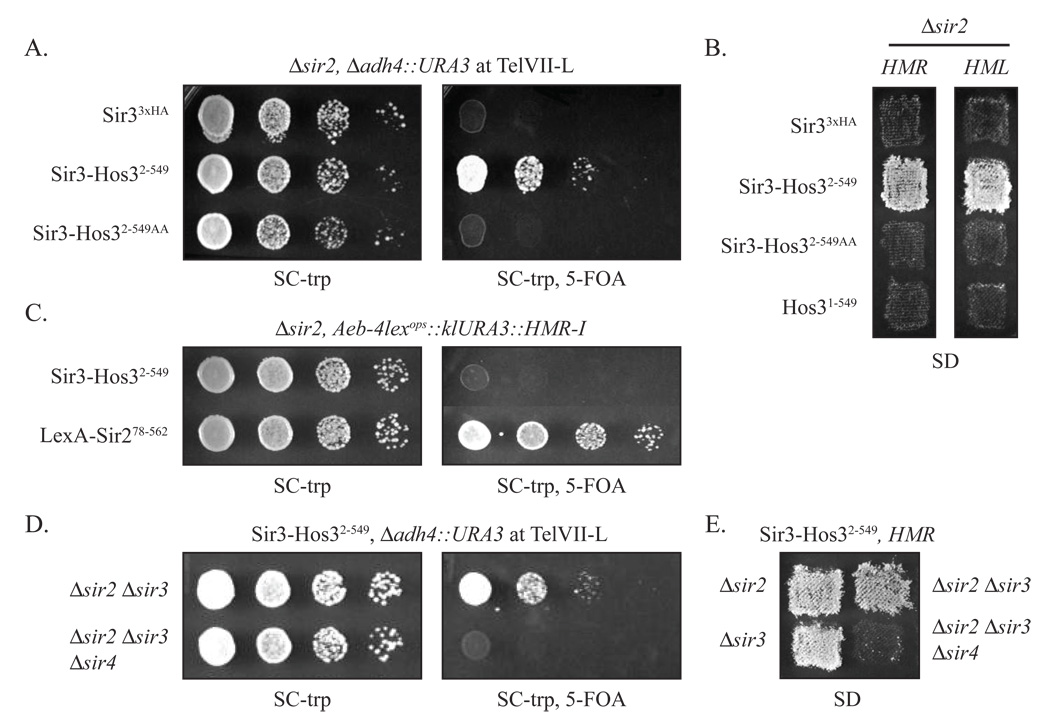
Figure 3. Silencing by Sir3-Hos32–549.
A) Silencing of a telomeric URA3 gene by Sir3-Hos32–549. Strain CCC1 (MATα Δsir2 Δadh4::URA3) was transformed with plasmids that express either Sir33xHA (pSIR3-HA TRP1) , Sir3-Hos32–549 (pCC10) or Sir3-Hos32–549AA (pCC11). B) Silencing of HMR by Sir3-Hos32–549. Strains CCC1 and GCY16 (MATa Δsir2) expressing Sir33xHA (pSIR3-HA TRP1), Sir3-Hos32–549 (pCC10), Sir3-Hos32–549AA (pCC11) or Hos31–549 (pCC21) were patch mated to K125 (MATa) and K126 (MATα), respectively. C) HMR silencing by Sir3-Hos32–549 requires the HMR-E silencer. Strain CCC44 (Aeb-4lexops::klURA3::HMR-I Δsir2) with a mutant HMR-E silencer (designated Aeb-4lexops) was transformed with plasmids expressing either Sir3-Hos32–549 (pCC10) or LexA-Sir278–562 (pGLC117) and then spotted on media containing 5-FOA. D) Telomeric silencing by Sir3-Hos32–549 requires Sir4. Strains CCC12 (MATα Δsir2 Δsir3 Δadh4::URA3) and CCC21 (MATα Δsir2 Δsir3 Δsir4 Δadh4::URA3) were transformed with a plasmid expressing Sir3-Hos32–549 (pCC10). E) Silencing of HMR by Sir3-Hos32–549 requires Sir4. Strains CCC1, CCC11 (MATα Δsir3), CCC12 and CCC21 expressing Sir3-Hos32–549 were patch mated with K125.
To confirm that silencing by Sir3-Hos32–549 requires the deacetylase activity of Hos3, we mutated a pair of conserved histidines (H235, H236) within the putative active site to alanines. The deacetylase activity of the homologous Rpd3 enzyme is abolished when corresponding residues are mutated (Kadosh and Struhl, 1998). Although both mutant and wild-type chimeras were expressed equally (data not shown), the mutant chimera Sir3-Hos32–549AA did not silence the telomeric reporter gene or the mating-type loci (Figure 3A and 3B). We conclude that the deacetylase activity of Hos3 is required for the function of the Sir3-deacetylase hybrid.
Linkage to Sir3 was crucial for Hos3 function in silencing. When the Sir3 portion of the chimera was omitted repression of the mating-type loci could not be detected (Figure 3B). This experiment shows that the silencing attributes of the chimera are not due soley to Hos32–549.
The mating-type genes at HMR are flanked by a principal silencer element on the left (HMR-E) and an auxiliary silencer on the right (HMR-I) that does not function in the absence of HMR-E (Brand et al., 1985; Rusché et al., 2002). To assess the role of silencers in repression by Sir3-Hos32–549 we turned to an hmr mutant that contained LexA operators in place of critical HMR-E sequences (designated the Aeb-4lexops silencer).
Figure 3C shows that Sir3-Hos32–549 failed to silence a URA3reporter gene at the locus. By contrast, a positive control chimera that bypasses the silencer, LexA-Sir278–562, yielded robust repression. The results indicate that HMR-E nucleates silencing by the Sir3-Hos32–549 just as it does for the native Sir proteins.
Sir-dependence of silencing by the Sir3-Hos3 chimera
The dependence of Sir3-Hos32–549 on other silencing factors was determined using a series of sir mutant strains. Figure 3D shows that the chimera silenced a telomeric reporter gene in the absence of the SIR2 and SIR3 genes. This indicates that the chimera supports the necessary roles of both Sir2 and Sir3 in silencing. By contrast, silencing was abolished when SIR4 was deleted from the sir2 sir3 double mutant. Similarly, patch mating assays in figure 3E show that silencing of HMR by Sir3-Hos32–549 did not require SIR2 or SIR3. Silencing of the mating-type locus was lost, however, when SIR4 was also deleted.
Sir3-Hos3 silences efficiently
Quantitative mating assays were used to evaluate the relative efficiency of Sir3-Hos32–549 in HMR and HML silencing. sir mutants expressing the chimera were compared to strains bearing a wild-type complement of Sir proteins. Silencing of HML by Sir3-Hos32–549 in a sir2 sir3 double mutant approached 75% of the wild-type value (Figure 4A, left panel). Silencing of HMR was similarly exceptional at 62% (right panel). The Sir3 chimera silenced better in strains lacking the SIR2 and SIR3 genes than in strains lacking just SIR2 (data not shown). In strains lacking SIR2, SIR3 and SIR4, however, HML silencing dropped to 5% and HMR silencing was negligible. The experiments show that silencing by Sir3-Hos32–549 is strikingly robust. Moreover, the dependence on SIR4 shows that the chimera acts within the Sir-defined pathway.
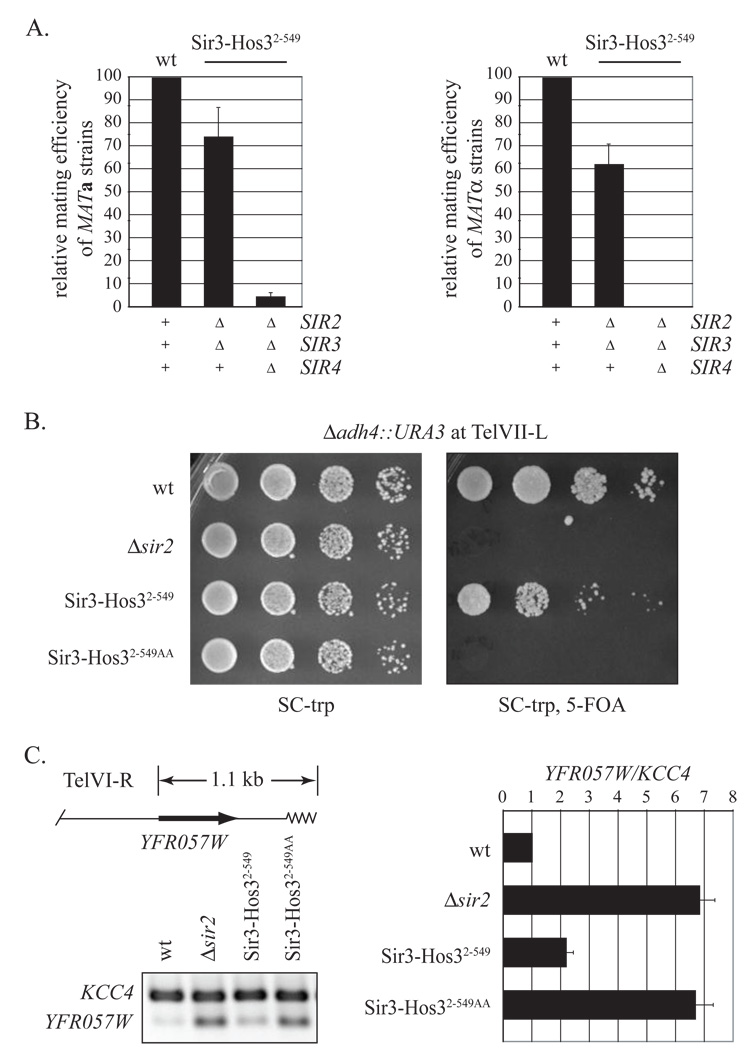
Figure 4. Quantitative measurements of silencing by Sir3-Hos32–549.
A) Quantitative mating assays. Strains GCY16 (MATa Δsir2) and CCC1 (MATα Δsir2 Δadh4::URA3) bearing plasmid-borne SIR2 (pCC7) served as wild-type standards to compare the mating of the following strains expressing Sir3-Hos32–549 (pCC10): CCC8 (MATa Δsir2 Δsir3), CCC20 (MATa Δsir2 Δsir3 Δsir4), CCC12 (MATα Δsir2 Δsir3 Δadh4::URA3), and CCC21 (MATα Δsir2 Δsir3 Δsir4 Δadh4::URA3). Strains K125 (MATa) and K126 (MATα) were used as mating testers. Reported values represent the mean and standard deviation of three independent trials. Mating of strain CCC21 with Sir3-Hos32–549 was ≤ 0.1%. B) Silencing of telomeric URA3. Strain CCC1 (MATα Δsir2 Δadh4::URA3) expressing SIR2 (pCC7) and strain CCC12 (MATα Δsir2 Δsir3 Δadh4::URA3) expressing Sir33xHA (pSIR3-HA TRP1) served as respective wild-type and Δsir2 controls to compare telomeric URA3 silencing in strain CCC12 expressing Sir3-Hos32–549 (pCC10) or Sir3-Hos32–549AA (pCC11). C) Silencing of a native telomeric gene. Multiplex RT-PCR was performed on RNA extracts from the strains described in (B). Primer sets to YFR057W, which resides within 1.1 kb of the right end of chromosome VI, and KCC4, which serves as an internal silencing-independent control are listed in Table S3. The YFR057W/KCC4 ratios for each strain were normalized to the wild-type control. Reported values represent the mean and standard deviation of three independent trials.
We used the TelVII-L URA3 reporter construct to compare telomeric silencing by Sir3-Hos32–549 to silencing by native Sir proteins. Figure 4B shows that the chimera in a sir2 sir3 double mutant is only one order of magnitude less effective than a strain bearing the wild-type complement of silencing proteins (each successive spot in a row represents a 10-fold dilution). Given that URA3 expression is exquisitely sensitive to silencing and that the spotting assay can measure differences spanning five orders of magnitude (van Leeuwen and Gottschling, 2002), the ten-fold difference between Sir3-Hos32–549 and native Sir proteins is not considerable.
We also examined the influence of Sir3-Hos32–549 on expression of a native telomeric gene, YFR057W, which resides near the right end of chromosome VI. In this case, steady-state RNA levels were measured relative to a silencing-independent internal control (KCC4 mRNA) by multiplex RT-PCR. YFR057W transcript levels were found to be roughly 7-fold higher in a strain lacking SIR2 than in a strain bearing the full complement of Sir proteins (Figure 4C). Expression of Sir3-Hos32–549 in a sir2 sir3 double mutant largely suppressed the increase in YFR057W mRNA. Expression of Sir3-Hos32–549AA, on the other hand, reduced YFR057W transcript levels only marginally. These results show Sir3-Hos32–549 silences native telomeric genes in addition to the ectopic URA3 reporter construct described above. Moreover, the results demonstrate that silencing by the chimera occurs at the level of mRNA, as expected for a transcriptional repressor, rather than by some other indirect mechanism.
Sir3-Hos3 binds HMR and deacetylates histones
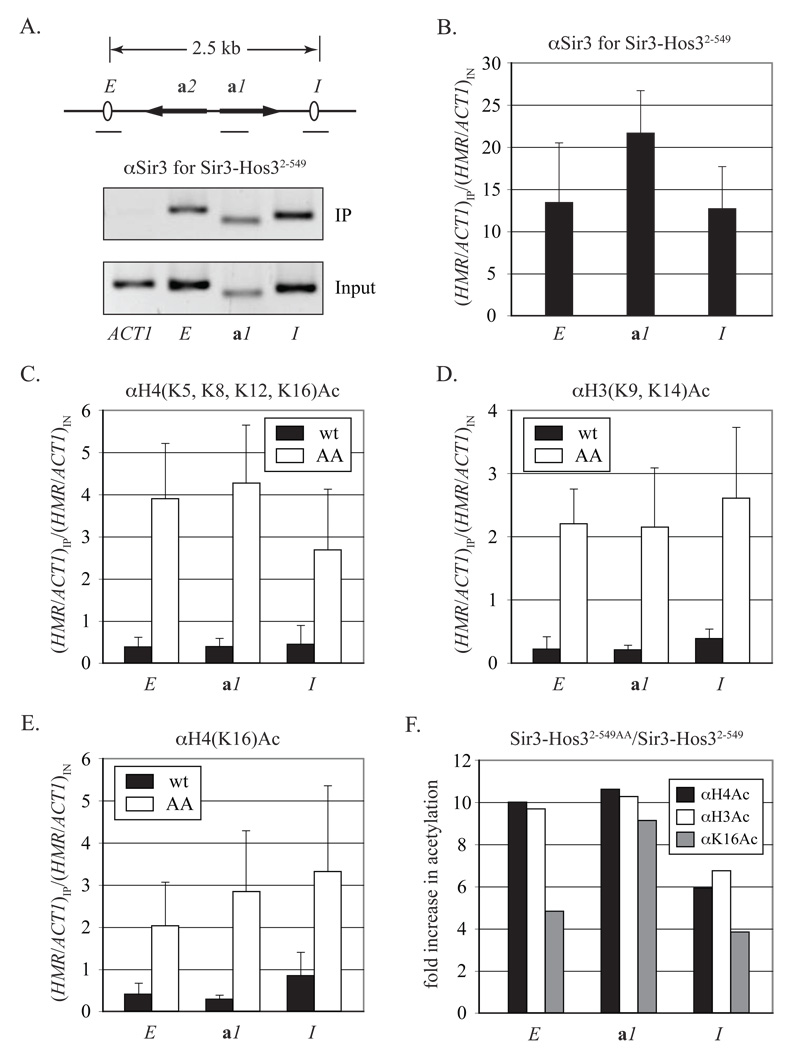
Chromatin immunoprecipitation (ChIP) was used to probe the distribution of the Sir3 chimeras at HMR in a sir2 sir3 null strain. Pull-downs were performed with polyclonal Sir3 antibodies and PCR was used to detect coimmunoprecipitation at HMR-E, HMR-I and a site within the a1 gene that lies between HMR-E and HMR-I (Figure 5A). An additional primer set corresponding to the Sir3-free ACT1 gene provided an internal negative control. Figures 5A and B show that Sir3-Hos32–549 associated well with the three HMR landmarks tested but not ACT1. Sir3-Hos32–549AA, by contrast, associated poorly with HMR (data not shown). Only residual binding was detected at HMR-E, as predicted for a protein that nucleates but does not spread. Our data indicate that Sir3-Hos32–549 assembles silent chromosomal domains.
Figure 5. Sir3-Hos32–549 binds and deacetylates histones throughout the silenced domain.
A) A map of HMR showing the sites examined by ChIP (see PCR primers #17–24 in Table S3) and gels of a representative experiment with Sir3 antibody (αSir3). ACT1 served as a silent chromatin-free control. Experiments were performed with strain CCC12 (MATα Δsir2 Δsir3) expressing either Sir-Hos32–549 (from plasmid pCC10 and labeled wt) or Sir3-Hos32–549AA (from plasmid pCC11 and labeled AA). Reported values represent the mean and standard deviation of at least three independent trials. B) Quantitation of Sir3 ChIP for the wt Sir3-Hos32–549 chimera. C) Quantitation of the H4(K5, K8, K12, K16)Ac ChIP. D) Quantitation of the H3(K9, K14)Ac ChIP. E) Quantitation of the H4K16Ac ChIP. F) Fold increase in acetylation in figures 5C–E by replacing Sir3-Hos32–549 with Sir3-Hos32–549AA. Black αH4Ac = αH4(K5, K8, K12, K16)Ac; white αH3Ac = αH3(K9, K14)Ac, grey αK16Ac = αH4K16Ac.
The ChIP experiments were repeated with antibodies specific for multi-acetylated tails of either histone H4 (residues K5, K8, K12, K16) or H3 (residues K9 and K14). Figures 5C, D and F show that Sir3-Hos32–549AA elevated acetylation of both histone tails 6–10 fold relative to Sir3-Hos32–549 at all sites tested. The experiments were also performed with an antibody specific for acetylated form of H4K16. Here too acetylation levels increased 4–9 fold (Figures 5E and F). Taken together, the data show that transcriptional repression by Sir3-Hos32–549 is accompanied by loss of H3 and H4 acetylation across the locus, including K16 of H4, a residue known to be critical for the Sir-mediated silencing pathway.
Nicotinamide does not inhibit silencing by Sir3-Hos3
Nicotinamide, a byproduct of the Sir2 reaction, binds and inhibits the enzyme non-competitively (Landry et al., 2000; Bitterman et al., 2002). Yeast cells exposed to high concentrations of nicotinamide phenocopy a sir2 null mutant. If the only inhibitory target of the small molecule in Sir-mediated silencing is Sir2, then the Sir3-Hos32–549 chimera should produce nicotinamide-resistant repression. Figure 6A shows that this is indeed the case. In a Δsir2 Δsir3 strain bearing the Sir3-Sir2243–562 chimera, silencing of a telomeric reporter gene was derepressed by 5 mM nicotinamide. Silencing by Sir3-Hos32–549, on the other hand, persisted. The results indicate that Sir2 is the only meaningful inhibitory target for nicotinamide in telomeric silencing.
Figure 6. Silencing by Sir3-Hos32–549 is nicotinamide resistant and does not require any sirtuins.
A) Sir3-Hos32–549 produces nicotinamide-resistant silencing. Strain CCC12 (MATα Δsir3 Δsir2 adh4::URA3) bearing plasmids expressing Sir33xHA (pSIR3-HA TRP1), Sir3-Hos32–549 (pCC10) and Sir3-Sir2243–562 (pCC4) was spotted on SC-trp plates containing 5-FOA and 5 mM nicotinamide (NAM), SC-trp plates containing 5-FOA alone and YPDA plates as a loading control. B) Silencing by Sir3-Hos32–549 without Hst1 and/or Hst2. Strains CCC46 (MATα Δsir2 Δsir3 Δhst1), CCC37 (MATα Δsir2 Δsir3 Δhst2) and CCC50 (MATα Δsir2 Δsir3 Δhst1 Δhst2) expressing Sir33xHA (pSIR3-HA TRP1), Sir3-Hos32–549 (pCC10) or Sir3-Hos32–549AA (pCC11) were patch mated to K125 (MATa). C) Silencing by Sir3-Hos32–549 in a strain lacking all sirtuins. Strain YCB496 (MATα Δsir2 Δhst1 Δhst2 Δhst3 Δhst4) expressing Sir2 (pCC32), Sir3-Hos32–549 (pCC31) or empty vector (pRS412) was patch mated to K125 (MATa).
Exogenous nicotinamide should lower the intracellular concentration of OAADPr since the drug inhibits all sirtuins to some extent (Sauve et al., 2006). The ability of Sir3-Hos32–549 to silence genes under these conditions suggests that the other yeast sirtuins, Hst1-4, do not compensate for the loss of Sir2 by producing OAADPr in trans. To examine this issue more closely, we tested the ability of Sir3-Hos32–549 to silence HMR in hst null mutants. Patch mating assays in figure 6B show that HMR remains silent in Δsir2 Δsir3 strains lacking HST1, HST2, or both HST genes together. More strikingly, the chimera silenced the mating-type locus in a strain lacking all five yeast sirtuins (Figure 6C). As sirtuins are the only known source of OAADPr in vivo, our results indicate that transcriptional silencing can occur without the metabolite.
Silencing by Sir2-Hos3 chimeras
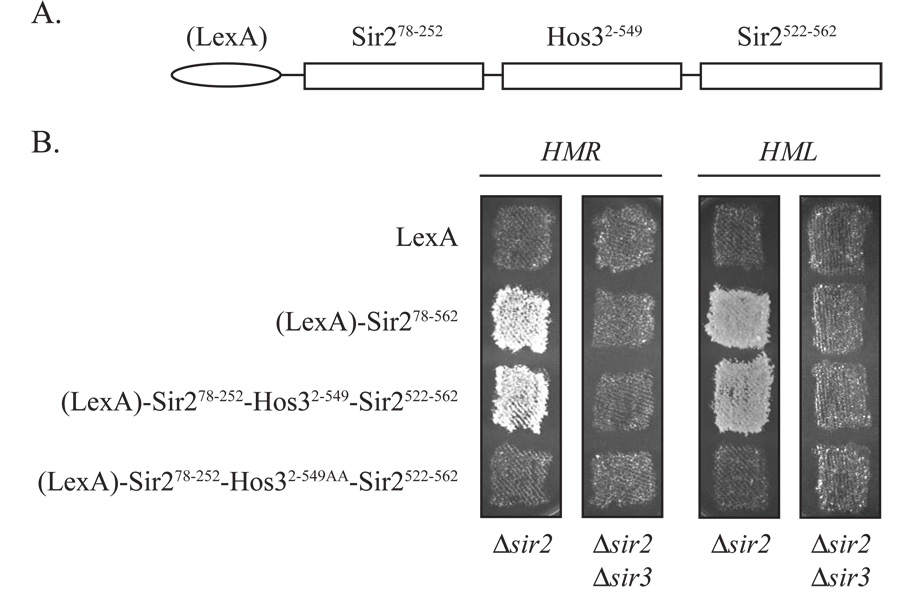
Previous work showed that the domains flanking the Sir2 enzymatic core interact with other members of the Sir complex to determine the specificity of the deacetylase for mating-type and telomeric loci. Hybrid proteins bearing one or both of these domains linked to either yeast Hst1 or human SirT2 substitute for Sir2 in mating-type and telomeric silencing (Mead et al., 2007; Hickman and Rusche, 2007; Sherman et al., 1999). We reasoned that the flanking domains of Sir2 might be similarly manipulated to target Hos3. To this end, we replaced the core of Sir278–562 (amino acids 253 to 521) with the Hos32–549 fragment described above (Figure 7A). The construct was also linked to LexA but subsequent studies revealed that LexA was not essential (data not shown). Patch mating assays in figure 7B show that the Sir278–252-Hos32–549-Sir2522–562, like the Sir278–562 positive control, silenced HMR and HML in sir2 null strains. Silencing by both constructs was abolished in strains lacking both SIR2 and SIR3, as expected for a Sir2 chimera operating within the Sir pathway. Silencing was also lost when the Hos3 active site of Sir21–252-Hos32–549AA-Sir2522–562 was mutated, indicating that deacetylation by Hos3 was required. These results indicate that the Sir2 N- and C-termini are sufficient to target a heterologous deacetylase for function within silent chromatin. Importantly, the experiment also shows that targeting of Sir278–252-Hos32–549-Sir2522–562 does not require that OAADPr be produced in cis.
Figure 7. Silencing of the mating-type loci by a Sir2-Hos3 chimera.
A) A map of the Sir2-Hos3 hybrid protein. LexA is appended to the N-terminus of all Sir2 chimeras used here but the bacterial protein was subsequently removed and found not to be relevant (data not shown). B) Patch mating assays with Sir278–252-Hos32–549-Sir2522–562. Strains CCC1 (MATα Δsir2), CCC12 (MATα Δsir2 Δsir3), GCY16 (MATa Δsir2) and CCC8 (MATa Δsir2 Δsir3) expressing LexA (pBTM116), LexA-Sir278–562 (pGLC117), LexA-Sir278–252-Hos32–549-Sir2522–562 (pCC29), or LexA-Sir278–252-Hos32–549AA-Sir2522–562 (pCC30) were patch mated to either K125 (MATa) and K126 (MATα).
Discussion
In this study we examined the roles of Sir2 in transcriptional silencing using a protein chimera approach. We fused Sir3 to a heterologous NAD+-independent deacetylase, Hos3, and found that the hybrid efficiently substituted for both Sir2 and Sir3 in Sir4-dependent silencing. More importantly, the hybrid functioned efficiently in a strain lacking all of the NAD+-dependent deacetylases, and thus one completely devoid of OAADPr. Taken together, our data demonstrate that protein deacetylation is the only necessary function of Sir2 in creation of silenced chromosomal domains.
Deacetylation and silencing
Hos3 deacetylates histone H2B preferentially at position K11 as part of the apoptotic response to oxidative stress (Ahn et al., 2006). The enzyme displays site preferences toward acetylated lysines on each of the histone tails in vitro but no overt activity for H4K16 (Carmen et al., 1999). How then does the heterologous enzyme yield deacetylated H4K16 for silencing (Figure 5D)? We speculate that in this instance targeting drives Hos3 substrate choice. Tethering the enzyme to chromatin via Sir3 maintains a high localized concentration in the vicinity of histones at HMR. Under these conditions, Hos3 may deacetylate sub-optimal substrates, as well as preferred ones. We note by analogy that despite the preference of Sir2 for acetylated H4K16 in vitro, all of the histone tails are deacetylated within silent chromatin where it is tethered in vivo (Suka et al., 2001).
A growing body of evidence indicates that deacetylation of H4K16 is not the only role for Sir2 in silencing. The Grunstein and Horokoshi labs identified the predominant H4K16 acetyltransferase, Sas2, and showed that silencing required Sir2 even in its absence (Kimura et al., 2002; Suka et al., 2002). The results implied that H4K16 deacetylation was not sufficient for silencing but could not discern whether the additional critical functions for Sir2 were enzymatic or structural. Subsequently, Yang and Kirchmaier showed that Sir proteins spread in an enzymatically-inactive Sir2 mutant if selected histone residues were replaced with arginine, a deacetylated lysine mimetic (Yang and Kirchmaier, 2006). Little silencing was observed under these conditions, however, suggesting that the enzymatic activity of Sir2 was required for the deacetylation of additional lysines. One substrate might be acetylated K56 of histone H3 on the surface of the histone octamer core. Sir2 deacetylates the lysine within silent chromatin and silencing defects occur when the residue is mutated to arginine (Xu et al., 2007). An advantage to the protein chimera approach used here is that the deacetylase delivered by Sir3 might act on a spectrum of acetylated lysines, including those residues directly responsible for silencing. While it is certain that histones are critical Sir2 substrates, we cannot rule out the possibility that other non-histone substrates are involved.
Possible roles for OAADPr in vivo
Human Sir2 homologues act on histones, histone modifiers, transcriptional regulators, as well as non chromatin targets to regulate diverse processes ranging from aging to apoptosis (see (Vaquero et al., 2007) and references in (Michan and Sinclair, 2007)). The conserved requirement for NAD+ in deacetylation reactions, as well as the inhibition of deacetylation by nicotinamide, positions these enzymes as metabolic sensors, capable of responding to changes in the energy state of the cell. The discovery of OAADPr as a byproduct of histone deacetylation raised speculation that the novel metabolite could also serve a physiological role, perhaps by acting locally to facilitate heterochromatin assembly, as suggested by biochemical studies (Liou et al., 2005; Onishi et al., 2007), or acting distally to signal that sirtuins have acted. OAADPr binding targets have been identified in human cells (Kustatscher et al., 2005; Grubisha et al., 2006), and when microinjected into oocytes OAADPr causes disturbances in maturation and subsequent cell cycle progression (Borra et al., 2002). An enzyme that cleaves the O-acetyl linkage of OAADPr efficiently has also been identified, suggesting that a mechanism to turnover the metabolite exists (Ono et al., 2006). Our work suggests that if OAADPr is a signal transducer in yeast, it is not one required to achieve silencing.
One caveat remains: OAADPr could influence silencing via Sir2. In a Sir2 bypass experiment, according to this line of reasoning, the metabolite would no longer appear to be relevant. For example, bound OAADPr could stabilize a protein-protein interaction or conformational change necessary for the enzyme to participate in silent chromatin assembly (posited in (Liou et al., 2005). That Sir278–252-Hos32–549-Sir2522–562 generates silencing (Figure 7B), however, indicates that the Sir2 N and C-terminal targeting domains remain functional even in the absence of the OAADPr.
Broader applications of the protein chimera approach
The protein chimera approach has allowed us to identify protein fragments that are essential for silencing and to identify those domains that can be substituted or bypassed. One can envision extending this approach to create specialized synthetic chimeras for technological applications. Protein modules containing histone modification activities and cognate histone binding activities could be combined (like Sir3/Hos3 here) with modules for targeting, such as DNA binding domains or domains that associate with DNA bound factors. In this way, it might be possible to shut down the expression of specific genes or families of genes by robust chromatin-based mechanisms in yeast or in higher eukaryotes.
Materials and methods
Strain construction
Complete ORF deletions were made by PCR-mediated gene replacement, unless specified otherwise, using plasmids or yeast DNA extracts as PCR templates. The TRP1 coding sequence at HMR in strain GA2050 was replaced with the URA3 coding sequence (klURA3) from K. lactis. Δsir3::HIS3 deletions were generated with pSIR3::HIS3 (Moretti and Shore, 2001). A cross between strains CCC45 and CCC46 produced segregant CCC48. All strain modifications were confirmed by PCR and/or functional tests.
Plasmid construction and confirmation
Table S1–Table S3 provide detailed information on plasmid constructions including parent vectors, PCR templates and oligonucleotide primers. The SIR3 promoter and terminator flanked all chimeras produced in this study, except where noted otherwise. Gene fusions were created by PCR-Mediated Plasmid Gap Repair (P-MPGR) in yeast. Deletions within gene fusion constructs were generated by Oligonucleotide-Mediated Plasmid Gap Repair (O-MPGR). Sequences of the constructs are available upon request.
A fortuitous PCR error during the construction of a functional Sir3-Hos32–549 introduced a stop codon at position 550, eliminating the terminal 148 amino acids of Hos3. A construct lacking the mutation did not produce silencing and was not pursued further.
Phenotypic silencing assays
Telomeric ura3 repression
Plasmid-bearing strains were grown overnight to saturation in selective media and spotted in 10-fold serial dilutions on SC-trp containing 0.1% 5-FOA to measure URA3 expression and on SC-trp as a control for loading and cell growth, unless noted otherwise.
Patch mating
Plasmid-bearing strains were pre-grown on selective medium and then patched together with mating tester strains on YPDA plates. After at least 5 hours, the cells were replica plated to SD agar.
Quantitative mating
Overnight cultures of plasmid-bearing strains were diluted and grown to mid-log phase in SC-trp media (all plasmids possess a TRP1 marker). They were mixed with a 5-fold excess of cells of mating tester strains K125 or K126, concentrated onto sterile nitrocellulose membranes and placed on YPDA plates for 5 hours at 30°C. Membranes were vortexed in sterile water to create a cell suspension that was spread on SD and SC plates to determine the fractional degree of mating. The use of SC rather than SC-trp plates may lead to an underestimate of mating efficiency because not all cells bear plasmids. However, colony formation by SIR4 cells bearing the Sir3-Hos3 chimera is mildly hindered on SC-trp plates, which leads to an overestimate of mating efficiency (data not shown).
Nicotinamide treatment
Cultures were grown overnight in selective media to saturation and spotted on indicator plates that were overlayed with nicotinamide (Cf = 5 mM).
Multiplex RT-PCR assays
Qiagen OneStep RT-PCR kits were used to analyze RNA isolated from mid-log cells by the hot acid phenol extraction procedure. Primer pairs for both YFR057W and KCC4 were used simultaneously with 200 ng of RNA and Q-solution according to the manufacturer’s protocol. Both PCR products were within the linear range after 29 amplification cycles. Gels were stained with ethidium bromide and then destained in water before digital photography and quantitation by lane densitometry (Alpha Innotech, San Leandro, CA).
ChIP assays
ChIP was performed with polyclonal antibodies to Sir3 (a gift from the Kamakaka lab) and acetylated histone antibodies (Upstate/Millipore #06–599, #06–866 and #07–329) according to (Li et al., 2001). PCR reactions were performed with individual pairs of primers to produce products within the linear range. Gels were imaged as described above. The intensity of each HMR ChIP band was recorded relative to the intensity of the ACT1 ChIP band and then normalized to the same ratio of input material.
Supplementary Material
Figure S1 – LexA-Sir2242–562 does not impart targeted silencing. Previous work showed that tethering Sir proteins to DNA can nucleate silent chromatin, which spreads and represses nearby genes (Chien et al., 1993; Cuperus et al., 2000; Cockell et al., 2000). Strain YCL73 (MATα Δhmr::6lexops ssEB-a2a1 Δsir2 Δsir1) with LexA operators (lexops) at a synthetic HMR-E silencer (labeled ssEB, (Li et al., 2001)) was patch mated with the K125 tester strain. The strain was transformed with plasmids that express Sir2242–562 (pCC18), LexA fused to either Sir278–562 (pCC27), or LexA alone (pJK1521). LexA-Sir278–562 produced robust targeted silencing (also see figure 3C). LexA-Sir2242–562 and LexA alone did not silence at all.
Figure S2 – Representative examples of raw ChIP data. Extracts were prepared from the strains described in Figure 4. A) PCR reactions of Input and samples immunoprecipitated with either anti-H3(K9, K14)Ac or anti-H4(K5, K8, K12, K16)Ac. B) PCR reactions of Input and samples immunoprecipitated with anti-H4K16Ac.
Acknowledgements
We thank Mike Hampsey, Jef Boeke, Susan Gasser, Rohinton Kamakaka, David Shore, Rolf Sternglanz, David Stillman, Drew Vershon and Jenel Nixon for strains, plasmids and antibodies. We thank Drew Vershon and Rolf Sternglanz for thoughtful discussions and Danesh Moazed for suggesting Hos3 as the heterologous deacetylase. This work was funded by grant GM51402 from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SirT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a 'silencer' in yeast: a DNA sequence with properties opposite those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Griffin PR, Calaycay JR, Rundlett SE, Suka Y, Grunstein M. Yeast Hos3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N-terminus regulates its binding to heterochromatin protein Sir3. J. Biol. Chem. 2001;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- Chien C-T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Cockell MM, Perrod S, Gasser SM. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics. 2000;154:1069–1083. doi: 10.1093/genetics/154.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus G, Shafaatian R, Shore D. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 2000;19:2641–2651. doi: 10.1093/emboj/19.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud AL, Scharenberg AM, Denu JM. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J. Biol. Chem. 2006;281:14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- Hickman M, McCullough K, Woike A, Raducha-Grace L, Rozario T, Dula ML, Anderson E, Margalit D, Holmes SG. Isolation and characterization of conditional alleles of the yeast SIR2 gene. J. Mol. Biol. 2007;367:1246–1257. doi: 10.1016/j.jmb.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Hickman MA, Rusche LN. Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 2007;3:e126. doi: 10.1371/journal.pgen.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci.USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Li Y-C, Cheng T-H, Gartenberg MR. Establishment of transcriptional silencing in the absence of DNA replication. Science. 2001;291:650–653. doi: 10.1126/science.291.5504.650. [DOI] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead J, McCord R, Youngster L, Sharma M, Gartenberg MR, Vershon AK. Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases. Mol. Cell. Biol. 2007;27:2466–2475. doi: 10.1128/MCB.01641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell. Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a Sir2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. USA. 2006;103:16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Ann. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Ann. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Sherman JM, Stone EM, Freeman-Cook LL, Brachmann CB, Boeke JD, Pillus L. The conserved core of a human SIR2 homologue functions in yeast silencing. Mol. Biol. Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gottschling DE. Assays for gene silencing in yeast. Methods Enzymol. 2002;350:165–186. doi: 10.1016/s0076-6879(02)50962-9. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Kirchmaier AL. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:5287–5297. doi: 10.1091/mbc.E06-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – LexA-Sir2242–562 does not impart targeted silencing. Previous work showed that tethering Sir proteins to DNA can nucleate silent chromatin, which spreads and represses nearby genes (Chien et al., 1993; Cuperus et al., 2000; Cockell et al., 2000). Strain YCL73 (MATα Δhmr::6lexops ssEB-a2a1 Δsir2 Δsir1) with LexA operators (lexops) at a synthetic HMR-E silencer (labeled ssEB, (Li et al., 2001)) was patch mated with the K125 tester strain. The strain was transformed with plasmids that express Sir2242–562 (pCC18), LexA fused to either Sir278–562 (pCC27), or LexA alone (pJK1521). LexA-Sir278–562 produced robust targeted silencing (also see figure 3C). LexA-Sir2242–562 and LexA alone did not silence at all.
Figure S2 – Representative examples of raw ChIP data. Extracts were prepared from the strains described in Figure 4. A) PCR reactions of Input and samples immunoprecipitated with either anti-H3(K9, K14)Ac or anti-H4(K5, K8, K12, K16)Ac. B) PCR reactions of Input and samples immunoprecipitated with anti-H4K16Ac.