Genetically encoded sensors of neural activity enable visualization of circuit-level function in the central nervous system. Although our understanding of the molecular events that regulate neuronal firing, synaptic function, and plasticity has expanded rapidly over the past fifteen years, an appreciation for how cellular changes are functionally integrated at the circuit level has lagged. A new generation of tools that employ fluorescent sensors of neural activity promises unique opportunities to bridge the gap between cellular and system-levels analysis.
This review will focus on genetically-encoded sensors. A primary advantage of these indicators is that they can be non-selectively introduced to large populations of cells using either transgenic or viral-mediated approaches. This ability removes the non-trivial obstacles of how to get chemical indicators into cells of interest, a problem that has dogged investigators who have been interested in mapping neural function in the intact CNS. Five different types of approaches and their relative utility will be reviewed here: 1) reporters of immediate-early gene (IEG) activation using promoters such as c-fos and arc, 2) voltage-based sensors, such as GFP-coupled Na+ and K+ channels, 3) Cl− based sensors, 4) Ca++-based sensors, such as Camgaroo and the troponin-based TN-L15, and 5) pH-based sensors, which have been particularly useful for examining synaptic activity of highly convergent afferents in sensory systems in vivo. Particular attention will be paid to reporters of IEG expression, since these tools employ the built-in threshold function that occurs with activation of gene expression, provoking new experimental questions by expanding the timescale of analysis for circuit and system-level functional mapping.
Fluorescent reporters of inducible gene expression
Immediate-early gene expression (IEG) can be a reliable marker of elevated neuronal firing, reporting activity over time scales that range from minutes to hours compared to ms and seconds for voltage- or ion-based sensors. In ion-based approaches, indicators are constitutively present in the cell, and the fluorescent protein must be engineered to be a sensor with rapid onset and offset kinetics to achieve temporal fidelity with the process being measured. In contrast, monitoring IEG expression can be directly coupled to transcription of the reporter gene. However, because maturation of the fluorophore is required for detection, a temporal delay may be introduced into reporter detection. This delay in reporting activity is advantageous in that it expands the types of experimental questions that can be addressed. For example, do IEG-expressing cells remain more excitable after stimulus cessation? Are cells activated by two temporally distinct stimuli (i.e. training and recall in a memory task) overlapping or distinct?
It was apparent from the early 1990’s that transgenic reporters of gene expression could be highly useful in functionally mapping the CNS, providing high-throughput mechanism to examine IEG expression (compared to immunohistochemistry, an expensive and labor-intensive procedure). Transgenic mice carrying LacZ reporters driven by the fos promoter [1] or a virally derived CREB-sensitive promoter [2] laid the groundwork for fluorescence-based approaches. A number of early attempts to create fluorescence-based reporters of neuronal activity (many of which were never reported), were unsuccessful. In some cases, this was due to gene knock-in strategies, where a single copy of the fluorescent reporter in to the c-fos locus did not yield sufficient GFP expression to be detected in living neurons [3]. In another effort using the egr-1/zif268 promoter to drive expression of a destabilized GFP in transgenic rats, fluorescence microscopy failed to detect any GFP signal, despite the presence of reporter transcript [4]. The GENSAT project, a consortium-based effort to create a library of GFP-transgenic mice driven by brain specific promoters, has also not been successful at producing animals showing detectable fluorescence that can be visualized without immunohistochemical amplification, most likely because levels of fluorescent protein expression remain too low for direct visualization [5]. However, this project has created egr1/zif268, arc, and homer-GFP transgenic mice.
GFP reporters of Fos-expression
The first transgenic mice expressing a fluorescent reporter of IEG expression, where fluorescence could be detected in living cells without signal amplification, used the fos promoter to drive expression of a fos-GFP fusion gene [6]. Transgenic animals carried multiple copies (in some cases, >10) of the reporter construct, which enhanced levels of GFP and facilitated fluorescence detection. Although the duration of fosGPF fluorescence with respect to endogenous Fos protein was slightly prolonged (~2 hrs for early detection, versus 30 minutes for the endogenous Fos protein, and ~6 hrs for degradation versus 90–120 minutes for endogenous Fos), this has not proved to be a significant disadvantage in using this research tool.
Because maturation of the fluorophore requires several hours, Fos induction due to slice preparation [6] is not readily observed in vitro. This has enable the fosGFP mice to be useful for identifying and analyzing cells activated by in vivo experience in reduced brain slice preparations, experiments which have shed light into the molecular events underlying the progression of plastic changes during sensory plasticity in vivo [7] (Figure 1). Fluorescent cells can be detected in vivo as well as in vitro using both 2-photon (A. Barth, L. Yassin, and J. Crowley, unpublished data) and microendoscope methods (R. Barretto and M. Schnitzer, personal communication).
Figure 1.
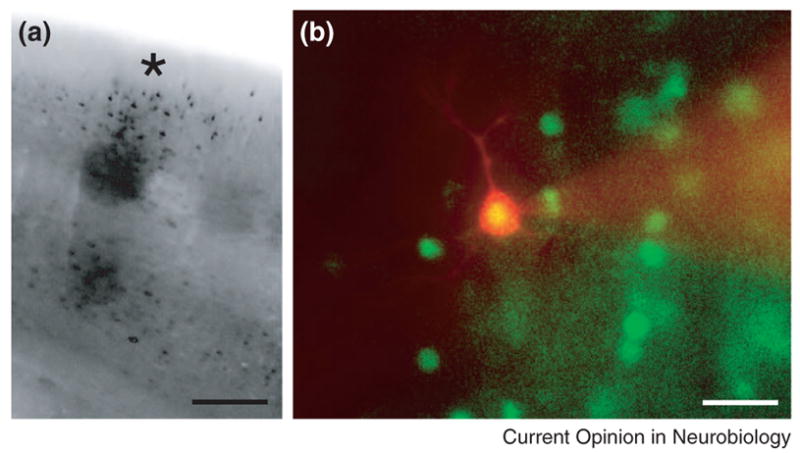
(A) FosGFP expression induced by in vivo sensory stimulation can be visualized in a single cortical column in vitro. Animals were subjected to removal of all but a single whisker from both sides of the mouse face for 24 hrs, a stimulus that results in robust fos expression. Cells expressing fosGFP can be detected in the single barrel column (*) representing this whisker in acute brain slices prepared from these animals. Figure modified from [6]. Scale ~150 μm. (B) A fosGFP+ Alexa-568 (red) filled cell after whole-cell recording. FosGFP+ neurons possess normal electrophysiological responses properties compared to neighboring, unlabeled cells [6]. Scale ~15 μm.
Arc-GFP transgenic mice
An important goal for IEG-reporters is that the fluorescence signal should be visible when endogenous expression levels are high and should be reduced as endogenous protein levels fall. Because fluorescent proteins are typically slow to mature and are relatively stable, this presents both challenges and opportunities. A second line of IEG transgenic mice expressing GFP knocked into the arc locus has been created to monitor arc expression in vivo [8]. These animals were made with a destabilized GFP (d2EGFP; [9]) to prevent the accumulation of the reporter gene over time, in contrast to the fosGFP transgene – a fusion protein that retains both mRNA and protein signals to control accumulation and degradation of the transgene. Regulation of the arc-GFP transgene is precise and maps directly on to arc-immunoreactive cells in heterozygote animals, although as in the fosGFP animals the GFP reporter appears to linger in the cell for slightly longer time periods (4+ hrs) than would be expected for the endogenous arc protein (2 hrs).
Although the fluorescence signal from these animals is weak, direct fluorescence can be observed in vivo using 2-photon or confocal microscopy. Arc-GFP transgenic animals have been used to show that sensory experience leads to the refinement of activated neuronal assemblies, and analysis of homozygote (arc-knock-out) animals has revealed a role for arc in enhancing orientation tuning.
The ability of the fosGFP and arc-GFP transgenic mice to report cell activity after an in vivo stimulus affords new ways to probe neural circuits both in vivo and in acute brain slices. Although in vivo imaging is a powerful tool, it remains technically challenging and has a number of experimental limitations (for example, artifacts induced by surgical preparation, restricted access to deep brain areas). Slice electrophysiology offers superior pharmacological and anatomical access, and is widely used within the neuroscience community; thus, it is likely that use of IEG-reporter transgenics will be most widely adopted by cellular and systems neuroscientists who hope to link in vivo stimuli with in vitro analysis of specific neuronal assemblies. Because both fosGFP and the arc-driven GFP reporter exhibit slightly prolonged signal half-life compared to endogenous proteins, fluorescence is maintained in vitro for many hours (particularly at room temperature). This feature significantly facilitates targeted whole-cell recording of activated cells and has been particularly useful for analysis of sensory-induced changes in neural circuits in the neocortex, where fosGFP expression in a single activated whisker column can be used to focus investigations into the relevant brain area [7].
Circuit analysis over time
IEG reporters are particularly useful for bridging cellular and molecular investigations with system-level questions about neural circuits activated by in vivo experience. Especially for reporters of IEG expression, transgenic (as opposed to transfection-based) approaches are particularly important because they report neural activation in an unbiased manner. Since all neurons within the CNS can conceivably activate transgene expression, visualization of GFP-expressing cells provides insight into specific neuronal ensembles that have been activated by in vivo stimuli.
The relatively long duration between cell firing and rise in fluorescence transforms the kinds of questions that can be asked using fast sensors of voltage, Ca++ influx, or vesicle release, events whose onset occurs with millisecond precision but leave no “trace” of prior history between detection events.
The delay between IEG induction and reporter gene detection can be particularly useful in tracing the activity history of neuronal ensembles. For example, Mark Mayford and colleagues recently developed an inducible (tet-Off) fosLacZ transgenic mouse, where LacZ expression was permitted only during a short window of time during acquisition of a fear memory [10]. Neuronal reactivation during retrieval of the fear memory days later was then examined by determining whether LacZ-expressing cells (activated during memory acquisition) also expressed a different IEG, zif268 (activated minutes before during memory retrieval). Delays in detection of IEG-coupled reporter genes are ideal for bridging in vivo and in vitro analysis of neural function. Such animals allow in vivo stimuli can be “recorded” in precise neural subsets by IEG-GFP expression and later analyzed using sophisticated histological, electrophysiological, and pharmacological techniques in vitro.
Channel-based voltage sensors
The K+ channel based sensor FlaSh, a fusion of GFP and the Drosophila K channel shaker, was the first genetically encoded protein that enabled direct visualization of changes in membrane potential [11]. The entire K+ channel was used in the construction of the indicator with the aim of preserving the fidelity of voltage-dependent changes in conformation, although mutations in the pore-forming region of the protein prevented the fusion protein from actively conducting. Similarly, a Na+ channel sensor named SPARC coupled GFP between the second and third transmembrane domains and was reversibly occluded by a mutation introduced into the pore-forming region of the channel[12]. The activation kinetics of this channel were significantly faster than that observed for FlaSh, however, the fractional change in fluorescence was low (~0.5% per 100 mV) in oocytes where the construct was overexpressed. In both cases, these constructs have not been especially useful for imaging activity in mammalian neurons, because the large fraction of intracellular versus plasma membrane FlaSh and SPARC makes signal detection difficult[13]. Because an important goal of ion channel-coupled sensors is imaging neural activity in real time, i.e. ms timescales, signal to noise issues are critical. Although FlaSh has been modified and improved for signal detection in mammalian systems, it has not been widely adopted [14]. Hybrid approaches that combined a membrane-bound GFP-sensor with a chemical fluorophore have yielded larger fractional fluorescence changes (up to 35% per 100 mV), but require delivery of exogenous and toxic dyes to the imaged cells [15]. Recently, a fluorescent voltage sensor derived from a non-ion channel phosphatase protein has been developed that shows good membrane targeting in mammalian cells, and as much as an 8% change/100 mV in fluorescence at physiological temperatures; however, it remains too slow to follow individual action potentials [16]. It is unclear whether this construct can be expressed at high enough levels in vivo to become widely used by systems neuroscientists. A nontrivial problem in the use of voltage sensors is that changes in cell voltage are not restricted to cell bodies but also occur in axonal and dendritic processes. Thus, signal changes occur over large areas of neuropil, and do not allow identification of individual, active neurons that are embedded within large networks.
Cl− based sensors
Clomeleon, a Cl− sensor developed in 2000 [17], is an effective FRET-based approach for detecting physiologically relevant Cl− concentration in cultured hippocampal neurons. Clomeleon has been used to examine somato-dendritic gradients of Cl− in ON-and OFF-bipolar cells in the retina in thy1-Clomeleon transgenic mice [18] and ischemia-induced changes in intracellular Cl−. However, Clomeleon transgenic mice have not been widely used to monitor Cl− influx during signal integration during neuronal firing in vitro or in vivo, in part because the kinetics of fluorescence change are slow (~1 sec) compared to a single action potential (AP). During normal firing activity in vivo, both excitation and inhibition are activated. Monitoring Cl− concentration across populations of cells is likely to reveal interesting and dynamic properties of neuron assemblies over longer timecourses, for example, during paroxysmal firing or Upstates in the neocortex.
Ca++-based sensors
Chemically-based Ca++-indicators have been enormously useful in studying the activity of neuronal ensembles both in acute brain slices and in vivo. Because action potential firing induces large Ca++ transients in neuronal cell bodies, Ca++ indicators provide excellent cellular resolution. Although there are a large number of protein Ca++-sensors, including Camgaroo-1 and 2 [19], Cameleon [20], GCaMP1.3 [21], Flash- and Inverse Pericam [22], and the troponin C-based sensor TN-L15 [23], the majority of these sensors have not been widely employed in vivo because Ca++ sensitivity is reduced when these proteins are expressed in the mammalian CNS.
In a rigorous, side-by-side comparison of many of these Ca++-sensors in Drosophila, TN-L15 showed the fastest signal at low firing rates and the least photobleaching [24]. This troponin-C based biosensor has been used to make transgenic mice (using a Thy1 promoter to randomly drive expression in neuronal subpopulations throughout the CNS) and can be used to visualize ensemble activity in acute brain slices and in vivo [25]. Since the sensor is cytoplasmically localized, these mice may be useful to examine Ca++ transients in specified neuronal compartments, including dendrites.
A significant problem for all Ca++-based indicators is their buffering of intracellular Ca++ that can alter intracellular signaling pathways, especially when indicators are present at high intracellular concentrations. In addition, the long decay time of the signal (for TN-L15, ~2–10 ms, depending on the number of APs) makes it difficult to temporally resolve individual spikes. However, the development of Ca++ sensors with a physiologically appropriate dynamic range in transgenic animals promises that these animals will become extremely useful for monitoring neuronal circuit activity under a variety of experimental conditions.
pHluorins, pH-based sensors
Synaptic vesicles undergo a dramatic change in pH upon vesicle fusion with the plasma membrane, a property that has been employed for the synaptic vesicle fusion sensor synaptopHluorin [26]. Both ratiometric and ecliptic pHlourins have been engineered [26], although the superecliptic synaptopHluorin has been most widely employed to address important biological questions in mammalian systems. For example, synaptic vesicle recycling dynamics in neurons from Thy1-driven synaptopHluorin transgenic mice have been examined, providing results that challenged current models about constraints on vesicle reuse [27].
The broad distribution of active release sites in the CNS – providing a diffuse haze of synapses to image – challenges widespread use of synpatopHluorin as a tool to examine the activity of neural circuits. One prominent exception this is in the olfactory bulb, where thousands of odorant-specific olfactory sensory neurons fasciculate to target individual glomeruli. The olfactory marker protein (OMP) promoter has been used to drive synaptopHluorin expression across olfactory sensory neurons (OSN), providing a functional map of OSN responsiveness to odorant presentation in vivo [28]. Intriguingly, results from this imaging study were different from those obtained using optical imaging or 2-deoxyglucose analysis, possibly because these methods are lower resolution, requiring greater signal averaging within and between animals, or because these methods may be more sensitive to post-synaptic activity (rather than presynaptic release). These animals have also been used to test hypotheses about inter- and intra-glomerular inhibition in vitro and in vivo [29].
Conclusion
Transgenic reporters of neural activity have been developed as indicators of IEG expression, sensors of voltage changes or ion flux, or detectors of presynaptic release. Despite great interest and initial enthusiasm for voltage- and ion based-sensors, they have been the least promising of all activity-reporters for CNS analysis in mammalian systems. IEG-reporters offer great promise in detecting neuronal activity, especially for questions requiring cellular resolution over longer timescales.
Acknowledgments
Special thanks to Ehud Isacoff and Vincent Pieribone for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, Morgan JI. fos-lacZ transgenic mice: mapping sites of gene induction in the central nervous system. Neuron. 1992;8:13–23. doi: 10.1016/0896-6273(92)90105-m. [DOI] [PubMed] [Google Scholar]
- 2.Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slade JP, Man PS, Wells T, Carter DA. Stimulus-specific induction of an Egr-1 transgene in rat brain. Neuroreport. 2002;13:671–674. doi: 10.1097/00001756-200204160-00027. [DOI] [PubMed] [Google Scholar]
- 5.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 6.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. The first demonstration that IEG expression could drive detectable GFP fluorescence in living cells. This report characterized the inducibility and expression patterns of fosGFP in multiple lines of transgenic mice using pharmacological, physiological, and behavioral stimuli. In addition, fosGFP expression was used to identify specific GFP+ neurons that had been activated by a prior in vivo sensory stimulus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clem RL, Barth A. Pathway-specific trafficking of native AMPARs by in vivo experience. Neuron. 2006;49:663–670. doi: 10.1016/j.neuron.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. The first report of an Arc-driven GFP reporter transgenic mouse. Transgenic animals were generated using a knock-in approach, but high levels of arc expression yielded detectable GFP fluorescence. Patterns of Arc-GFP expression were used to examine experience-dependent refinement of neuronal activation in visual cortex, using 2-photon imaging. A major tour-de-force of this study was the repeated identification of the same neurons over multiple imaging sessions. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 10.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 11.Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. This landmark paper initiated the development of the field of genetically-encoded sensors of neural activity. The authors created a fusion protein of the K channel Shaker and GFP and used the voltage-sensitive properties of the potassium channel as a way to control changes in GFP fluorescence. Maximal fractional fluorescence changes were comparable to some of the best voltage-sensitive dyes available at the time. [DOI] [PubMed] [Google Scholar]
- 12.Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker BJ, Lee H, Pieribone VA, Cohen LB, Isacoff EY, Knopfel T, Kosmidis EK. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods. 2007;161:32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Guerrero G, Siegel MS, Roska B, Loots E, Isacoff EY. Tuning FlaSh: redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys J. 2002;83:3607–3618. doi: 10.1016/S0006-3495(02)75361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci. 2005;8:1619–1626. doi: 10.1038/nn1558. [DOI] [PubMed] [Google Scholar]
- 16.Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akemann W, Knopfel T. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS ONE. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuner T, Augustine GJ. A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron. 2000;27:447–459. doi: 10.1016/s0896-6273(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 18.Duebel J, Haverkamp S, Schleich W, Feng G, Augustine GJ, Kuner T, Euler T. Two-photon imaging reveals somatodendritic chloride gradient in retinal ON-type bipolar cells expressing the biosensor Clomeleon. Neuron. 2006;49:81–94. doi: 10.1016/j.neuron.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 21.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 22.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J Biol Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- 24.Reiff DF, Ihring A, Guerrero G, Isacoff EY, Joesch M, Nakai J, Borst A. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. A major issue for genetically-encoded indicators is their relative efficacy in living animals. In this excellent report, the authors systematically evaluated the performance of 10 different Ca++ indicators as well as synapto-pHluorin at the Drosophila neuromuscular junction. The troponin-C based sensor TN-15 showed the fastest activation kinetics, and the circularly permuted GFP GCaMP1.3 and Yellow Cameleon 3.3 showed good linear fluorescence changes in response to increasing neural activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, Kovalchuk Y, Konnerth A, Griesbeck O. Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nat Methods. 2007;4:127–129. doi: 10.1038/nmeth1009. [DOI] [PubMed] [Google Scholar]
- 26.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Burrone J, Tyler WJ, Hartman KN, Albeanu DF, Murthy VN. Synaptic vesicle recycling studied in transgenic mice expressing synaptopHluorin. Proc Natl Acad Sci U S A. 2005;102:6131–6136. doi: 10.1073/pnas.0501145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozza T, McGann JP, Mombaerts P, Wachowiak M. In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 2004;42:9–21. doi: 10.1016/s0896-6273(04)00144-8. [DOI] [PubMed] [Google Scholar]
- 29.McGann JP, Pirez N, Gainey MA, Muratore C, Elias AS, Wachowiak M. Odorant representations are modulated by intra- but not interglomerular presynaptic inhibition of olfactory sensory neurons. Neuron. 2005;48:1039–1053. doi: 10.1016/j.neuron.2005.10.031. [DOI] [PubMed] [Google Scholar]


