Abstract
Genome-wide scans in Icelandic, Australian/New Zealand and Finnish pedigrees have provided evidence for maternal susceptibility loci for pre-eclampsia on chromosome 2, although at different positions (Iceland: 2p13 and 2q23, Australia/New Zealand: 2p11–12 and 2q22, Finland: 2p25). In this project, a large population-based (n=65 000) nested case–control study was performed in Norway to further explore the association between positional candidate genes on chromosome 2q and pre-eclampsia, using single-nucleotide polymorphisms (SNPs). DNA samples from 1139 cases (women with one or more pre-eclamptic pregnancies) and 2269 controls (women with normal pregnancies) were genotyped using the Applied Biosystems SNPlex high-throughput genotyping assay. In total, 71 SNPs within positional candidate genes at 2q22–23 locus on chromosome 2 were genotyped in each individual. Genotype data were statistically analysed with the sequential oligogenic linkage analysis routines (SOLAR) computer package. Nominal evidence of association was found for six SNPs (rs1014064, rs17742134, rs1424941, rs2161983, rs3768687 and rs3764955) within the activin receptor type 2 gene (ACVR2A) (all P-values <0.05). The non-independence of statistical tests due to linkage disequilibrium between SNPs at a false discovery rate of 5% identifies our four best SNPs (rs1424941, rs1014064, rs2161983 and rs3768687) to remain statistically significant. The fact that populations with different ancestors (Iceland/Norway–Australia/New Zealand) demonstrate a common maternal pre-eclampsia susceptibility locus on chromosome 2q22–23, may suggest a general role of this locus, and possibly the ACVR2A gene, in pre-eclampsia pathogenesis.
Keywords: pre-eclampsia, genetic association, chromosome 2q22, ACVR2
Introduction
Pre-eclampsia is a complex and potentially life-threatening disorder affecting approximately 3% of all pregnant women in the western world.1 Pre-eclampsia typically develops in the latter half of pregnancy and is a rapidly progressive condition characterized by hypertension and proteinuria. Although the condition has been known for a long time, pre-eclampsia is still one of the major pregnancy disorders and a leading cause of maternal and fetal morbidity and mortality.2 There is no effective treatment, and delivery of the baby is the only alternative if the life of the pregnant woman is threatened.
Epidemiological research through decades has demonstrated that pre-eclampsia has a familial association, and this has led to the conclusion that genetic control and inheritance has a major function in the pathology of pre-eclampsia.3 Genetic factors are suggested to be responsible for more than 50% of the liability to pre-eclampsia,4, 5 but the pattern of inheritance is still unclear. Although the tendency for pre-eclampsia to be passed on from mother to daughter was the first pattern to be observed,3, 6, 7 it is now clear that fathers also contribute to pre-eclampsia susceptibility in their partners, possibly through paternal transmission of genes on to the fetus.8, 9
The fact that genetic factors have a major function in the development of pre-eclampsia has encouraged several research groups to search for maternal pre-eclampsia susceptibility genes. New molecular technologies and better bioinformatics tools have become important approaches in the search for genetic clues in ‘complex traits' such as pre-eclampsia. Genome-wide linkage scans of pre-eclampsia pedigrees have pointed to several maternal pre-eclampsia susceptibility loci.10, 11, 12, 13, 14 Three independent and distinct studies from Iceland, Australia/New Zealand and Finland have shown linkage to chromosome 2 although at different positions.10, 13, 14 By the use of a variance components-based linkage approach, the Australian research group resolved a susceptibility quantitative trait locus for pre-eclampsia on chromosome 2q22, and an objective prioritization strategy assigned the highest priority to the activin receptor type 2 gene (ACVR2A),4 which is a receptor for the cell signalling protein activin A. Activin A is a multifunctional protein with roles in cell differentiation, implantation15 and decidualization.16 In addition, recent studies suggest that activin A is involved in pathogenesis of inflammatory diseases17, 18, 19 and atherogenesis.20, 21
In this study, the association between the positional candidate genes at the 2q22 susceptibility locus, among them ACVR2A, has been explored further in a large Norwegian case–control cohort, using association analysis of SNPs. In total, 1139 cases (women with one or more pre-eclamptic pregnancies) and 2269 controls (women with normal pregnancies) were genotyped.
Materials and methods
Study population
All women in this study were retrospectively recruited from Nord-Trøndelag County in Norway as part of a large multipurpose health survey conducted during 1995–1997 (the HUNT2 study). More than 65 000 inhabitants participated. The people living in Nord-Trøndelag County are considered representative for the Norwegian population, and is well suited for genetic studies because of its ethnic homogeneity (<3% non-Caucasians).22, 23 The study population is described in detail elsewhere.24
Pre-eclampsia diagnosis and identification of cases
Pre-eclampsia was defined as the onset of persistent hypertension (exceeding 140/90 mmHg), in combination with proteinuria (exceeding 300 mg/day) after the 20th week of gestation. The women with pre-eclamptic and normal pregnancies in the HUNT cohort were identified by linking the HUNT database to the database at the Medical Birth Registry of Norway (MBRN), using diagnosis codes ICD-8 (before 1998) and ICD-10 (after 1998).
We identified 1179 women registered with pre-eclampsia in one or more pregnancies (cases) and 2358 women with a history of normal pregnancies as controls. Among these, blood samples were available from 1139 women from the case group and 2269 women from the control group at the HUNT biobank. Finally, cases were subdivided into subgroups; 1003 women were registered with one pre-eclamptic pregnancy, 606 of these had pre-eclampsia in their first pregnancy, whereas 397 had pre-eclampsia in a later pregnancy. One hundred and thirty-six women had experienced pre-eclampsia in more than one pregnancy.
Chromosome 2q candidate genes and SNP prioritization
The identification and objective prioritization of positional candidate genes at the Australian/New Zealand chromosome 2q pre-eclampsia susceptibility locus has been described previously.4 In this study, seven positional candidate genes at this locus were selected for SNP-specific association analysis on the basis of their GeneSniffer ‘hit-scores' (www.genesniffer.org), differential gene expression levels, preliminary SNP association and linkage disequilibrium (LD) data and/or scientific literature pertaining to their potential function in reproductive biology.4, 25, 26, 27 The seven positional candidate genes selected are: chemokine (C-X-C motif) receptor 4 (CXCR4), histamine N-methyltransferase (HNMT), kynureninase (-kynurenine hydrolase) (KYNU), reprimo (RPRM), activin A receptor type I (ACVR1), activin A receptor type IC (ACVR1C) and activin A receptor type 2 (ACVR2A). Seventy-one validated and/or coding sequence SNPs within these genes were either selected from the NCBI SNP database (dbSNP) (human build 36.2), identified by re-sequencing of candidate genes in the Australian/New Zealand pre-eclampsia pedigrees (E Fitzpatrick, personal communication) or obtained from the published literature28 (Table 1).
Table 1. Objectively prioritized chromosome 2q positional candidate SNPs.
| Gene | SNP | Location (bp) | Genetic location | Function | Allele frequency | P-value (MG) | OR (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| CXCR4 | rs2734871 | 136 586 343 | Genomic | C 0.7957 | T 0.2043 | NS | ||
| rs2228014 | 136 589 554 | Exonic | C 0.9746 | T 0.0254 | NS | |||
| rs1051557 | 136 589 623 | Exonic | Synonymous | NP | ||||
| rs2680880 | 136 590 019 | Intronic | T 0.6125 | A 0.3875 | NS | |||
| R334Xa | 136 590 140 | Exonic | Nonsense | NP | ||||
| E343Xa | 136 590 167 | Exonic | Nonsense | NP | ||||
| rs3806501 | 136 593 539 | Genomic | NP | |||||
| rs4954391 | 136 600 293 | Genomic | T 0.7932 | C 0.2068 | NS | |||
| HNMT | rs1471003 | 138 441 472 | Intronic | NP | ||||
| rs1982494 | 138 445 583 | Intronic | T 0.6269 | C 0.3731 | NS | |||
| rs3791244 | 138 448 235 | Genomic | C 0.8625 | T 0.1375 | NS | |||
| rs3100725 | 138 459 636 | Intronic | C 0.7951 | T 0.2049 | NS | |||
| rs3113222 | 138 472 267 | Genomic | NP | |||||
| rs1801105 | 138 476 119 | Genomic | C 0.8668 | T 0.1332 | NS | |||
| rs3100719 | 138 480 404 | Genomic | C 0.7685 | T 0.2315 | NS | |||
| rs4954941 | 138 486 437 | Genomic | G 0.7902 | A 0.2098 | NS | |||
| KYNU | rs16858172 | 143 352 974 | Intronic | C 0.9597 | A 0.0403 | NS | ||
| rs1439875 | 143 355 039 | Intronic | T 0.9515 | G 0.0485 | NS | |||
| rs16858196 | 143 356 117 | Intronic | NP | |||||
| rs4662304 | 143 358 120 | Genomic | G 0.5119 | A 0.4881 | NS | |||
| rs16858205 | 143 359 529 | Exonic | Synonymous | NP | ||||
| rs1438266 | 143 363 789 | Intronic | A 0.5138 | G 0.4862 | NS | |||
| rs6429992 | 143 379 037 | Intronic | T 0.5112 | C 0.4888 | NS | |||
| rs351675 | 143 428 514 | Intronic | G 0.9927 | C 0.0073 | NS | |||
| rs2304705 | 143 431 735 | Exonic | Missense | NP | ||||
| rs1050951 | 143 463 434 | Exonic | 3′UTR | G 0.8966 | A 0.1034 | NS | ||
| rs2304700 | 143 507 484 | Genomic | G 0.8907 | T 0.1093 | NS | |||
| rs9013 | 143 514 659 | Genomic | T 0.8435 | C 0.1565 | NS | |||
| ACVR2A | rs1424954 | 148 317 264 | 5′UTR | G 0.7247 | A 0.2753 | 0.387 | 0.94 (0.82–1.07) | |
| rs13224 | 148 319 154 | 5′UTR | NP | |||||
| rs7572676 | 148 320 092 | Intronic | NP | |||||
| LF002b | 148 328 072 | Intronic | NP | |||||
| rs1014064 | 148 328 624 | Intronic | A 0.7176 | G 0.2824 | 0.0184 | 0.86 (0.77–0.98) | ||
| rs17741978b | 148 333 850 | Intronic | C 0.8267 | G 0.1733 | 0.0689 | 1.14 (0.99–1.32) | ||
| rs1895694 | 148 336 216 | Intronic | A 0.5240 | G 0.4760 | 0.918 | 1.00 (0.90–1.11) | ||
| rs2113794 | 148 339 447 | Intronic | A 0.5235 | C 0.4765 | 0.998 | 1.01 (0.90–1.12) | ||
| rs17742134b | 148 343 866 | Intronic | C 0.8252 | T 0.1748 | 0.0214 | 1.17 (1.02–1.35) | ||
| rs1424941 | 148 359 588 | Intronic | G 0.8235 | A 0.1765 | 0.0171 | 1.18 (1.03–1.36) | ||
| rs2161983 | 148 365 856 | Intronic | C 0.7139 | T 0.2861 | 0.0196 | 0.86 (0.77–0.98) | ||
| rs2288190 | 148 374 059 | Intronic | C 0.6428 | T 0.3572 | 0.0554 | 0.89 (0.79–0.99) | ||
| rs10497025b | 148 378 672 | Intronic | C 0.7618 | G 0.2382 | 0.0586 | 0.88 (0.78–1.00) | ||
| rs1227307 | 148 378 775 | Intronic | NP | |||||
| rs3768687 | 148 388 490 | Intronic | G 0.7162 | A 0.2838 | 0.0214 | 0.86 (0.76–0.98) | ||
| LF020b | 148 390 323 | Intronic | A 0.9946 | G 0.0054 | 0.787 | 0.81 (0.29–2.24) | ||
| rs3764955 | 148 391 267 | Intronic | C 0.7208 | G 0.2792 | 0.0327 | 0.87 (0.77–0.98) | ||
| rs7601098 | 148 393 084 | Intronic | G 0.9380 | C 0.0620 | 0.538 | 1.08 (0.86–1.34) | ||
| rs2303392 | 148 396 897 | Intronic | Failed | |||||
| RPRM | rs3770303 | 154 041 809 | 3′ near gene | C 0.8265 | T 0.1735 | NS | ||
| rs1052581 | 154 042 352 | 3′UTR | G 0.9141 | A 0.0859 | NS | |||
| rs1063728 | 154 042 745 | 3′UTR | G 0.5289 | C 0.4711 | NS | |||
| rs2033764 | 154 044 214 | Genomic | A 0.8134 | G 0.1866 | NS | |||
| ACVR1C | rs6751595 | 158 097 764 | 3′UTR | NP | ||||
| rs7594480 | 158 098 714 | Exonic | Missense | T 0.9349 | C 0.0651 | NS | ||
| rs6746788 | 158 109 351 | Exonic | Synonymous | NP | ||||
| rs4556933 | 158 152 135 | Exonic | Synonymous | G 0.6425 | A 0.3575 | NS | ||
| ACVR1 | rs3738927 | 158 302 306 | Exonic | Synonymous | NP | |||
| rs6729964 | 158 330 839 | Exonic | Synonymous | NP | ||||
| rs1146031 | 158 335 226 | Exonic | Synonymous | A 0.9949 | G 0.0051 | NS | ||
| rs2227861 | 158 345 156 | Exonic | Synonymous | A 0.7437 | G 0.2563 | NS | ||
| rs13406336 | 158 364 208 | Exonic | Missense | G 0.9970 | C 0.0030 | NS | ||
| rs16842103 | 158 367 073 | Intronic | NP | |||||
| rs16842105 | 158 367 916 | Intronic | NP | |||||
| rs7561419 | 158 370 035 | Intronic | NP | |||||
| rs17798043 | 158 371 523 | Intronic | C 0.9130 | T 0.0870 | NS | |||
| rs16842106 | 158 371 886 | Intronic | NP | |||||
| rs1036739 | 158 373 442 | Intronic | C 0.7790 | T 0.2210 | NS | |||
| rs10497191 | 158 375 463 | Intronic | C 0.8717 | T 0.1283 | NS | |||
| rs4380178 | 158 376 691 | Intronic | Failed | |||||
| rs4294956 | 158 376 700 | Intronic | Failed | |||||
| rs12327948 | 158 377 663 | Intronic | C 0.8047 | G 0.1953 | NS | |||
| rs1965812 | 158 379 544 | Intronic | C 0.8071 | T 0.1929 | NS | |||
| rs10497192 | 158 379 946 | Intronic | T 0.7024 | C 0.2976 | NS |
NP; non-polymorphic, P-value (MG); P-value for the measured genotype test, OR; odds ratio, CI; confidence interval, NS; not significant, SNPs; single-nucleotide polymorphisms.
Hernandez et al.28
SNP discovery in Australian/New Zealand pre-eclampsia cohort (E Fitzpatrick, personal communication).
SNP genotyping
DNA for genotyping was extracted from blood samples stored in the HUNT biobank as described elsewhere.24 The Applied Biosystems SNPlex™ Genotyping System (Applied Biosystems, CA, USA) was used for SNP genotyping. All samples were electrophoretically separated on a 3730 DNA Genetic Analyzer (Applied Biosystems), and automated allele calls and genotype clustering of each individual sample was performed by Applied Biosystems' GeneMapper® Software (version 4.0), as described in more detail elsewhere.24
Statistical analysis
Positional candidate SNPs were analysed in SOLAR29 using a measured genotype test.30 The analysis used a standard threshold model assuming an underlying normal distribution of liability. The threshold model and its assumptions are virtually identical to those used in standard logistic regression but benefits from the ease of interpretation with regard to genetic effects. The measured genotype test of associations assesses the extent of genotypic mean differences (on the liability or risk scale) between case and control singletons assuming a model of additive gene action. Odds ratios (ORs) with 95% confidence intervals were calculated using SPSS 13.0 (for Mac OS® X) under the assumption of an additive genetic model. LD parameters among intragenic SNPs were estimated using basic correlation methods to assess all disequilibria jointly using SOLAR. To accommodate for multiple hypothesis testing, we used the approach of Li and Ji31 to determine the effective number of independent SNPs (and tests) in combination with applying a false discovery rate (FDR) of 5%.32
Ethics
The study was approved by the Regional Committee for Medical Research Ethics, Norway and approved by the National Data Inspectorate and The Directorate of Health and Social Welfare. Ethical approval for genotyping and statistical analysis of the Norwegian cohort was obtained from The University of Texas Health Science Center at San Antonio, Institutional Review Board.
Results
Statistical power analysis
Formal power calculations using SOLAR determined that for the case and control sample available for study, there was an 80% likelihood of identifying a SNP accounting for at least 2% of the total variation in the dichotomous pre-eclampsia phenotype (where affected is scored as 1 and unaffected as 0).
SNP genotyping and association
Twenty of the 71 gene-centric SNPs (15 coding, 56 non-coding) were non-polymorphic and three additional SNPs failed the SNPlex genotyping assay (Table 1). All successfully genotyped polymorphic SNPs conformed to Hardy–Weinberg proportions (P>0.05).
Of the seven genes tested, we observed nominal SNP associations pertaining to ACVR2A only (Table 1). There were a total of 19 SNPs tested in ACVR2A with six SNPs (rs1014064, rs17742134, rs1424941, rs2161983, rs3768687 and rs3764955) exhibiting nominal association with the pre-eclampsia group (n=1139) (P<0.05). An additional three SNPs (rs17741978, rs2288190 and rs10497025) exhibited borderline significance (P<0.07), whereas another five SNPs were nonsignificant (Table 1). The remainder of the tested ACVR2A SNPs were either non-polymorphic (n=4) or failed the SNPlex genotyping assay (n=1). Analysis of the ACVR2A genotypes of subgroups of cases (women registered with one pre-eclamptic pregnancy (n=1003), women with pre-eclampsia in their first and only registered pre-eclamptic pregnancy (n=606), women with pre-eclampsia once but in a later pregnancy (n=397) and women experienced pre-eclampsia in more than one pregnancy (n=136)) were compared to controls but there were no significant differences between the subgroups (data not shown). Thus, there is no evidence of locus-specific genetic heterogeneity associated with variations in the definition of the disease.
Of the SNPs displaying a nominal or borderline association with pre-eclampsia, we observed discordant OR either greater than one (rs17741978, rs17742134 and rs1424941) or less than one (rs1014064, rs2161983, rs2288190, rs10497025, rs3768687 and rs3764955) (Table 1). In our calculations an OR>1 is indicative of pre-eclamptic women more likely than control women to carry the minor allele at a tested ACVR2A SNP locus, whereas an OR<1 is indicative of pre-eclamptic women less likely than control women to carry the minor allele, hence favouring the major allele, at a tested ACVR2A SNP locus.
All polymorphic ACVR2A SNPs were re-tested in SOLAR with the inclusion of maternal age as a covariate effect. Nominal and borderline associated ACVR2A SNPs did not differ when adjusted for maternal age (data not shown). OR for nominal and borderline ACVR2A SNPs associated with pre-eclampsia did not differ either when adjusted for maternal age (data not shown).
The re-evaluation of our statistical results for ACVR2A to take into account multiple testing and the non-independence of the tests due to LD between SNPs utilized 13 of the 14 tested SNPs. The LF020 SNP was eliminated because of the observed low number of copies preventing valid use of our statistical test. Of the 13 remaining SNPs, we identified the effective number of independent SNPs and therefore statistical tests to be five using the approach by Li and Ji.31 Five tests is a substantial reduction from the original 13 and when a modified FDR approach is incorporated we find that the four best SNPs (rs1424941, rs1014064, rs2161983 and rs3768687) meet an FDR of 0.05. Therefore, allowing for the total experiment, these four SNPs are significant and we have only a 1 in 20 chance of having made a false discovery. FDR approaches can be substantially more powerful than classical approaches (eg, Bonferroni) for multiple testing.32
None of the tested SNPs within the other six genes (CXCR4, HNMT, KYNU, RPRM, ACVR1 or ACVR1C) exhibited an association with pre-eclampsia (Table 1).
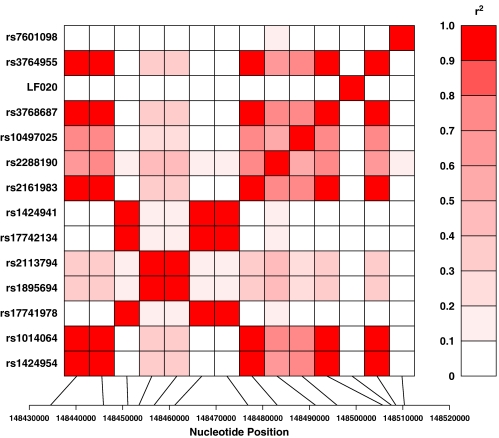
ACVR2A SNP LD
Figure 1 portrays the overall pattern of LD exhibited among ACVR2A SNPs. The patterns of LD are measured by the squared value of the pairwise correlation among intragenic genotypes (r2). Hence, observed patterns of LD are herein referred to as isocorrelated redundant variant (IRV) sets.33 Among the typed ACVR2A SNPs, we observe three very strong (r2>0.7) IRV sets, namely, IRV set 1 (IRV1); rs1424954, rs1014064, rs2161983, rs2288190, rs10497025, rs3768687 and rs3764955, IRV2; rs17741978, rs17742134 and rs1424941, IRV3; rs1895694 and rs2113794. The SNP loci within IRV1 all exhibit an OR<1, hence pre-eclamptic women are more likely than control women to carry the major allele at these ACVR2A loci. The SNP loci within IRV2 all exhibit an OR>1, hence pre-eclamptic women are more likely than control women to carry the minor allele at these ACVR2A loci.
Figure 1.
The pattern of linkage disequilibrium in ACVR2A. The intensity of red colour depicts the magnitude of the squared value of the pairwise correlation among intragenic genotypes. The more intense colour (0.0≥r2≥1.0) the stronger the correlation.
Discussion
In the present large positional candidate gene-based association study, the existence of a maternal susceptibility locus for pre-eclampsia at chromosome 2 was verified in Norwegian women with nominal association being obtained for multiple SNPs within the activin receptor gene ACVR2A on 2q22. When we accounted for multiple hypothesis testing and the non-independence of genotyped SNPs, our four best ACVR2A SNPs (rs1424941, rs1014064, rs2161983 and rs3768687) remained significantly associated with pre-eclampsia. For some of these SNPs it was the minor allele that was associated with pre-eclampsia, whereas for other SNPs it was the major allele, most likely reflecting naturally occurring evolutionary processes such as frequency differences, history and origins of each ACVR2A SNP locus.
Although replication of an association with the ACVR2A gene across non-related populations supports the hypothesis that genes influencing maternal susceptibility for development of pre-eclampsia are located at chromosome 2q22, we could not see concordance between the tested ACVR2A SNPs showing association in our Norwegian and Australian/New Zealand pre-eclampsia cohorts. However, we do observe strong LD between the SNP showing the strongest association in the Australian study (rs1424954) and four SNPs (rs1014064, rs2161983, rs3768687, rs3764955) showing nominal association with pre-eclampsia in the Norwegian cohort. Unfortunately, this SNP was relatively poorly typed in the Norwegian sample and the reduced sample size (n=2274) is a likely reason for the weaker association result. Nonetheless, these data continue to bring focus on a possible role for the ACVR2A gene in pre-eclampsia pathogenesis.
Activin belongs to the transforming growth factor-β (TGF-β) family that can regulate cell differentiation, proliferation and apoptosis,34, 35 and utilizes two types of cell surface receptors (ACVR1 and ACVR2). The receptors consist of an extracellular domain that specifically binds activin A, a membrane-spanning domain and an intracellular kinase domain.36 The ACVR2A gene encodes the activin A type 2 receptor.36 Potential ligands for ACVR2 include activin A, activin B and inhibin A, but not TGF-β.37 Activin A is secreted by most cell types, and control of expression of both receptors appears to occur in a tissue- and gene-specific manner during human development.38 Activin receptors are expressed in human placental tissue (trophoblasts and vascular endothelial cells) from early pregnancy.39 Activin A promotes trophoblast proliferation and differentiation15 and appears to have a fundamental function during implantation and decidualization.40 In addition, placental hormonogenesis41 and uterotonin secretion42 are modulated by activin A.
The elevated levels of activin A have been found in pre-eclamptic women.43, 44, 45, 46, 47, 48 Placenta is probably the main source of activin A in maternal circulation.49 Altered expression of ACVR2A, as suggested by the gene expression analysis performed by Moses et al,4 may influence activin A concentrations and action with consequences on trophoblast invasion and remodelling of the spiral arteries. However, it is not clear whether the increased activin A concentration/reduced ACVR2A expression is a cause or a consequence of placental dysfunction. Thus, the activin pathway is a plausible biological candidate for a role in mechanisms perpetuating pre-eclampsia pathogenesis. Recently, activin A has been suggested to have a significant function in inflammatory diseases17, 18, 19 and atherogenesis.20, 21 The elevated serum levels of activin A have been reported in patients with cardiovascular diseases (CVD), and a downregulated expression of ACVR2A has been shown in patients with unstable CVD, suggesting a dysregulated receptor expression.50 Women with pre-eclampsia have an increased morbidity and mortality of CVD.51 Thus, the hypothesis may be raised that the linkage between ACVR2A and pre-eclampsia may be ascribed to the role of activin A in endothelial activation and systemic endovascular inflammatory responses. Supporting the inflammatory response hypothesis, we recently reported a significant association between the SEPS1 gene (which is involved in stress response in the endoplasmic reticulum and inflammation control) and pre-eclampsia in our Norwegian cohort.24
It is also plausible that our association signals are not the result of a causal variation involving the ACVR2A gene but are acting as markers for causal variation in a nearby gene(s). To investigate this possibility we assessed other genes within 500 kb of ACVR2A. The origin recognition complex, subunit 4-like (yeast) (ORC4L) gene is the only gene within this chromosomal region and resides telomeric to ACVR2A with ∼300 kb separating their respective 3′UTRs. Utilizing the Australian/New Zealand data set, there is evidence of strong LD (0.74≥r2≥1.00) between typed ACVR2A and ORC4L SNPs (unpublished data). The protein encoded by ORC4L is a subunit of the ORC complex, which is essential for DNA replication in mammalian cells.52 Without complete deep re-sequencing of ACVR2A and ORC4L in the Norwegian sample, followed by functional assessment of the prioritized variants showing association (which is beyond the scope of this current study), we can only speculate as to the potential role of the genetic signals observed in this genomic region, either by ACVR2A and/or ORC4L, in reference to pre-eclampsia susceptibility.
In conclusion, we have shown evidence for association between the ACVR2A gene and pre-eclampsia in a Norwegian population. By this, we have confirmed data obtained in the Australian/New Zealand family linkage study.4 The determination of the actual functional polymorphisms in ACVR2A and nearby genes that confer susceptibility to pre-eclampsia remains the challenge ahead. The Australian group performed whole genome microarray analysis of decidual tissues collected from pre-eclamptic and normotensive pregnant women. They observed 15 of the 17 transcripts at 2q with at least a 2.5-fold change in expression being downregulated in the pre-eclamptic samples, compared to normotensive tissue.4 Among these 15 transcripts, the ACVR genes (both ACVR2 and ACVR1C) demonstrated a more than 10-fold difference. However, expression analysis was performed on a rather small collection of samples (seven cases and seven controls), and the individual samples of each group were pooled before expression analysis was performed.4 We propose to perform further transcriptional profiling on a large Norwegian collection of decidual samples from pre-eclampsia cases and controls, with associated genotyping of the ACVR2A gene in individuals that show differential decidual expression of the gene. By using this approach, we hope to gain information about a possible causal link between ACVR2A genotype(s) and the expression of this gene. In addition, transcription-based approaches may imply a more appropriate prioritization of candidate genes in future epidemiological studies of genetics in pre-eclampsia.
Acknowledgments
The Nord-Trøndelag Health Study (the HUNT study) is a collaboration between the HUNT Research Centre, Faculty of Medicine, NTNU, the Norwegian Institute of Public Health and the Nord-Trøndelag County Council. We thank Drs Harald HH Göring and Åsa Johansson for their constructive review of the manuscript. This study was supported by grants from the Central Norway Regional Health Authority and the Medical Faculty and Office of International Relations at NTNU (Linda T Roten), the Cowles Post-Doctoral Fellowship (Matthew P Johnson), the Elizabeth Huth Coates Charitable Foundation and grants from the National Institutes of Health. (Eric K Moses, R01HD049847; John Blangero, MH59490). Genotyping and statistical analysis were conducted in facilities constructed with support for the Research Facilities Improvement Program Grant Numbers C06 RR013556 and C06 RR017515 from the National Center for Research Resources, NIH.
References
- Saftlas AF, Olson DR, Franks AL, Atrash HK, Pokras R. Epidemiology of preeclampsia and eclampsia in the United States, 1979–1986. Am J Obstet Gynecol. 1990;163:460–465. doi: 10.1016/0002-9378(90)91176-d. [DOI] [PubMed] [Google Scholar]
- Taylor D. The epidemiology of hypertension during pregnancy. Amsterdam: Elsevier; 1988. [Google Scholar]
- Arngrimsson R, Bjornsson S, Geirsson RT, Bjornsson H, Walker JJ, Snaedal G. Genetic and familial predisposition to eclampsia and pre-eclampsia in a defined population. Br J Obstet Gynaecol. 1990;97:762–769. doi: 10.1111/j.1471-0528.1990.tb02569.x. [DOI] [PubMed] [Google Scholar]
- Moses EK, Fitzpatrick E, Freed KA, et al. Objective prioritization of positional candidate genes at a quantitative trait locus for pre-eclampsia on 2q22. Mol Hum Reprod. 2006;12:505–512. doi: 10.1093/molehr/gal056. [DOI] [PubMed] [Google Scholar]
- Salonen Ros H, Lichtenstein P, Lipworth L, Cnattingius S. Genetic effects on the liability of developing pre-eclampsia and gestational hypertension. Am J Med Genet. 2000;91:256–260. [PubMed] [Google Scholar]
- Chesley LC, Cooper DW. Genetics of hypertension in pregnancy: possible single gene control of pre-eclampsia and eclampsia in the descendants of eclamptic women. Br J Obstet Gynaecol. 1986;93:898–908. doi: 10.1111/j.1471-0528.1986.tb08006.x. [DOI] [PubMed] [Google Scholar]
- Cooper DW, Hill JA, Chesley LC, Bryans CI. Genetic control of susceptibility to eclampsia and miscarriage. Br J Obstet Gynaecol. 1988;95:644–653. doi: 10.1111/j.1471-0528.1988.tb06524.x. [DOI] [PubMed] [Google Scholar]
- Esplin MS, Fausett MB, Fraser A, et al. Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med. 2001;344:867–872. doi: 10.1056/NEJM200103223441201. [DOI] [PubMed] [Google Scholar]
- Lie RT, Rasmussen S, Brunborg H, Gjessing HK, Lie-Nielsen E, Irgens LM. Fetal and maternal contributions to risk of pre-eclampsia: population based study. BMJ. 1998;316:1343–1347. doi: 10.1136/bmj.316.7141.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arngrimsson R, Sigurard ttir S, Frigge ML, et al. A genome-wide scan reveals a maternal susceptibility locus for pre-eclampsia on chromosome 2p13. Hum Mol Genet. 1999;8:1799–1805. doi: 10.1093/hmg/8.9.1799. [DOI] [PubMed] [Google Scholar]
- Harrison GA, Humphrey KE, Jones N, et al. A genomewide linkage study of preeclampsia/eclampsia reveals evidence for a candidate region on 4q. Am J Hum Genet. 1997;60:1158–1167. [PMC free article] [PubMed] [Google Scholar]
- Lachmeijer AM, Arngrimsson R, Bastiaans EJ, et al. A genome-wide scan for preeclampsia in the Netherlands. Eur J Hum Genet. 2001;9:758–764. doi: 10.1038/sj.ejhg.5200706. [DOI] [PubMed] [Google Scholar]
- Moses EK, Lade JA, Guo G, et al. A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia, on chromosome 2. Am J Hum Genet. 2000;67:1581–1585. doi: 10.1086/316888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laivuori H, Lahermo P, Ollikainen V, et al. Susceptibility loci for preeclampsia on chromosomes 2p25 and 9p13 in Finnish families. Am J Hum Genet. 2003;72:168–177. doi: 10.1086/345311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniggia I, Lye SJ, Cross JC. Activin is a local regulator of human cytotrophoblast cell differentiation. Endocrinology. 1997;138:3976–3986. doi: 10.1210/endo.138.9.5403. [DOI] [PubMed] [Google Scholar]
- Jones RL, Salamonsen LA, Findlay JK. Activin A promotes human endometrial stromal cell decidualization in vitro. J Clin Endocrinol Metab. 2002;87:4001–4004. doi: 10.1210/jcem.87.8.8880. [DOI] [PubMed] [Google Scholar]
- Gribi R, Tanaka T, Harper-Summers R, Yu J. Expression of activin A in inflammatory arthropathies. Mol Cell Endocrinol. 2001;180:163–167. doi: 10.1016/s0303-7207(01)00517-2. [DOI] [PubMed] [Google Scholar]
- Hubner G, Brauchle M, Gregor M, Werner S. Activin A: a novel player and inflammatory marker in inflammatory bowel disease. Lab Invest. 1997;77:311–318. [PubMed] [Google Scholar]
- Phillips DJ, Jones KL, Scheerlinck JY, Hedger MP, de Kretser DM. Evidence for activin A and follistatin involvement in the systemic inflammatory response. Mol Cell Endocrinol. 2001;180:155–162. doi: 10.1016/s0303-7207(01)00516-0. [DOI] [PubMed] [Google Scholar]
- Engelse MA, Neele JM, van Achterberg TA, et al. Human activin-A is expressed in the atherosclerotic lesion and promotes the contractile phenotype of smooth muscle cells. Circ Res. 1999;85:931–939. doi: 10.1161/01.res.85.10.931. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Akishita M, Eto M, et al. Role of activin-A and follistatin in foam cell formation of THP-1 macrophages. Arterioscler Thromb Vasc Biol. 1997;17:2389–2394. doi: 10.1161/01.atv.17.11.2389. [DOI] [PubMed] [Google Scholar]
- Holmen J, Kjelsaas M, Krüger Ø, et al. Attitudes to genetic epidemiology–illustrated by questions for re-consent to 61 426 participants at HUNT. Norsk Epidemiologi (Nor J Epidemiol) 2004;14:27–31. [Google Scholar]
- Holmen J, Midthjell K, Krüger Ø, et al. The Nord-Trøndelag Health Study 1995–97 (HUNT 2): Objectives, contents, methods and participation. Norsk Epidemiologi. 2003;13:19–32. [Google Scholar]
- Moses EK, Johnson MP, Tommerdal L, et al. Genetic association of preeclampsia to the inflammatory response gene SEPS1. Am J Obstet Gynecol. 2008;198:336.e1–336.e5. doi: 10.1016/j.ajog.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Masuda J, Takayama E, Satoh A, et al. Levels of annexin IV and V in the plasma of pregnant and postpartum women. Thromb Haemost. 2004;91:1129–1136. doi: 10.1160/TH03-12-0778. [DOI] [PubMed] [Google Scholar]
- Jaleel MA, Tsai AC, Sarkar S, Freedman PV, Rubin LP. Stromal cell-derived factor-1 (SDF-1) signalling regulates human placental trophoblast cell survival. Mol Hum Reprod. 2004;10:901–909. doi: 10.1093/molehr/gah118. [DOI] [PubMed] [Google Scholar]
- Manuelpillai U, Ligam P, Smythe G, Wallace EM, Hirst J, Walker DW. Identification of kynurenine pathway enzyme mRNAs and metabolites in human placenta: up-regulation by inflammatory stimuli and with clinical infection. Am J Obstet Gynecol. 2005;192:280–288. doi: 10.1016/j.ajog.2004.06.090. [DOI] [PubMed] [Google Scholar]
- Hernandez PA, Gorlin RJ, Lukens JN, et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet. 2003;34:70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Blangero J, Goring HH, Kent JW, Jr, et al. Quantitative trait nucleotide analysis using Bayesian model selection. Hum Biol. 2005;77:541–559. doi: 10.1353/hub.2006.0003. [DOI] [PubMed] [Google Scholar]
- Cocolakis E, Lemay S, Ali S, Lebrun JJ. The p38 MAPK pathway is required for cell growth inhibition of human breast cancer cells in response to activin. J Biol Chem. 2001;276:18430–18436. doi: 10.1074/jbc.M010768200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhao Y, Batres Y, Lin MF, Ying SY. Regulation of growth and prostatic marker expression by activin A in an androgen-sensitive prostate cancer cell line LNCAP. Biochem Biophys Res Commun. 1997;234:362–365. doi: 10.1006/bbrc.1997.6649. [DOI] [PubMed] [Google Scholar]
- Donaldson CJ, Mathews LS, Vale WW. Molecular cloning and binding properties of the human type II activin receptor. Biochem Biophys Res Commun. 1992;184:310–316. doi: 10.1016/0006-291x(92)91194-u. [DOI] [PubMed] [Google Scholar]
- Mathews LS, Vale WW. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell. 1991;65:973–982. doi: 10.1016/0092-8674(91)90549-e. [DOI] [PubMed] [Google Scholar]
- Hilden K, Tuuri T, Eramaa M, Ritvos O. Expression of type II activin receptor genes during differentiation of human K562 cells and cDNA cloning of the human type IIB activin receptor. Blood. 1994;83:2163–2170. [PubMed] [Google Scholar]
- Schneider-Kolsky ME, Manuelpillai U, Waldron K, Dole A, Wallace EM. The distribution of activin and activin receptors in gestational tissues across human pregnancy and during labour. Placenta. 2002;23:294–302. doi: 10.1053/plac.2002.0787. [DOI] [PubMed] [Google Scholar]
- Jones RL, Findlay JK, Salamonsen LA. The role of activins during decidualisation of human endometrium. Aust N Z J Obstet Gynaecol. 2006;46:245–249. doi: 10.1111/j.1479-828X.2006.00581.x. [DOI] [PubMed] [Google Scholar]
- Petraglia F, Vaughan J, Vale W. Inhibin and activin modulate the release of gonadotropin-releasing hormone, human chorionic gonadotropin, and progesterone from cultured human placental cells. Proc Natl Acad Sci USA. 1989;86:5114–5117. doi: 10.1073/pnas.86.13.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraglia F, Anceschi MM, Calza L, et al. Inhibin and activin in human fetal membranes: evidence for a local effect on prostaglandin release. J Clin Endocrinol Metab. 1993;77:542–548. doi: 10.1210/jcem.77.2.8345060. [DOI] [PubMed] [Google Scholar]
- Bersinger NA, Smarason AK, Muttukrishna S, Groome NP, Redman CW. Women with preeclampsia have increased serum levels of pregnancy-associated plasma protein A (PAPP-A), inhibin A, activin A and soluble E-selectin. Hypertens Pregnancy. 2003;22:45–55. doi: 10.1081/PRG-120016794. [DOI] [PubMed] [Google Scholar]
- D′Antona D, Reis FM, Benedetto C, et al. Increased maternal serum activin A but not follistatin levels in pregnant women with hypertensive disorders. J Endocrinol. 2000;165:157–162. doi: 10.1677/joe.0.1650157. [DOI] [PubMed] [Google Scholar]
- Laivuori H, Kaaja R, Turpeinen U, Stenman UH, Ylikorkala O. Serum activin A and inhibin A elevated in pre-eclampsia: no relation to insulin sensitivity. Br J Obstet Gynaecol. 1999;106:1298–1303. doi: 10.1111/j.1471-0528.1999.tb08185.x. [DOI] [PubMed] [Google Scholar]
- Muttukrishna S, Knight PG, Groome NP, Redman CW, Ledger WL. Activin A and inhibin A as possible endocrine markers for pre-eclampsia. Lancet. 1997;349:1285–1288. doi: 10.1016/s0140-6736(96)09264-1. [DOI] [PubMed] [Google Scholar]
- Petraglia F, Aguzzoli L, Gallinelli A, et al. Hypertension in pregnancy: changes in activin A maternal serum concentration. Placenta. 1995;16:447–454. doi: 10.1016/0143-4004(95)90102-7. [DOI] [PubMed] [Google Scholar]
- Silver HM, Lambert-Messerlian GM, Star JA, Hogan J, Canick JA. Comparison of maternal serum total activin A and inhibin A in normal, preeclamptic, and nonproteinuric gestationally hypertensive pregnancies. Am J Obstet Gynecol. 1999;180:1131–1137. doi: 10.1016/s0002-9378(99)70606-x. [DOI] [PubMed] [Google Scholar]
- Petraglia F, Sawchenko P, Lim AT, Rivier J, Vale W. Localization, secretion, and action of inhibin in human placenta. Science. 1987;237:187–189. doi: 10.1126/science.3299703. [DOI] [PubMed] [Google Scholar]
- Smith C, Yndestad A, Halvorsen B, et al. Potential anti-inflammatory role of activin A in acute coronary syndromes. J Am Coll Cardiol. 2004;44:369–375. doi: 10.1016/j.jacc.2004.03.069. [DOI] [PubMed] [Google Scholar]
- Bellamy L, Casas JP, Hingorani AD, Williams DJ. Pre-eclampsia and risk of cardiovascular disease and cancer in later life: systematic review and meta-analysis. Bmj. 2007;335:974. doi: 10.1136/bmj.39335.385301.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J Biol Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]